Back to Journals » Clinical, Cosmetic and Investigational Dermatology » Volume 16
Serum miR-342-3p Acts as a Biomarker for Systemic Lupus Erythematosus and Participates in the Disease Progression
Authors You M, Zhang L, Ding J
Received 17 June 2022
Accepted for publication 13 October 2022
Published 5 January 2023 Volume 2023:16 Pages 39—46
DOI https://doi.org/10.2147/CCID.S378985
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jeffrey Weinberg
Min You, Long Zhang, Junxiao Ding
Department of Dermatology, Affiliated Hospital of Gansu Medical College, Pingliang, Gansu, People’s Republic of China
Correspondence: Junxiao Ding, Department of Dermatology, Affiliated Hospital of Gansu Medical College, No. 296, Kongtong East Road, Kongtong District, Pingliang, Gansu, 744000, People’s Republic of China, Tel +86-0933-8613787, Fax +86-0933-8613787, Email [email protected]
Purpose: The aim of this study was to investigate the diagnostic ability and prognostic value of miR-342-3p in SLE patients by detecting the expression level of serum miR-342-3p in SLE patients.
Patients and Methods: The expression level of serum miR-342-3p in all subjects was determined by qRT-PCR technology, and the correlation of miR-342-3p with SLEDAI scores was evaluated using Spearman correlation coefficient. The diagnostic value of miR-342-3p in SLE was assessed using ROC curve. The prognostic value of miR-342-3p in SLE patients was analyzed by Kaplan–Meier survival curve and multivariate COX regression.
Results: The qRT-PCR results showed that the expression level of serum miR-342-3p in SLE patients was significantly lower than that in healthy controls (P < 0.001). Serum miR-342-3p expression level was negatively correlated with SLEDAI scores (r = - 0.810, P < 0.001). The ROC curve suggested that serum miR-342-3p expression level was discriminative between SLE patients and healthy controls. Survival analysis results indicated that SLE patients with low serum miR-342-3p expression had a higher probability of poor prognosis of SLE (Log rank P = 0.003).
Conclusion: The expression level of serum miR-342-3p was valuable in the diagnosis of SLE. Meanwhile, the low expression level of serum miR-342-3p was associated with the poor prognosis of SLE.
Keywords: miR-342-3p, systemic lupus erythematosus, diagnosis, prognosis
Introduction
Systemic lupus erythematosus,1 a multi-origin autoimmune disease, is a disease whose etiological factor is still unknown.2 Currently, it is widely believed that SLE is a series of clinical symptoms caused by the abnormal activation of the human immune system, which attacks the body’s own tissues and leads to organ and tissue damage.3,4 SLE can affect any organ. Kidney involvement, also called lupus erythematosus nephritis, can significantly increase the incidence and mortality of lupus erythematosus. Once lupus nephritis occurs, a lot of immunosuppressants are needed to combat it. At the same time, the increased side effects of the drug make it difficult for patients to survive this checkpoint.1,5 The complexity of this disease is also manifested in the difficulty in identifying patients with SLE in the early stages of the disease. As we all know, the delay of disease treatment has a great relationship with the prognosis of the disease and shortening the time between diagnosis and treatment is of great significance for improving the quality of life of patients and increasing the survival rate of patients. Therefore, the early diagnosis of systemic lupus erythematosus is of great significance to SLE patients.6
MiRNA is a short non-coding RNA molecule that negatively regulates gene expression at the post-transcriptional level by targeting messenger RNA (mRNA).7 One miRNA molecule may target hundreds of messenger RNAs, and it has been suggested that up to 50% of mammalian genes are hidden under miRNA fine-tuning.8 miRNAs are mainly involved in the regulation of a variety of cells and developmental processes, including the differentiation of immune cells and the result of immune responses. Studies in recent years have shown that miRNAs play a vital role in the occurrence and development of different autoimmune diseases, including rheumatoid arthritis (RA) and SLE.9,10 A study showed that miR-142-5p is considered important in the pathogenesis of SLE. miR-142-5p expression level was significantly down-regulated in T cells of SLE patients. This phenomenon contributes to the over-activation of T cells, thereby mediating T/B cell interaction and promoting the formation of B cell germinal center and the production of antibodies, and thus exacerbating the development of SLE.11 The study by Stagakis et al confirmed that miR-21 expression levels were different in patients with active and inactive SLE. MiR-21 expression was up-regulated in SLE active patients and significantly decreased during disease remission.12 Carlsen et al studied the miRNA expression profile associated with SLE, and they found that the expression of miR-342-3p in the serum of patients with SLE combined with active nephritis was significantly down-regulated.13 Although it has been documented that miR-342-3p is associated with the occurrence of SLE, there are relatively few studies in this field in diagnosis and prognosis in SLE.
In this paper, the expression of miR-342-3p in SLE patients and healthy subjects were investigated through a series of experiments, and the factors influencing the prognosis of SLE patients were evaluated through follow-up analysis.
Materials and Methods
Study Population and Sample Collection
In this study, 90 SLE patients (mean age: 62.73 ± 7.03, SLEDAI scores: 8.21 ± 3.21) and 85 healthy controls (mean age: 63.45 ± 7.49) were recruited. All the SLE patients met the criteria for the classification of SLE as specified by the 2019 European League Against Rheumatism/American College of Rheumatology Classification Criteria.14 Exclusion criteria are as follows: Patients with malignant tumors, infectious diseases, other autoimmune diseases, as well as thyroid, adrenal, liver, kidney, and severe cardiovascular diseases were excluded. The samples from the healthy control group met the criteria for clinical blood donation through interviews and clinical tests. They did not have any of the diagnostic criteria for SLE and had no history of major diseases. Blood samples were collected from all subjects and stored at −80°C after centrifugation. This study protocol was approved by the Clinical Research Ethics Committee of Affiliated Hospital of Gansu Medical College, and all the recruiters had signed written informed consent (Ethics approval number: 2019–0325).
RNA Extraction and Quantitative Determination
Blood samples were collected for total RNA extraction and microRNA determination by qRT-PCR technology as previously described with slight modification. Total RNA was extracted from serum samples using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and then cDNA was obtained via reverse transcription which performed by a PrimeScript RT Reagent kit (Takara Biotechnology Co., Ltd., Dalian, China). With U6 as the internal reference, the expression of miR-342-3p was measured by SYBR Premix Ex Taq™ II commercial kit (Takara, Dalian, China) and Applied Biosystems 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA). The reactions were performed in duplicate, and the relative expression level was calculated by the method of 2−ΔΔCt method.
Follow-Up Plan
Ninety patients with SLE were divided into two groups according to the expression level of serum miR-342-3p and performed a 2-year follow-up plan at fixed time, and the poor prognosis of the patients was as follows: 1) the patient died; 2) the appearance of persistent active disease; 3) the appearance of new or worsening clinical lupus symptoms.15,16 Kaplan–Meier survival curve was plotted according to the adverse prognosis of the patients. Log-rank method was used to compare the survival rate of different groups. Multivariate COX regression analysis was used to determine the independent factors affecting survival.
Statistical Analysis
All statistical analysis was performed using GraphPad Prism 7.0 (GraphPad Software, Inc., CA, USA) and SPSS 20.0 (SPSS Inc., Chicago, USA). The difference among groups was tested by Student’s t-test. The Spearman correlation coefficient was used to assess the correlation between continuous variables and an ROC curve was used to evaluate the diagnostic ability of miR-342-3p in SLE. The Kaplan–Meier analysis and log-rank method were used to analyze the relationship between the expression level of miR-342-3p and the poor prognosis of SLE, and multivariate regression analysis was performed using COX regression model. P < 0.05 was considered as a statistically significant difference.
Results
Clinical Characteristic of Subjects
The age, gender, body mass index (BMI), and other data of all subjects are shown in Table 1. The mean SLEDAI score and anti-dsDNA value of SLE patients were 8.21 ± 3.21 and 132.75 ± 60.94, respectively. Compared with healthy controls, the erythrocyte sedimentation rate (ESR) and C-reactive protein (CRP) values in SLE group were all significantly increased (P < 0.001). In addition, there were no significant differences in gender, age, BMI between SLE patients and healthy controls (P > 0.05), indicating that the two groups were comparable.
 |
Table 1 Clinical Data of the Study Population |
Serum miR-342-3p Levels in SLE Patients
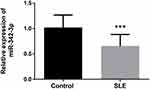
The serum expression levels of miR-342-3p in all participants were detected by qRT-PCR technology. The results showed that the expression level of miR-342-3p in SLE patients was significantly lower than that in healthy controls (P < 0.001, Figure 1), which suggested that the differences in serum miR-342-3p expression levels between SLE patients and healthy controls may be a new method for the diagnosis of SLE, and miR-342-3p is expected to be a new biomarker for the diagnosis of SLE in the future.
Correlation of miR-342-3p Expression Level with SLEDAI Scores in SLE Patients
Systemic lupus erythematosus disease activity index (SLEDAI) scores are clinically recognized as a reliable and effective evaluation tool for determining disease activity of SLE in research and clinical applications. Different doses of hormones and immunosuppressive agents can be selected by SLEDAI scores. Different scores represent different levels of activity in SLE disease, with a score of 0 to 4 indicating almost no activity, a score of 5 to 9 indicating mild activity, a score of 10 to 14 indicating moderate activity, and a score of ≥15 indicating severe activity. In this study, the mean SLEDAI score of SLE patients was 8.21 ± 3.21, ranging from 3 to 18, which included patients with different levels of SLE activity. The correlation between serum miR-342-3p expression level and SLEDAI scores was evaluated and analyzed in this study. As shown in Figure 2, it could be seen that serum miR-342-3p expression level was negatively correlated with SLEDAI scores (r = −0.810, P < 0.001), suggesting that the level of miR-342-3p was significantly related to disease activity.
 |
Figure 2 The correlation of serum miR-342-3p level with SLEDAI scores in SLE patients. Serum miR-342-3p levels were negatively correlated with SLEDAI scores (r = - 0.810, P < 0.001) in SLE patients. |
Evaluation of the Diagnostic Ability of miR-342-3p in SLE
An ROC curve was established to assess the ability of serum miR-342-3p to differentiate SLE patients from healthy controls based on the expression levels of serum miR-342-3p in all subjects. As shown in Figure 3, the area under the curve was 0.870 and when the cut-off value was 0.745, the sensitivity and specificity were 70.0% and 90.6%, respectively. The above results indicated that serum miR-342-3p has a high accuracy in identifying and distinguishing SLE patients.
Kaplan–Meier Survival Curve Analysis
Ninety patients with SLE were divided into two groups according to the expression level of serum miR-342-3p, including miR-342-3p low-expression group (n = 44) and miR-342-3p high-expression group (n = 46). During the two-year follow-up, the number of patients with poor prognosis in the miR-342-3p high-expression group and in miR-342-3p low-expression group was 6 and 21, respectively, and the probability of poor prognosis in the miR-342-3p low-expression group was significantly higher than that in the miR-342-3p high-expression group (Log Rank P = 0.003, Figure 4). After adjusting the influence of confounding factors, multivariate Cox regression analysis was used to analyze the factors influencing the prognosis of SLE patients. Table 2 reveals that serum miR-342-3p level (HR: 0.188, 95% CI: 0.067–0.526, P = 0.001) and SLEDAI score (HR: 2.828, 95% CI: 1.149–6.963, P = 0.014) were the independent risk factors influencing the prognosis of SLE patients. Meanwhile, we also found from Table 3 that the SLEDAI score of the low miR-342-3p expression group was significantly higher than that of the high miR-342-3p expression group (P < 0.001), while there was no significant difference in other indicators (P > 0.05). The above results indicated that the low expression of serum miR-342-3p predicted a higher probability of poor prognostic events in SLE patients.
 |
Table 2 Multivariate Cox Regression Analysis for the Event-Free Survival of SLE Patients |
 |
Table 3 Clinical Data of the SLE Group |
Discussion
SLE is a chronic autoimmune disease that affects different organs and systems. Although the etiology of the disease has not been fully clarified, the multifactorial pathogenesis is widely accepted and both genetic environment and hormonal factors contribute to its development.17,18 As SLE is a heterogeneous disease, no single clinical feature or laboratory test can diagnose it. Therefore, the discovery of an effective and reliable tool to better diagnose SLE is critical for disease surveillance and prognosis. A biomarker can be defined as a physical signal or cell, biochemical, molecular or genetic change, through which normal or abnormal biological processes can be identified and measured. Biomarkers can be used for diagnosis, prognosis, prediction, pharmacodynamics or substitution.
In the present study, we detected the expression level of serum miR-342-3p in SLE patients and the healthy controls by qRT-PCR technology and found that the expression level of serum miR-342-3p in SLE patients was significantly lower than that in the healthy controls, which suggested that the difference in the expression level of serum miR-342-3p might reflect whether the subjects had SLE.
Since the introduction of miRNAs in recent decades, a series of related studies have demonstrated the emerging role of miRNAs in regulating the maturation of the immune system and the pathological mechanisms of autoimmune diseases including SLE. In addition, miRNAs can also be used to monitor the severity of SLE and can be used as a new potential target for the treatment of SLE.19,20 MiR-342-3p plays a crucial role in autoimmune diseases. Wang et al found that miR-342-3p was specifically down-regulated in the serum of rheumatoid arthritis (RA) patients compared with healthy controls.21 Jeannie et al used microarray technology to study the differential miRNA expression in peripheral blood mononuclear cells (PBMCs) in patients with lupus nephritis and healthy controls. They found a total of 29 miRNAs with abnormal expression, among which 5 miRNAs, including miR-342-3p, were down-regulated in patients with lupus nephritis.22 This result is consistent with our study on the serum miR-342-3p expression level in SLE patients and healthy controls. Therefore, based on this study, we evaluated the correlation between the serum miR-342-3p expression level and SLEDAI scores through Spearman correlation coefficient analysis, and the results showed that miR-342-3p was significantly negatively correlated with SLEDAI scores. SLEDAI score has been developed as an indicator in clinic for evaluating the activity of SLE and has been used as a universal approach in assessing the SLE activity since its introduction in 1985.23,24 This indicator has been used successfully by clinical experts and trainees and has been proven to be valuable in both research and clinical applications.25 These results further proved that miR-342-3p played a vital role in the development of SLE. In addition, we assessed the diagnostic value of miR-342-3p in SLE by establishing ROC curve in this study, and finally concluded that miR-342-3p has good differentiating power in SLE patients and healthy controls. Then, the subjects were followed up for two years and the survival curve was drawn. The results showed that patients with low miR-342-3p expression had a poor prognosis, indicating that patients with low miR-342-3p expression had a higher probability of poor prognosis.
Conclusion
In summary, this study proved that the serum miR-342-3p in SLE patients was lower than in healthy controls, and low serum miR-342-3p expression level may indicate the occurrence of SLE. Meanwhile, low expression of serum miR-342-3p was often associated with poor prognosis of SLE. Therefore, the expression level of serum miR-342-3p can be used as a new potential biomarker for the diagnosis and prognosis assessment of SLE.
Ethics Statement
This study protocol was approved by the Clinical Research Ethics Committee of Affiliated Hospital of Gansu Medical College and confirmed to the Helsinki Declaration standards for human medical research. All the recruiters had signed written informed consent.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Hanly JG, Su L, Urowitz MB, et al. A longitudinal analysis of outcomes of lupus nephritis in an international inception cohort using a multistate model approach. Arthritis Rheumatol. 2016;68(8):1932–1944. doi:10.1002/art.39674
2. Durcan L, O’Dwyer T, Petri M. Management strategies and future directions for systemic lupus erythematosus in adults. Lancet. 2019;393(10188):2332–2343. doi:10.1016/S0140-6736(19)30237-5
3. Liu S, Cheng Y, Xie Z, et al. A conscious resting state fMRI study in SLE patients without major neuropsychiatric manifestations. Front Psychiatry. 2018;9:677. doi:10.3389/fpsyt.2018.00677
4. Flynn A, Gilhooley E, O’Shea F, Wynne B. The use of SLICC and ACR criteria to correctly label patients with cutaneous lupus and systemic lupus erythematosus. Clin Rheumatol. 2018;37(3):817–818. doi:10.1007/s10067-018-3999-0
5. Satish S, Deka P, Shetty MS. A clinico-pathological study of lupus nephritis based on the international society of nephrology-renal pathology society 2003 classification system. J Lab Physicians. 2017;9(3):149–155. doi:10.4103/JLP.JLP_44_16
6. Sebastiani GD, Prevete I, Iuliano A, Minisola G. The importance of an early diagnosis in systemic lupus erythematosus. Isr Med Assoc J. 2016;18(3–4):212–215.
7. Shumnalieva R, Kachakova D, Shoumnalieva-Ivanova V, Miteva P, Kaneva R, Monov S. Whole peripheral blood miR-146a and miR-155 expression levels in Systemic lupus erythematosus patients. Acta Reumatol Port. 2018;43(3):217–225.
8. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi:10.1016/j.cell.2004.12.035
9. O’Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10(2):111–122. doi:10.1038/nri2708
10. Tang Y, Luo X, Cui H, et al. MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum. 2009;60(4):1065–1075. doi:10.1002/art.24436
11. Ding S, Liang Y, Zhao M, et al. Decreased microRNA-142-3p/5p expression causes CD4+ T cell activation and B cell hyperstimulation in systemic lupus erythematosus. Arthritis Rheum. 2012;64(9):2953–2963. doi:10.1002/art.34505
12. Stagakis E, Bertsias G, Verginis P, et al. Identification of novel microRNA signatures linked to human lupus disease activity and pathogenesis: miR-21 regulates aberrant T cell responses through regulation of PDCD4 expression. Ann Rheum Dis. 2011;70(8):1496–1506. doi:10.1136/ard.2010.139857
13. Carlsen AL, Schetter AJ, Nielsen CT, et al. Circulating microRNA expression profiles associated with systemic lupus erythematosus. Arthritis Rheum. 2013;65(5):1324–1334. doi:10.1002/art.37890
14. Aringer M, Costenbader K, Daikh D, et al. 2019 European league against rheumatism/American College of Rheumatology classification criteria for systemic lupus erythematosus. Ann Rheum Dis. 2019;78(9):1151–1159. doi:10.1136/annrheumdis-2018-214819
15. Yuan Y, Zhao L, Ye Z, Ma H, Wang X, Jiang Z. Association of toll-like receptor 9 expression with prognosis of systemic lupus erythematosus. Exp Ther Med. 2019;17(4):3247–3254. doi:10.3892/etm.2019.7290
16. Pamuk ON, Akbay FG, Donmez S, Yilmaz N, Calayir GB, Yavuz S. The clinical manifestations and survival of systemic lupus erythematosus patients in Turkey: report from two centers. Lupus. 2013;22(13):1416–1424. doi:10.1177/0961203313499956
17. Maselli A, Conti F, Alessandri C, et al. Low expression of estrogen receptor beta in T lymphocytes and high serum levels of anti-estrogen receptor alpha antibodies impact disease activity in female patients with systemic lupus erythematosus. Biol Sex Differ. 2016;7:3. doi:10.1186/s13293-016-0057-y
18. Ceccarelli F, Perricone C, Cipriano E, et al. Joint involvement in systemic lupus erythematosus: from pathogenesis to clinical assessment. Semin Arthritis Rheum. 2017;47(1):53–64. doi:10.1016/j.semarthrit.2017.03.022
19. Honarpisheh M, Kohler P, von Rauchhaupt E, Lech M. The involvement of MicroRNAs in modulation of innate and adaptive immunity in systemic lupus erythematosus and lupus nephritis. J Immunol Res. 2018;2018:4126106. doi:10.1155/2018/4126106
20. Qu B, Shen N. miRNAs in the pathogenesis of systemic lupus erythematosus. Int J Mol Sci. 2015;16(5):9557–9572. doi:10.3390/ijms16059557
21. Wang W, Zhang Y, Zhu B, et al. Plasma microRNA expression profiles in Chinese patients with rheumatoid arthritis. Oncotarget. 2015;6(40):42557–42568. doi:10.18632/oncotarget.6449
22. Te JL, Dozmorov IM, Guthridge JM, et al. Identification of unique microRNA signature associated with lupus nephritis. PLoS One. 2010;5(5):e10344. doi:10.1371/journal.pone.0010344
23. Bombardier C, Gladman DD, Urowitz MB, Caron D, Chang CH. Derivation of the SLEDAI. A disease activity index for lupus patients. The Committee on Prognosis Studies in SLE. Arthritis Rheum. 1992;35(6):630–640. doi:10.1002/art.1780350606
24. Gladman DD, Ibanez D, Urowitz MB. Systemic lupus erythematosus disease activity index 2000. J Rheumatol. 2002;29(2):288–291.
25. Hawker G, Gabriel S, Bombardier C, Goldsmith C, Caron D, Gladman D. A reliability study of SLEDAI: a disease activity index for systemic lupus erythematosus. J Rheumatol. 1993;20(4):657–660.
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.