Back to Journals » Journal of Asthma and Allergy » Volume 11
Severe bronchial asthma in children: a review of novel biomarkers used as predictors of the disease
Authors Uwaezuoke SN , Ayuk AC , Eze JN
Received 20 August 2017
Accepted for publication 14 December 2017
Published 15 January 2018 Volume 2018:11 Pages 11—18
DOI https://doi.org/10.2147/JAA.S149577
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Amrita Dosanjh
Video abstract presented by Samuel N Uwaezuoke
Views: 306
Samuel N Uwaezuoke, Adaeze C Ayuk, Joy N Eze
Department of Pediatrics, University of Nigeria Teaching Hospital, Ituku-Ozalla, Nigeria
Abstract: Severe asthma or therapy-resistant asthma in children is a heterogeneous disease that affects all age-groups. Given its heterogeneity, precision in diagnosis and treatment has become imperative, in order to achieve better outcomes. If one is thus able to identify specific patient phenotypes and endotypes using the appropriate biomarkers, it will assist in providing the patient with more personalized and appropriate treatment. However, there appears to be a huge diagnostic gap in severe asthma, as there is no single test yet that accurately determines disease phenotype. In this paper, we review the published literature on some of these biomarkers and their possible role in bridging this diagnostic gap. We also highlight the cellular and molecular mechanisms involved in severe asthma, in order to show the basis for the novel biomarkers. Some markers useful for monitoring therapy and assessing airway remodeling in the disease are also discussed. A review of the literature was conducted with PubMed to gather baseline data on the subject. The literature search extended to articles published within the last 40 years. Although biomarkers specific to different severe asthma phenotypes have been identified, progress in their utility remains slow, because of several disease mechanisms, the variation of biomarkers at different levels of inflammation, changes in relying on one test over time (eg, from sputum eosinophilia to blood eosinophilia), and the degree of invasive tests required to collect biomarkers, which limits their applicability in clinical settings. In conclusion, several biomarkers remain useful in recognizing various asthma phenotypes. However, due to disease heterogeneity, identification and utilization of ideal and defined biomarkers in severe asthma are still inconclusive. The development of novel serum/sputum-based biomarker panels with enhanced sensitivity and specificity may lead to prompt diagnosis of the disease in the future.
Keywords: therapy-resistant asthma, heterogeneous respiratory disorder, phenotypes, diagnostic markers
Introduction
Bronchial asthma is a chronic inflammatory disease characterized by bronchial hyperreactivity, reversible airway obstruction, and excessive mucus production that arises from an inappropriate stimulation of the immune system, especially by environmental aeroallergens.1,2 Bronchial asthma more closely resembles a complex of clinical diseases than a single condition. Within the past few years, it has emerged as the most common incommunicable respiratory disease affecting children worldwide.3 Its genetic basis is well established,2 although nongenetic (nonatopic) forms are also recognized, especially in adult subjects.4 Asthma is further distinguished by multiple phenotypes, which may differ based on age of onset, trigger factors, and patterns of severity, as reflected by variably reversible loss of pulmonary function.5
Severe asthma, which occurs in all age-groups, is a clinically and etiologically heterogeneous respiratory disorder that affects about 5%–10% of patients with bronchial asthma.6 It is associated with reduced quality of life, with substantial impact on health-care expenditure.7 Severe asthma refers to “asthma which requires treatment with high-dose inhaled corticosteroids . . . plus a second controller (and/or systemic corticosteroids) to prevent it from becoming ‘uncontrolled’ or which remains ‘uncontrolled’ despite this therapy.”8,9 However, there exists an age-specific variation in the definition of high-dose inhaled corticosteroids (ICSs) for children aged <6 years, 6–12 years, and >12 years.8
Severe asthma, which is now considered a global challenge, consists of multiple phenotypes, such as the inflammatory phenotypes that include severe allergic asthma, eosinophilic asthma, and neutrophilic asthma. This may imply that identifying patients according to phenotypes based on observable clinical characteristics can provide targeted treatment options. Studies are thus beginning to define phenotypic biomarkers, while phenotype-targeted biological therapies are increasingly showing efficacy in the management of severe asthma.8
Given its heterogeneity, precision in diagnosis and treatment has become imperative to achieve a better outcome. If one is thus able to identify specific patient phenotypes and endotypes (biologically distinct groups) using the appropriate biomarkers, it will assist in providing the patient with more personalized and appropriate treatment.
Obviously, there appears to be a huge diagnostic gap in severe asthma, as there is no single test yet that accurately determines disease phenotype. There is also a shift in the value of assessment of one single biomarker to a combination of various markers, considering the heterogeneity of the disease spectrum. Although biomarkers that are specific to different severe asthma phenotypes have been identified, progress in their utility remains slow, because of several disease mechanisms: variation in biomarkers at different levels of inflammation, the change in relying on one test over time (eg, from sputum eosinophilia to blood eosinophilia), and the degree of invasive tests required to collect these biomarkers, which limits their applicability in clinical settings.6,10
This paper reviews the published literature on some of these novel biomarkers and their possible role in bridging the diagnostic gap in severe asthma. The cellular and molecular mechanisms involved in the disease are also highlighted, in order to establish the basis for these markers. Some markers useful for monitoring therapy and assessing airway remodeling in the disease are also discussed.
Literature search: strategy and outcome
In line with the review objectives, a combination of search terms was used to gather articles from the PubMed database between January and July 2017. Articles published within the last 40 years were included. The search terms “severe asthma and biomarkers” yielded 754 publications, “severe asthma phenotypes and biomarkers” gave 231 publications, “severe asthma phenotypes in children and biomarkers” yielded 56, “severe asthma and molecular mechanisms” gave 198, and “severe asthma and cellular mechanisms” yielded 507 publications. However, several studies were either duplicated or failed to meet the review objectives. After exclusion of these publications, 256 articles formed the list from which original and review articles were selected for the current narrative review.
Molecular and cellular mechanisms involved in severe asthma phenotypes
Several published reports have suggested that severe asthma phenotypes are closely linked to genetic factors, age of asthma onset, duration of disease, flare ups, comorbid sinus disease, and inflammatory characteristics.11–15 Initial evaluations of patients with asthma using bronchoalveolar lavage and endobronchial biopsies revealed distinct inflammatory subtypes.6 Following the concept of molecular phenotyping, molecular pathways have been linked to clinical and physiological characteristics of the disease.16 Subsequently, molecular and cellular mechanisms have been described for different disease phenotypes. The asthma phenotype was predominantly eosinophilic in children with acute asthma (about 50% of cases), but neutrophilic in adults with acute asthma (about 82%); paucigranulocytic asthma was the most common phenotype in both adults and children with stable asthma.17 Nevertheless, for severe asthma, the recognized inflammatory phenotypes are eosinophilic, noneosinophilic and paucigranulocytic asthma (see Table 1).18 The identification of these phenotypes has propelled the search for novel biomarkers to help in patient classification, targeting therapies, and predicting different mechanisms of disease pathogenesis.19
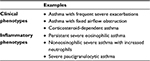  | Table 1 Phenotypes of uncontrolled severe asthma Note: Adapted with permission from Campo P, Rodríguez F, Sánchez-García S, et al. Phenotypes and endotypes of uncontrolled severe asthma: new treatments. J Investig Allergol Clin Immunol. 2013;23:76–88.18 |
Studies based on bronchoalveolar lavage and endobronchial biopsies conducted in children have shown that inflammation and remodeling, especially of the epithelium and distal airways, constitute the hallmarks of the pathobiology of severe asthma.20 Characteristically, airway remodeling is associated with local matrix deposition, vascularization, epithelial hyperplasia, and changes in the submucosa, such as hyperplasia of smooth-muscle cells and proliferation of fibroblasts.21
Severe asthma is characterized by neutrophilic inflammation (neutrophilic asthma). This phenotype can occur in both the presence of eosinophilic inflammation or absence of it (severe noneosinophilic asthma with increased neutrophils).18,22 Neutrophilic inflammation is induced by type 2 T-helper (TH2) cells.22,23 Two distinct endotypes are currently recognized – TH2-high (TH2-positive) and TH2-low asthma – based on expression levels of genes encoding IL13, IL5, and IL4, cytokine expression, and responsiveness to ICSs.16 In addition, recruitment of neutrophils can also be mediated by TH17 cells, which have been implicated in asthma pathogenesis, particularly in asthmatics who fail to respond to CS therapy and exhibit airway hyperresponsiveness and decreased posttherapeutic improvement in airflow limitation or forced expiratory volume in 1 second.24,25 In children with the phenotype of severe noneosinophilic asthma characterized by increased neutrophils, neutrophilic inflammation may result from comorbidities, such as bronchiolitis obliterans.18 Other molecular mechanisms that contribute to neutrophil recruitment in the airways and thus neutrophilic airway inflammation include the action of reactive oxygen species,26 the effect of the imbalance between RAGE and its soluble forms,27 and the role of DAM8.28,29
Secondly, severe asthma has also been characterized by eosinophilic recruitment (eosinophilic asthma), and included in the mechanism are mucus plugging and epithelial denudation in the large-airway wall and lumen. It is associated with increased expression of TGFβ and reticular basement membrane (RBM) thickness.11 The RBM is formed by the fusion of alveolar basal lamina and pulmonary capillaries, and constitutes the interface for oxygen and carbon dioxide diffusion. Active recruitment of eosinophils is largely mediated by proteins secreted by the epithelia: the most potent chemoattractant for eosinophils being eotaxin 1 (CCL11), which is responsible for 80% of TGFβ expression in asthma.30–32
The other subphenotype (severe paucigranulocytic asthma) has a poorly understood mechanism and is better described for adult populations.33 Its inflammation may be localized to the distal airways only, where it is difficult to obtain biopsy specimens for further analysis. It has been described as having no thickening of the subepithelial BM and lacking the classic inflammation with activation of other cell types.18 Airway remodeling is an important mechanism in severe asthma. The phenotype with fixed airway obstruction resulting from airway remodeling has been highlighted as one of the key features of many severe asthmatics.
In highlighting the role of distal lung inflammation and remodeling in the disease, some investigators have reported that rennin-positive mast cells are increased in the distal airways of patients compared with a normal autopsied distal lung.34 Furthermore, inflammation and remodeling appear to differ between the proximal and distal lung, as another study had reported that fibroblasts (FGF2) from the distal lung proliferated more rapidly at baseline and in response to TGFβ than those from the proximal airway.35 Galectin 3 has been recognized as a marker of airway remodeling, while it is now known that the gene-regulating molecule IRF4 also plays a key role in the development of TH9 lymphocytes, which are mainly implicated in the development of this chronic inflammatory airway disease.36
Research findings have further revealed that the assessment of by-products of oxidative stress is a novel way to monitor disease severity, a concept applicable to severe asthma in terms of biomarkers. Protein–S-glutathionylation is an oxidative modification of reactive cysteines that has emerged as an important player in pathophysiological processes, and it has been shown that genetic ablation of glutaredoxin 1 enhances the resolution of airway hyperresponsiveness and mucus metaplasia in mice with allergic airway disease.37 Glutaredoxin 1/S-glutathionylation redox status in mice has thus been noted as a critical regulator of airway hyperresponsiveness, suggesting that avenues to increase S-glutathionylation of specific target proteins may be beneficial to attenuate airway hyperresponsiveness.38 Monitoring glutaredoxin 1 protein levels in sputum may thus be an important biomarker for assessing therapy in paucigranulocytic severe asthma. Despite myriad potential biomarkers in severe asthma, they can still be grouped as predictors of airway remodeling, epithelium-derived biomarkers, tissue-associated biomarkers, biomarkers of mixed origin (epithelium- and tissue-associated markers), and exhaled breath condensate (EBC), as shown in Table 2.
Novel biomarkers in severe asthma
Reliable markers in severe asthma in children are predicated upon the prospect of using noninvasive and inexpensive methods to obtain and estimate ideal biomarkers for identifying either clinical phenotypes or treatment-response phenotypes, evaluating changes in disease activity, or confirming a diagnosis.6 These biomarkers, which are useful for diagnostic purposes, may be obtained from blood, bronchoalveolar lavage, bronchial biopsies, induced sputum, and EB (eg, such gases as fractional exhaled nitric oxide [FENO] and EBC) (Table 2). Induced sputum and EB are noninvasive lower-airway sampling techniques that are easily applicable in children. Gases in EB and other molecules in EBC also stand out as biomarkers of disease. Biomarkers that are found in more easily accessible samples in the pediatric population are summarized in the following sections.
Tissue-associated biomarkers
Basophils, eosinophils, and neutrophils are considered fundamental effector cells that infiltrate the asthmatic airway in conjunction with platelets.39 Sputum neutrophil percentages are highly reproducible in patients with moderate–severe asthma, and can be used to evaluate novel anti-inflammatory therapies; expectedly, targeting neutrophils has been suggested as a therapeutic option for these patients.40,41 Although neutrophilic inflammation has been established as a common occurrence in severe asthma, the use of this specific biomarker in confirming severe asthma is not yet a standard practice, as the functional role of these cells in disease progression remains unclear. Nevertheless, several novel surface biomarkers of neutrophils have been reported as predictors of established disease and airway remodeling (Table 2). Some of these surface markers are CD11b, CD62L, CD274,42 CD35,43 CD14, TLR2, TLR4,44 CXCR2, and IL8Rβ.45 Some of the more easily accessible ones have also been highlighted in Table 2.
As shown in Table 2, eosinophils and their many surface biomarkers have similar roles in severe asthma. They are quite numerous, but a few examples with direct correlation are worthy of note: C3AR,46 CD13,47 CD25,48 CD29,49 CD32,50 CD44/HCAM,51 and galectin 3.52 Similarly, multiple mediators derived from platelets, such as leukotrienes, CXCL4, β-thromboglobulin, CCL5, RANTES, thromboxane, and serotonin, have also been reported as potential markers.49,53–56 These mediators are directly correlated with severe asthma, and thus CXCL4 may specifically serve as a disease biomarker for severe asthma.55 Despite the use of eosinophil count in induced sputum as the method of choice to assess eosinophilic lung inflammation (possibly making it a reliable biomarker of airway inflammation),57–59 the current recommendation is rather in favor of utilizing blood eosinophils as biomarkers, as this could help to personalize asthma management in patients with severe allergic asthma.6 In fact, some investigators have shown a relationship between blood eosinophil counts (but not sputum eosinophilia) and response to an asthma medication.60 Nevertheless, asthma severity might be linked to a relationship between TGFβ expression and the presence of submucosal eosinophils.6 This is supported by the observation of eosinophilic expression of TGFβ2 isoforms in severe allergic asthma, where this cytokine promotes fibrotic responses and modulates mucin production.61 Other tissue-associated biomarkers that have equally been reported as predictors of disease and airway remodeling are CRP,62 ECP,63 IFNα,64 IFNγ,65 TNFα,66 IL2, IL4, IL18, and IL22.65,67–69
Epithelium-derived biomarkers
Again, several studies have reported the potential role of some epithelium-derived biomarkers in severe asthma, especially due to the relative ease of obtaining samples from the respiratory epithelium. Prominent among them are periostin,70,71 fibrinogen,72 MMPs,73–76 eotaxins,77 and TIMPs74 (Table 2). Specifically, periostin has attracted much scientific interest and elucidation with respect to its role in allergic asthma.78–80 It is secreted in bronchial epithelial cells and lung fibroblasts following induction by IL4 and IL13, and its expression is directly related to RBM thickness.81–83 Periostin is essentially involved in amplifying and maintaining chronic inflammation arising from allergic diseases,84 as well as initiating the process of subepithelial fibrosis in bronchial asthma.81 Serum periostin, which is a good marker of eosinophilic asthma, is considered a promising biomarker in severe asthma, because of its easy movement from inflamed tissues to blood circulation, making its serum levels a reflection of its local production in lesions induced by TH2-type immunoresponses.85,86 (Table 2).
Epithelium- and tissue-associated biomarkers (biomarkers of mixed origin)
Several of these biomarkers have been reported, but the following are highlighted: IL1β,87 osteopontin,88 TGF,89 VEGF,90 and YKL40 (CHI3L1)91 (Table 2). Osteopontin is a cytokine, originally described as a structural component of the extracellular matrix, which is capable of binding to proteins and most types of collagen.92 This biomarker is produced by most cells of the immune system, including T cells, B cells, macrophages, neutrophils, eosinophils, natural killer cells, and mast cells, as well as structural cells, including fibroblasts and smooth-muscle and epithelial cells.93 In the study that aimed to estimate osteopontin levels in the serum, bronchoalveolar lavage fluid, and bronchial tissue of healthy controls and asthmatics, serum and bronchoalveolar lavage-fluid levels were significantly elevated in all asthmatics in the steady state, whereas serum levels decreased during exacerbations.88 In addition, osteopontin was upregulated in the bronchial tissue of all patients, and expressed by epithelial, airway, and vascular smooth-muscle cells, myofibroblasts, T lymphocytes, and mast cells, and its expression correlated with RBM thickness and was more prominent in subepithelial inflammatory cells in severe asthma compared to mild–moderate asthma.88
YKL40 is a chitinase-like protein that regulates the formation of collagen fibrils and increases smooth-muscle proliferation.94 It has also been linked with asthma and airway remodeling.95 In fact, some authors have suggested that this biomarker with biological plausibility for airway remodeling (when corrected for genotype) differentiates severe treatment-resistant asthma from controlled persistent asthma and correlates well with other markers for asthma control.91
VEGF is highly expressed in the airway of patients with bronchial asthma, and as it increases airway vascular permeability, the resultant thickening of the airway-wall mucosa may result in narrowing of the airway lumen. This finding has been corroborated by investigators who noted that the VEGF levels in induced sputum were not only increased in asthmatic subjects but were also associated with the degree of airway narrowing and airway vascular permeability90 (Table 2).
Exhaled breath condensate
EB mainly contains aerosolized, involatile particles of fluid from airway lining, water-vapor condensation, inorganic (O2, N2) and organic (CO2) atmospheric volatile water-soluble gases, and endogenous and exogenous volatile organic compounds.96 EBC is obtained after condensation of EB with a commercially available refrigerating device based on American Thoracic Society/European Respiratory Society guidelines.97 Generally, it has been documented that the following molecules in EBC are elevated in severe asthma: leukotrienes, lipoxins, arachidonic acid metabolites, nitrogen oxides and related products, cytokines, ammonia, oxidative stress markers, hydrogen peroxide, and adenosine.6 Interestingly, few studies that have evaluated and compared its composition in patients with severe asthma and their healthy counterparts show dissimilarities. For example, some authors reported that such eicosanoids as proinflammatory leukotriene LTB4 and anti-inflammatory LXA4 were elevated in asthmatics when compared to healthy subjects, while the LXA4:LTB4 ratio decreased remarkably in EBC in correlation with asthma severity.98 In addition, eotaxin 1 (CCL11) has been evaluated in EBC (among other specimens like blood, sputum and bronchoalveolar lavage fluid) and has been proposed as a tool for assessment of asthma severity.99 Although collection of EBC is a procedure potentially influenced by several endogenous and exogenous factors, its analysis may thus emerge as a promising, safe, and noninvasive method for monitoring severe asthmatics on treatment.97
Fractional exhaled nitric oxide
FENO levels in the respiratory tract are mainly determined by epithelial inducible nitric oxide synthase.100 Measurement of nitric oxide concentration in EB is a quantitative, noninvasive, simple, and safe method to evaluate airway inflammation, to monitor responsiveness to ICS therapy in adults, and to predict asthma-control status in children.101,102 Other studies conducted in adults have also shown that FENO levels correlate more with asthma control than disease severity.103,104 Given the markedly hyperplastic bronchial mucosa, susceptibility of the epithelium to destruction, and flaking and detachment from the RBM seen in severe asthma, FENO levels may be underrepresented.105 Because of this limitation, elevated FENO values are not considered an effective biomarker in the management of severe asthma, but rather as a marker of TH2 inflammation and atopy.8
Conclusion
The identification and utilization of ideal and defined biomarkers in severe asthma remains inconclusive. Apart from disease heterogeneity, the reasons for this include the diverse and overlapping biological aspects of each prospective biomarker, its changing concentrations depending on disease pathobiology, and methods of sample collection/evaluation, as well as the invasiveness of sampling techniques that need specific tools and skills. In children, the challenge of sampling has been circumvented by the use of less invasive methods of sample collection. Therefore, the development of novel serum/sputum-based biomarker panels with enhanced sensitivity and specificity may lead to prompt diagnosis of severe asthma in future.
Acknowledgment
The authors acknowledge the invaluable information obtained from the articles by Chiappori et al6 and Zissler et al.21
Disclosure
The authors report no conflicts of interest in this work.
References
Martinez FD, Vercelli D. Asthma. Lancet. 2013;382:1360–1372. | ||
Bierbaum S, Heinzmann A. The genetics of bronchial asthma in children. Respir Med. 2007;101:1369–1375. | ||
World Health Organization. Asthma. 2017. Available from: http://www.who.int/mediacentre/factsheets/fs307/en/index.html. Accessed March 7, 2017. | ||
Ozdemir C, Ceyhan B, Yazi D, et al. Non-atopic asthma in children is related to maternal bronchial hyper-reactivity. Pediatr Allergy Immunol. 2008;19:248–254. | ||
Lemanske RF, Busse WW. Asthma: clinical expression and molecular mechanisms. J Allergy Clin Immunol. 2010;125:S95–S102. | ||
Chiappori A, De Ferrari L, Folli C, Mauri P, Riccio AM, Canonica GW. Biomarkers and severe asthma: a critical appraisal. Clin Mol Allergy. 2015;13:20. | ||
Walsh LJ, Wong CA, Cooper S, Guhan AR, Pringle M, Tattersfield AE. Morbidity from asthma in relation to regular treatment: a community based study. Thorax. 1999;54:296–300. | ||
Chung KF, Wenzel SE, Brozek JL, et al. International ERS/ATS guidelines on definition, evaluation and treatment of severe asthma. Eur Respir J. 2014;43:343–373. | ||
American Thoracic Society. Proceedings of the ATS workshop on refractory asthma: current understanding, recommendations, and unanswered questions. Am J Respir Crit Care Med. 2000;162:2341–2351. | ||
Arron JR, Izuhara K. Asthma biomarkers: what constitutes a ‘gold standard’? Thorax. 2015;70:105–107. | ||
Haldar P, Pavord ID, Shaw DE, et al. Cluster analysis and clinical asthma phenotypes. Am J Respir Crit Care Med. 2008;178:218–224. | ||
Miranda C, Busacker A, Balzar S, Trudeau J, Wenzel SE. Distinguishing severe asthma phenotypes: role of age at onset and eosinophilic inflammation. J Allergy Clin Immunol. 2004;113:101–108. | ||
Wenzel SE, Balzar S, Ampleford E, et al. IL4Rα mutations are associated with asthma exacerbations and mast cell/IgE expression. Am J Respir Crit Care Med. 2007;175:570–576. | ||
Li X, Howard TD, Zheng SL, et al. Genome-wide association study of asthma identifies RAD50-IL13 and HLA-DR/DQ regions. J Allergy Clin Immunol. 2010;125:328–335. | ||
Haselkorn T, Zeiger RS, Chipps BE, et al. Recent asthma exacerbations predict future exacerbations in children with severe or difficult-to-treat asthma. J Allergy Clin Immunol. 2009;124:921–927. | ||
Woodruff PG, Modrek B, Choy DF, et al. T-helper type 2-driven inflammation defines major subphenotypes of asthma. Am J Respir Crit Care Med. 2009;180:388–395. | ||
Wang F, He XY, Baines KJ, et al. Differentiating inflammatory phenotypes in adults and children with acute asthma. Eur Resp J. 2011;38:567–574. | ||
Campo P, Rodríguez F, Sánchez-García S, et al. Phenotypes and endotypes of uncontrolled severe asthma: new treatments. J Investig Allergol Clin Immunol. 2013;23:76–88. | ||
Moore WC, Meyers DA, Wenzel SE, et al. Identification of asthma phenotypes using cluster analysis in the severe asthma research program. Am J Respir Crit Care Med. 2010;181:315–323. | ||
Wenzel SE, Busse WW. Severe asthma: lessons from the Severe Asthma Research Program. J Allerg Clin Immunol. 2007;119:14–21. | ||
Zissler UM, Esser-von Bieren J, Jakwerth CA, Chaker AM, Schmidt-Weber CB. Current and future biomarkers in allergic asthma. Allergy. 2016;71:475–494. | ||
Jatakanon A, Uasuf C, Maziak W, Lim S, Chung KF, Barnes PJ. Neutrophilic inflammation in severe persistent asthma. Am J Respir Crit Care Med. 1999;160:1532–1539. | ||
Gibson PG, Simpson JL, Saltos N. Heterogeneity of airway inflammation in persistent asthma: evidence of neutrophilic inflammation and increased sputum interleukin-8. Chest. 2001;119:1329–1336. | ||
Schleimer RP. Glucocorticoids suppress inflammation but spare innate immune responses in airway epithelium. Proc Am Thorac Soc. 2004;1:222–230. | ||
Barnes PJ, Adcock IM. Glucocorticoid resistance in inflammatory diseases. Lancet. 2009;373:1905–1917. | ||
Boldogh I, Bacsi A, Choudhury BK, Dharajiya N, Alam R, Hazra TK. ROS generated by pollen NADPH oxidase provide a signal that augments antigen-induced allergic airway inflammation. J Clin Invest. 2005;115:2169–2179. | ||
Sims GP, Rowe DC, Rietdijk ST, Herbst R, Coyle AJ. HMGB1 and RAGE in inflammation and cancer. Annu Rev Immunol. 2010; | ||
Gómez-Gaviro M, Domínguez-Luis M, Canchado J, et al. Expression and regulation of the metalloproteinase ADAM-8 during human neutrophil pathophysiological activation and its catalytic activity on L-selectin shedding. J Immunol. 2007;178:8053–8063. | ||
Oreo KM, Gibson PG, Simpson JL, Wood LG, McDonald VM, Baines KJ. Sputum ADAM-8 expression in increased in severe asthma and COPD. Clin Exp Allergy. 2013;44:342–352. | ||
Conroy DM, Williams TJ. Eotaxin and the attraction of eosinophils to the asthmatic lung. Respir Res. 2001;2:150–156. | ||
Kim CK, Kita H, Callaway Z, et al. The roles of a Th2 cytokine and CC chemokine in children with stable asthma: potential implication in eosinophil degranulation. Pediatr Allergy Immunol. 2010;21:e697–e704. | ||
Zietowski Z, Tomasiek-Lozowska MM, Skiepko R, Zietowska E, Bodzenta-Lukaszyk A. Eotaxin-1 in exhaled breath condensate of stable and unstable asthma patients. Respir Res. 2010;11:110. | ||
Wenzel S. Severe asthma in adults. Am J Resp Crit Care Med. 2005;172:149–160. | ||
Balzar S, Chu HW, Strand M, Wenzel S. Relationship of small airway chymase-positive mast cells and lung function in severe asthma. Am J Respir Crit Care Med. 2005;171:431–439. | ||
Kotaru C, Schoonover KJ, Trudeau JB, et al. Regional fibroblast heterogeneity in the lung: implications for remodeling. Am J Respir Crit Care Med. 2006;173:1208–1215. | ||
Staudt V, Bothur E, Klein M, et al. Interferon-regulating factor 4 is essential for the developmental program of T helper 9 cells. Immunity. 2010;33:192–202. | ||
Jiang L, Diaz PT, Best TM, Stumpf JN, He F, Zuo L. Molecular characterization of redox mechanisms in allergic asthma. Ann Allergy Asthma Immunol. 2014;113:137–142. | ||
Hoffman SM, Tully JE, Lahue KG, et al. Genetic ablation of glutaredoxin-1 causes enhanced resolution of airways hyperresponsiveness and mucus metaplasia in mice with allergic airways disease. Am J Physiol Lung Cell Mol Physiol. 2012;303:L528–L538. | ||
Idzko M, Pitchford S, Page C. Role of platelets in allergic airway inflammation. J Allergy Clin Immunol. 2015;135:1416–1423. | ||
Rossall MR, Cadden PA, Molphy SD, Plumb J, Singh D. Repeatability of induced sputum measurements in moderate to severe asthma. Respir Med. 2014;108:1566–1568. | ||
Nair P, Gaga M, Zervas E, et al. Safety and efficacy of a CXCR2 antagonist in patients with severe asthma and sputum neutrophils: a randomized, placebo-controlled clinical trial. Clin Exp Allergy. 2012;42:1097–1103. | ||
Tak T, Hilvering B, Tesselaar K, Koenderman L. Similar activation state of neutrophils in sputum of asthma patients irrespective of sputum eosinophilia. Clin Exp Immunol. 2015;182:204–212. | ||
Mann BS, Chung KF. Blood neutrophil activation markers in severe asthma: lack of inhibition by prednisolone therapy. Respir Res. 2006;7:59. | ||
Simpson JL, Grissell TV, Douwes J, Scott RJ, Boyle MJ, Gibson PG. Innate immune activation in neutrophilic asthma and bronchiectasis. Thorax. 2007;62:211–218. | ||
Nair P, Gaga M, Zervas E, et al. Safety and efficacy of a CXCR2 antagonist in patients with severe asthma and sputum neutrophils: a randomized, placebo-controlled clinical trial. Clin Exp Allergy. 2012;42:1097–1103. | ||
Krug N, Tschernig T, Erpenbeck VJ, Hohlfeld JM, Köhl J. Complement factors C3a and C5a are increased in bronchoalveolar lavage fluid after segmental allergen provocation in subjects with asthma. Am J Respir Crit Care Med. 2001;164:1841–1843. | ||
Braun RK, Foerster M, Workalemahu G, Haefner D, Kroegel C, Walker C. Differential regulation of aminopeptidase N (CD13) by transendothelial migration and cytokines on human eosinophils. Exp Lung Res. 2003;29:59–77. | ||
Bates ME, Liu LY, Esnault S, et al. Expression of interleukin-5- and granulocyte macrophage-colony-stimulating factor-responsive genes in blood and airway eosinophils. Am J Respir Cell Mol Biol. 2004;30:736–743. | ||
Johansson MW, Han ST, Gunderson KA, Busse WW, Jarjour NN, Mosher DF. Platelet activation, P-selectin, and eosinophil β1-integrin activation in asthma. Am J Respir Crit Care Med. 2012;185:498–507. | ||
Kanters D, ten Hove W, Luijk B, et al. Expression of activated FcγRII discriminates between multiple granulocyte priming phenotypes in peripheral blood of allergic asthmatic subjects. J Allergy Clin Immunol. 2007;120:1073–1081. | ||
Johansson MW. Activation states of blood eosinophils in asthma. Clin Exp Allergy. 2014;44:482–498. | ||
Gao P, Gibson PG, Baines KJ, et al. Anti-inflammatory deficiencies in neutrophilic asthma: reduced galectin-3 and IL-1RA/IL1 β. Respir Res. 2015;16:5. | ||
Tornhamre S, Ehnhage A, Kölbeck KG, Edenius C, Lindgren JA. Uncoupled regulation of leukotriene C4 synthase in platelets from aspirin-intolerant asthmatics and healthy volunteers after aspirin treatment. Clin Exp Allergy. 2002;32:1566–1573. | ||
Zietkowski Z, Bodzenta-Lukaszyk A, Tomasiak MM, Skiepko R, Mroczko B, Szmitkowski M. Changes in RANTES and β-thromboglobulin after intensive exercise in patients with allergic asthma. Int Arch Allergy Immunol. 2009;148:31–40. | ||
Knauer KA, Lichtenstein LM, Adkinson NF Jr, Fish JE. Platelet activation during antigen-induced airway reactions in asthmatic subjects. N Engl J Med. 1981;304:1404–1407. | ||
Dürk T, Duerschmied D, Müller T, et al. Production of serotonin by tryptophan hydroxylase 1 and release via platelets contribute to allergic airway inflammation. Am J Respir Crit Care Med. 2013;187:476–485. | ||
Wadsworth S, Sin D, Dorscheid D. Clinical update on the use of biomarkers of airway inflammation in the management of asthma. J Asthma Allergy. 2011;4:77–86. | ||
Eltboli O, Brightling CE. Eosinophils as diagnostic tools in chronic lung disease. Expert Rev Respir Med. 2013;7:33–42. | ||
Szefler SJ, Wenzel S, Brown R, et al. Asthma outcomes: biomarkers. J Allergy Clin Immunol. 2012;129:9–23. | ||
Pavord ID, Korn S, Howarth P, et al. Mepolizumab for severe eosinophilic asthma (DREAM): a multicentre, double-blind, placebo-controlled trial. Lancet. 2012;380:651–659. | ||
Harrop CA, Gore RB, Evans CM, Thornton DJ, Herrick SE. TGF-β2 decreases baseline and IL-13-stimulated mucin production by primary human bronchial epithelial cells. Exp Lung Res. 2013;39:39–47. | ||
Shimoda T, Obase Y, Kishikawa R, Iwanaga T. Serum high-sensitivity C-reactive protein can be an airway inflammation predictor in bronchial asthma. Allergy Asthma Proc. 2015;36:e23–e28. | ||
Kato M, Yamada Y, Maruyama K, Hayashi Y. Serum eosinophil cationic protein and 27 cytokines/chemokines in acute exacerbation of childhood asthma. Int Arch Allergy Immunol. 2010;152:62–66. | ||
Wright AK, Mistry V, Richardson M, et al. Toll-like receptor 9 dependent interferon-α release is impaired in severe asthma but is not associated with exacerbation frequency. Immunobiology. 2015;220:859–864. | ||
Pennino D, Bhavsar PK, Effner R, et al. IL-22 suppresses IFN-γ-mediated lung inflammation in asthmatic patients. J Allergy Clin Immunol. 2013;131:562–570. | ||
Thomas PS, Heywood G. Effects of inhaled tumour necrosis factor alpha in subjects with mild asthma. Thorax. 2002;57:774–778. | ||
Boonpiyathad S, Pornsuriyasak P, Buranapraditkun S, Klaewsongkram J. Interleukin-2 levels in exhaled breath condensates, asthma severity, and asthma control in non-allergic asthma. Allergy Asthma Proc. 2013;34:e35–e41. | ||
Bellini A, Marini MA, Bianchetti L, Barczyk M, Schmidt M, Mattoli S. Interleukin (IL)-4, IL-13, and IL-17A differentially affect the profibrotic and proinflammatory functions of fibrocytes from asthmatic patients. Mucosal Immunol. 2012;5:140–149. | ||
Rovina N, Dima E, Bakakos P, et al. Low interleukin (IL)-18 levels in sputum supernatants of patients with severe refractory asthma. Respir Med. 2015;109:580–587. | ||
Bobolea I, Barranco P, Del Pozo V, et al. Sputum periostin in patients with different severe asthma phenotypes. Allergy. 2015;70:540–546. | ||
Kanemitsu Y, Ito I, Niimi A, et al. Osteopontin and periostin are associated with a 20-year decline of pulmonary function in patients with asthma. Am J Respir Crit Care Med. 2014;190:472–474. | ||
Desai D, Gupta S, Siddiqui S, et al. Sputum mediator profiling and relationship to airway wall geometry imaging in severe asthma. Respir Res. 2013;14:17. | ||
Boulay ME, Prince P, Deschesnes F, Chakir J, Boulet LP. Metalloproteinase-9 in induced sputum correlates with the severity of the late allergen-induced asthmatic response. Respiration. 2004;71:216–224. | ||
Cataldo DD, Gueders M, Munaut C, et al. Matrix metalloproteinases and tissue inhibitors of matrix metalloproteinases mRNA transcripts in the bronchial secretions of asthmatics. Lab Invest. 2004;84:418–424. | ||
Han Z. Xu J, Zhong N. Expression of matrix metalloproteinases MMP-9 within the airways in asthma. Respir Med. 2003;97:563–567. | ||
Karakoc GB, Yukselen A, Yilmaz M, Altintas DU, Kendirli SG. Exhaled breath condensate MMP-9 level and its relationship with asthma severity and interleukin-4/10 levels in children. Ann Allergy Asthma Immunol. 2012;108:300–304. | ||
Isgro M, Bianchetti L, Marini MA, Bellini A, Schmidt M, Mattoli S. The C-C motif chemokine ligands CCL5, CCL11, and CCL24 induce the migration of circulating fibrocytes from patients with severe asthma. Mucosal Immunol. 2013;6:718–727. | ||
Jia G, Erickson RW, Choy DF, et al. Periostin is a systemic biomarker of eosinophilic airway inflammation in asthmatic patients. J Allergy Clin Immunol. 2012;130:647–654. | ||
Kanemitsu Y, Matsumoto H, Izuhara K, et al. Increased periostin associates with greater airflow limitation in patients receiving inhaled corticosteroids. J Allergy Clin Immunol. 2013;132:305–312. | ||
Parulekar AD, Mustafa AA, Hanania NA. Periostin, a novel biomarker of Th2-driven asthma. Curr Opin Pulm Med. 2014;20:60–65. | ||
Takayama G, Arima K, Kanaji T, et al. Periostin: a novel component of sub-epithelial fibrosis of bronchial asthma downstream of IL-4 and IL-13 signals. J Allergy Clin Immunol. 2006;118:98–104. | ||
Sidhu SS, Yuan S, Innes AL, et al. Roles of epithelial cell-derived periostin in TFG-β activation, collagen production, and collagen gel elasticity in asthma. Proc Natl Acad Sci U S A. 2010;107:14170–14175. | ||
Yuyama N, Davies DE, Akaiwa M, et al. Analysis of novel disease-related genes in bronchial asthma. Cytokine. 2002;19:287–296. | ||
Conway SJ, Izuhara K, Kudo Y, et al. The role of periostin in tissue remodeling across health and disease. Cell Mol Life Sci. 2014;71:1279–1288. | ||
Masuoka M, Shiraishi H, Ohta S, et al. Periostin promotes chronic allergic inflammation in response to Th2 cytokines. J Clin Invest. 2012;122:2590–2600. | ||
Matsumoto H. Serum periostin: a novel biomarker for asthma management. Allergol Int. 2014;63:153–160. | ||
Baines KJ, Simpson JL, Wood LG, et al. Sputum gene expression signature of 6 biomarkers discriminates asthma inflammatory phenotypes. J Allergy Clin Immunol. 2014;133:997–1007. | ||
Samitas K, Zervas E, Vittorakis S, et al. Osteopontin expression and relation to disease severity in human asthma. Eur Respir J. 2011;37:331–341. | ||
Torrego A, Hew M, Oates T, Sukkar M, Chung KF. Expression and activation of TGF-β isoforms in acute allergen induced remodeling in asthma. Thorax. 2007;62:307–313. | ||
Asai K, Kanazawa H, Kamoi H, Shiraishi S, Hirata K, Yoshikawa J. Increased levels of vascular endothelial growth factor in induced sputum in asthmatic patients. Clin Exp Allergy. 2003;33:595–599. | ||
Konradsen JR, James A, Nordlund B, et al. The chitinase-like protein YKL-40: a possible biomarker of inflammation and airway remodeling in severe pediatric asthma. J Allergy Clin Immunol. 2013;132:328–335. | ||
Butler WT. Structural and functional domains of osteopontin. Ann N Y Acad Sci. 1995;760:6–11. | ||
Koh A, da Silva AP, Bansal AK, et al. Role of osteopontin in neutrophil function. Immunology. 2007;122:466–475. | ||
Konradsen JR, James A, Nordlund B, et al. The chitinase-like protein YKL-40: a possible biomarker of inflammation and airway remodeling in severe pediatric asthma. J Allergy Clin Immunol. 2013;132:328–335. | ||
Ober C, Tan Z, Sun Y, et al. Effect of variation in CHI3L1 on serum YKL-40 level, risk of asthma, and lung function. N Engl J Med. 2008;358:1682–1691. | ||
Dent AG, Sutedja TG, Zimmerman PV. Exhaled breath analysis for lung cancer. J Thorac Dis. 2013;5:540–550. | ||
Horváth I, Hunt J, Barnes PJ, et al. Exhaled breath condensate: methodological recommendations and unresolved questions. Eur Respir J. 2005;26:523–548. | ||
Kazani S, Planaguma A, Ono E, Bonini M, Zahid M, Marigowda G. Exhaled breath condensate eicosanoid levels associate with asthma and its severity. J Allergy Clin Immunol. 2013;132:547–553. | ||
Wu D, Zhou J, Bi H, et al. CCL-11 as a potential diagnostic marker for asthma? J Asthma. 2014;51:847–854. | ||
Lane C, Knight D, Burgess S, et al. Epithelial inducible nitric oxide synthase activity is the major determinant of nitric oxide concentration in exhaled breath. Thorax. 2004;59:757–760. | ||
Dweik RA, Boggs PB, Erzurum SC, et al. An official ATS clinical practice guideline: interpretation of exhaled nitric oxide levels (FENO) for clinical applications. Am J Respir Crit Care Med. 2011;184:60–615. | ||
Yang S, Park J, Lee YK, Kim H, Hahn YS. Association of longitudinal fractional exhaled nitric oxide measurements with asthma control in atopic children. Respir Med. 2015;109:572–579. | ||
Gemicioglu B, Musellim B, Dogan I, Guven K. Fractional exhaled nitric oxide (FeNo) in different asthma phenotypes. Allergy Rhinol. 2014;5:157–161. | ||
Sippel JM, Holden WE, Tilles SA, et al. Exhaled nitric oxide levels correlate with measures of disease control in asthma. J Allergy Clin Immunol. 2000;106:645–650. | ||
Hamid Q. Gross pathology and histopathology of asthma. J Allergy Clin Immunol. 2003;111:431–432. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

