Back to Journals » OncoTargets and Therapy » Volume 14
Serum Exosomal miRNA-1226 as Potential Biomarker of Pancreatic Ductal Adenocarcinoma
Authors Wang C, Wang J, Cui W , Liu Y, Zhou H , Wang Y, Chen X , Chen X, Wang Z
Received 15 December 2020
Accepted for publication 8 February 2021
Published 26 February 2021 Volume 2021:14 Pages 1441—1451
DOI https://doi.org/10.2147/OTT.S296816
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Nicola Silvestris
Cheng Wang,1,2,* Jianhua Wang,1,* Wenjing Cui,1,* Yongkang Liu,1 Hao Zhou,1 Yajie Wang,1 Xin Chen,1 Xiao Chen,1 Zhongqiu Wang1
1Department of Radiology, The Affiliated Hospital of Nanjing University of Chinese Medicine, Nanjing, 210029, People’s Republic of China; 2Department of Radiology, Nanjing Drum Tower Hospital, The Affiliated Hospital of Nanjing University Medical School, Nanjing, 210029, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Zhongqiu Wang; Xiao Chen
Department of Radiology, Affiliated Hospital of Nanjing University of Chinese Medicine, No. 155 Hanzhong Road, Nanjing, Jiangsu Province, 210029, People’s Republic of China
Tel +8613905162963
; +8613816957732
Fax +862586619843
Email [email protected]; [email protected]
Background: Pancreatic ductal adenocarcinoma (PDAC) is the fourth leading cause of cancer-related mortality and it is urgent to find biomarkers for early detection of PDAC. Exosomal miRNAs are useful biomarkers for cancer detection. The aims of this study were to investigate the potential role of serum exosomal miRNA in detection of PDAC and to analyze the correlation between the levels of exosome miRNA and the tumor biological behaviors.
Materials and Methods: Thirteen serum samples were collected from five patients with PDACs, three healthy individuals (HIs) and five benign pancreatic lesions (BP) for a high throughput profiling analysis to identify an altered miRNA expression patterns in PDAC. Candidate exosomal miRNAs were filtered based on a second independent cohort that included 17 PDACs and 12 benign pancreatic lesions by quantitative real-time polymerase chain reaction (qRT-PCR). Four miRNAs were selected for miRNA validation as PDAC biomarkers in a subsequent set of samples. The association between candidate exosomal miRNA and tumor behavior (tumor invasion or metastases) was evaluated in 17 PDACs. In vitro studies were performed to evaluate the role of candidate exosomal miRNA on cell viability, apoptosis and cell migration in two PDAC cell lines.
Results: The expression of 11 miRNAs showed same trend between PDAC and BP, and between PDAC and HIs. Six of them were upregulated (miR-203b-5p, miR-342-5p, miR-337-5p, miR-149-5p, miR-877-5p, miR-203a-3p), and five were downregulated (miR-1226-3p, miR-3182, miR-625-3p, miR-624-5p, miR-664a-5p). miR-1226-3p was selected as the candidate exosomal biomarker for the PDAC detection. The expression of serum exosomal miRNA-1226-3p was downregulated in PDACs compared to the BPs (p = 0.025). miR-1226-3p had acceptable performance in predicting [area under the curve (AUC) = 0.74] PDAC. Exosomal miRNA-1226-3p level in PDAC with invasion or metastases was lower than that without invasion or metastases (p = 0.028). Transfection of miRNA-1226-3p significantly inhibited the proliferation of PANC-1 and BXP-3 cells, stimulated cell apoptosis and inhibited cell migration.
Conclusion: Serum exosomal miRNA-1226-3p is a potential biomarker in diagnosing and predicting the tumor invasion or metastases of PDAC.
Keywords: exosome, miRNA, pancreatic ductal adenocarcinoma, miRNA-1226-3p
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the third leading cause of cancer-related death with a five-year survival rate of nearly 9%.1 PDAC is expected to be the second leading cause of cancer-death by 2030 in the USA.1,2 Chemotherapy, radiotherapy and surgery are the treatment strategies for PDAC.3 However, more than 80% of the patients miss the chance of surgical resection.4–6 Detection of PDAC at early stage will lead to improved survival.7
CA19-9 has been used as a biomarker for PDAC detection. However, because of its low positive predictive value, serum CA 19–9 cannot be used as a screening marker.8 It is urgent to search for novel diagnostic biomarker that can detect the early PDAC and predict the clinical outcomes. Circulating protein, metabolite and miRNAs have been reported in previous studies.7,9–11 More and more studies demonstrate that exosome is a potential diagnostic biomarker for PDAC.12,13 Exosomes (Exo), 50–150 nm small membrane vesicles that derived from various body fluids,14,15 contain a lot of information-containing factors, including mRNAs, miRNA, lipids, and proteins.16–18 Kitagawa et al18 showed that circulating pancreatic cancer exosomal RNAs was useful tools for the early detection of PDAC. Melo et al19 reported that exosomal Glypican-1 may be a non-invasive diagnostic and screening tool to detect early stages of pancreatic cancer. Tumor-related miRNAs exit and play a key role in biological process.20 Exosomal miRNAs are relatively stable in the body fluids, especially in blood, because of the protective effect of the exosomal vesicles.21 This feature of stability suggests that exosomal miRNA may be a good potential biomarker.22 Some studies showed that the expression of miRNAs in blood was correlated with the prognostic of PDAC, which indicated that miRNAs may be the potential diagnostic biomarker or predict the tumor invasion.23 However, few studies have shown the role of exosomal miRNA in detecting early stages of pancreatic cancer.
In this study, we isolated exosomal miRNA from the serum and assessed the expression levels of exosomal miRNA in healthy people, PDAC patients, and patients with benign pancreatic lesion. The aim of this study was to explore the potential biomarker of exosomal miRNA for early detecting PDAC. In addition, we also investigated the association between candidate exosomal miRNA and tumor invasion or metastases in PDAC patients and in pancreatic cell lines.
Materials and Methods
Patients and Samples
Blood samples were collected from 3 healthy individuals (HIs), patients with PDAC (n = 27), patients with benign lesion of pancreas (BPs, n = 17) during April 2017 to April 2019. The peripheral blood samples were centrifuged at 5000 rmp for 10 min at 4°C. The serum was separated to EP tube and stored at −80°C until further use. The serum samples were divided into three groups: 5 PDAC vs 3 HIs, 5 PDAC vs 5 BPs, and 17 PDACs vs 12 PBs. The first two groups were set for next-generation sequencing analysis. Twenty-nine samples (17 PDACs vs 12 BPs) were used to verify the selected potential biomarker. Documented informed consent was obtained from each subject, and all aspects of the study were approved by the Ethics Committee of Nanjing University of Chinese Medicine (2017NL-137-05). The study was performed in accordance with the relevant guidelines, regulations and the Declaration of Helsinki.
Isolation and Identification of Exosomes
Serum Exosomes were isolated using exoEasy Maxi Kit (Qiagen, Hildesheim, Germany) according to the manufacturer’s instructions. The expression of exosome-derived cluster of differentiation CD63 (surface markers of exosom) was investigated to identify the exosomes by Western blot (WB) analysis. Separate exosome pellets isolated from serum were treated with RIPA lysis buffer. Transmission electron microscopy (TEM) and nanoparticle tracking analysis (NTA, NanoSight NS300, Malvern Instruments Ltd; UK) were also used to identify the exosomes.
Isolation of Exosomal RNA from Serum
Total RNA was extracted from serum exosome using a miRNeasy Serum/Plasma Kit (Qiagen, Hildesheim, Germany). RNA extraction was performed following the manufacturer’s instructions and RNA concentration was quantified using the NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). All RNA samples presented an OD 260/280 nm ratio ≥ 1.8.
Small RNA Library Preparation and Sequence Analysis
Small RNA libraries were constructed using the New England Biolabs (NEB) NEBNext Multiplex Small RNA Library Prep Set for Illumina sequencers and the NEB standard protocol had been reported previously.24 The qualified library preparations were sequenced on an Illumina Hiseq X platform (Illumina, San Diego, CA, USA). Adapter trimming and sequence analysis were conducted using Flow, v3.0 (Partek Incorporated, St. Lousi, Missouri, USA), Bowtie (v2.1.0) and miRbase v20 for alignment and annotation.
qRT-PCR Analysis for Evolution and Validation of Candidate Serum Exosomal miRNA
The candidate miRNAs were further confirmed with quantitative real-time polymerase chain reaction (qRT-PCR). qRT-PCR was performed using the QuantiFast® SYBR® Green PCR Master Mix in an ABI 9700 qPCR system (Applied Biosystems, Foster City, CA, USA). The relative gene expression values of the target miRNAs were normalized to that of RNU6B (U6) and the differences were calculated using the 2−∆∆Ct method.25
Construction of Overexpression Plasmid
GAPDH was chosen as the internal reference. The hairpin structure primers were designed to construct the miRNA-1226 overexpression plasmid using pCD316-ZsGreen-siRNA. The primer sequences were as follows: miRNA-1226, forward: 5′-ACACTCCAGCTGGGTCACCAGCCCTGTGTT-3′, reverse: 5′- TCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCTAGGG-3′; GAPDH, forward: 5′-CACATCGCTCAGACACCATG-3′, reverse: 5′- TGACGGTGCCATGGAATTTG −3′.
Cell Proliferation
CCK8 was used to assess the effects of overexpression of miRNA-1226 on cell proliferation. Briefly, PANC-1 and BXP-3 cells were seeded into 6-well plates. When the cells confluence reached 80%, the two cells were transfected with the control plasmid and miRNA-1226-3p overexpression plasmid. After 24 h, the cell was harvested and cultured in 96-well plates for 24 h. Then, add 10 μL of CCK8 solution to each well and the plate was incubated in an incubator for 1–4 hours. Then the wells were measured at 450 nm using a microplate reader.
Apoptosis, Proliferation and Migration Assays
We performed flow cytometry assays to analyze the effects of overexpression of miRNA-1226 on apoptosis or necrosis and proliferation.
PANC-1 and BXP-3 cells were transfected with the control plasmid and miRNA-1226-3p overexpression plasmid for 48h. Then, 500μL of trypsin without EDTA was added in each well to digest the cells for 2 min, and centrifuged at 1500 rpm for 5 min at 4°C to harvest the cells. Then, washed them three times with cold phosphate buffered saline (PBS) and stained with Annexin V-FITC/PI. Subsequently, 400 μL of binding buffer was added to each cell, and then filtered using the 300-mesh nylon mesh. Cells were then detected by flow cytometry two-channel wavelengths of FL1 and FL3.
Migration rate was measured by wound healing assay and transwell chamber method. Wound was made using a pipette tip and pictures were taken immediately and 24h after wounding. The distance migrated by the cell or the number of cell migration during the time period was measured.
Computed Tomography (CT) Imaging and Image Analysis
The CT images were obtained using the 64-channel multi-detector CT scanner (Brilliance 64, Philips Medical System, Netherland) or (Optima, GE Healthcare, Japan). The imaging parameters were reported previously.26 CT images were independently reviewed by two abdominal radiologists. If there was an inconsistency, consensus was decided by the third radiologist with >20 years’ experience in abdominal radiology. The variables including: size, tumor margin, shape, components, tumor attenuation, pancreatic or bile duct dilatation, metastases or invasion. The definition and classification of the variables were reported previously.26
Statistical Analyses
All statistical analyses were performed with the commercially available software (SPSS 22.0 Chicago, IL, USA). The quantitative data were presented as means ± SD. One-way ANOVA using Bonferroni correction or Independent-sample T test was performed for statistical analysis. The receiver operating characteristic (ROC) curve was used to assess the diagnostic performance in of candidate exosomal miRNA. P-values < 0.05 were regarded as statistical significance.
Result
The Characterization of Exosomes from the Serum
The results of TEM and NTA analysis showed that exosomes were cup-like vesicle with the double lipid layer (Figure 1A) and >90% of the recovered particles had a size of <200 nm (30–150 nm) diameter. WB showed that exosomes were positive for CD63 (Figure 1B).
Altered miRNA Expression Pattern
The WB showed that the small RNA libraries were qualified to sequence (Figure 1C). Ninety exosomal miRNAs showed altered expression between PDACs vs 3His (Figure 2A). Twenty-nine miRNAs were upregulated and 61 were downregulated. A total of 59 exosomal miRNAs showed altered expression between PDACs and BPs, including 28 upregulated miRNAs and 31 downregulated miRNAs (Figure 2B). Subsequently, those miRNAs showed same altered patterns between PDACs and HIS and between PDACs and BPs were figured out. We found that 6 miRNAs were upregulated (miR-203b-5p, miR-342-5p, miR-337-5p, miR-149-5p, miR-877-5p, miR-203a-3p) and five were downregulated (miR-1226-3p, miR-3182, miR-625-3p, miR-624-5p, miR-664a-5p). The data was deposited in the Sequence Read Archive (SRA) and the database accession number is SUB6747167.
Selection and Verification Serum Exosomal miRNA as the Diagnostic Biomarker
Four miRNAs (miR-203a-3p, miR-149-5p, miRNA-1226-3p, miR-624-5p) with largest differences in expression were selected for miRNA validation as PDACs biomarkers. No significant differences were observed in miR-203a-3p, miR-149-5p, and miR-624-5p between PDACs and BPs (Supplement Figure 1). The expression of serum exosomal miRNA-1226-3p (Figure 3A) was significantly higher in the BPs patients compared to the PDACs (p = 0.025). ROC curve analyses were constructed to evaluate the diagnostic value of miRNA-1226-3p for PDAC and the area under curve (AUC) was 0.74 (95% confidence interval: 0.55–0.92) (Figure 3B).
Relationship Between miRNA-1226-3p and CT Imaging Findings of PDAC
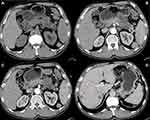
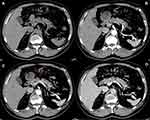
The associations between the exosomal miRNA-1226-3p and tumor components, pancreatic or bile duct dilatation, invasion or metastases were evaluated. There was a significantly negative correlation between the expression of the miRNA-1226-3p and invasion or metastases of PDACs (R= −0.516, p = 0.028) (Table 1, Figure 4 and 5). No significant correlations were found between miRNA-1226-3p and with tumor components, tumor enhancement, pancreatic or bile duct dilatation.
 |
Table 1 The Correlation Between the Expression of miRNA-1226-3P and CT Imaging Findings |
MiRNA-1226 Affects Biological Behaviors of PDAC Cells
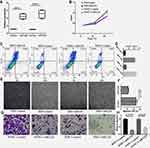
Subsequently, we observed the effects of miRNA-1226 on biological behaviors of PDAC in vitro. The levels of miRNA-1226 in PANC-1 and BXP-3 transfected with miRNA-1226 were significantly higher than the control (Figure 6A). The proliferation of PANC-1 and BXP-3 cells transfected with miRNA-1226-3p was lower than those transfected with control miRNA (Figure 6B). We also observed cell apoptosis (Figure 6C and D) and migration (Figure 6E–H). The apoptosis in PDACs cells with miRNA-1226 transfection was much lower than that in the control group. Moreover, the migration ability of PDACs cells transfected with miRNA-1226 was lower than in the control group.
Discussion
The detection of PDAC at early stage remains a challenge in clinical practice. More than 80% of the patients at diagnosis are locally advanced or metastatic disease and miss the chance for surgical resection. It would be valuable to find more sensitive diagnostic biomarker for detection PDAC at early stage and for prediction the invasion or metastases.27 Many efforts have been made by using circulating protein, metabolite and miRNAs. However, few studies showed the role of exosomal miRNA for PDAC detection. Our data showed that exosomal miRNA-1226 may have potential in PDAC detection. Moreover, our data showed that miRNA-1226 were associated with biological behavior of PDAC. Low miRNA-1226 expression was associated with worse biological behavior.
Recently, many studies have shown that miRNA can regulate tumor invasion and migration, and can also be used as biomarker for tumor diagnosis.28 Circulating miRNAs are associated with development and prognosis of PDAC.29,30 Bloomston et al reported that 30 miRNAs (including miRNA-203) were upregulated in PDAC.31 Ikenaga et al reported miRNA-203 was a new prognostic biomarker in PDACs, and it was also overexpressed in PDAC compared with chronic pancreatitis and normal pancreas samples.32 A previous report showed that miRNA-203 expressed in panc-1 cells and its exosomes.33 However, it is not clear whether exosomal miRNA is a useful marker for PDACs detection.
Exosomes have been noted as a novel source of noninvasive biomarker because exosome contain tumor-specific molecules.34,35 Many studies have demonstrated that exosome could be used as diagnostic biomarker for PDAC.12,13,36 In addition, some studies have reported that exosomal miRNA can improve the accuracy of cancer detection.28,37,38 Xu et al39 show that plasma exosome miR-196a and miR-1246 are two potential indicators of localized pancreatic cancer. A recent study indicated that exosomal miRNAs in pancreatic cyst juice could be biomarkers for detection of PDAC.40 In this study, we identified 11 miRNAs with altered expression between PDAC and HIs or BPs. Four miRNAs were validated by RT-PCR and miRNA-1226 was found to be a biomarker in differentiation between PDAC and BPs. Our data also indicated that low level of exosomal miRNA-1226-3P expression was associated with tumor invasion or metastases of PDAC. Jin et al showed that miRNA-1226 promoted the induction of cell death by inhibiting the expression of the MUC1 oncoprotein.41 Our data were consistent with this report. MUC1 was a histological biomarker of PDAC. A recent study indicated that specific anti-MUC1 antibody could inhibit pancreatic cancer progression in a mouse model.42 We speculate that miRNA-1226-3P may also inhibit PDAC by downregulating MUC1.
To the best of our knowledge, this is the first study to exam the correlation between the expression of serum exosomal miRNA and the CT imaging finding. However, no significant correlations were found between miRNA-1226-3p and size, tumor components, tumor enhancement, pancreatic or bile duct dilatation. Further studies are needed in the future. Exosomal miRNA from pancreatic cancer cells may promote invasion and metastasis or chemoresistance, such as miRNA-222 and miRNA-155.36 In the present study, we also performed in vitro study to test the role of miRNA-1226-3p on biological behaviors of PDAC. Interestingly, we found that high level of miRNA-1226-3p expression could inhibit PANC-1 and BXP-3 cell viability, promoted cell apoptosis and inhibit cell migration. Those effects may be related to the inhibition of MUC1 by miRNA-1226-3P.41 Our data indicated that miRNA-1226-3p may be a potential agent for PDAC treatment.
There are some limitations in the present study. First, the number of samples in the sequencing analysis and in the validation cohort was small. In addition, there are no healthy individuals in the validation cohort. Future studies with a large number of validation cohorts are necessary to assess the diagnostic and predictive value of exosomal miRNA-1226. Second, we only validated 4 exosomal miRNA. Further studies are needed to show the value of other miRNA with altered expression. Third, our study did not analyze the association between miRNA-1226 and TNM stage, PDAC differentiation or prognosis of PDAC. Finally, we did not perform the in vivo study to evaluate the role of miRNA-1226 on tumor growth.
Conclusion
In summary, this study showed that serum exosomal miRNA-1226-3p was downregulated in PDACs compared with healthy individuals and BPs. Exosomal miRNA-1226 may be a potential biomarker for PDAC detection. In addition, we also showed that the expression of miRNA-1226-3p was negatively correlated with the invasion or metastases of PDACs. Overexpression of miRNA-1226-3p could inhibit PDAC cell proliferation, cell migration and promote the apoptosis.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 81771899, 81773460), the Key Program of Research and Development of Jiangsu Province (Grant No. BE2017772). We thank all authors for their continuous and excellent support with patient data collection, imaging analysis, statistical analysis, and valuable suggestions for the article.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 81771899, 81773460), the Key Program of Research and Development of Jiangsu Province (Grant No. BE2017772), Jiangsu Provincial Hospital of Chinese Medicine Peak Academic Talent Project (NO: y2018rc04) and Jiangsu Provincial Traditional Chinese Medicine Science and Technology Development Plan(NO: ZD201907).
Disclosure
There are no conflicts of interest to declare.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi:10.3322/caac.21551
2. Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74(11):2913–2921. doi:10.1158/0008-5472.CAN-14-0155
3. Renz BW, Boeck S, Roeder F, Trumm C, Heinemann V, Werner J. Oligometastatic Disease in Pancreatic Cancer – How to Proceed?. Visc Med Mar. 2017;33(1):36–41.
4. Al-Hawary MM, Francis IR, Chari ST, et al. Pancreatic ductal adenocarcinoma radiology reporting template: consensus statement of the society of abdominal radiology and the American Pancreatic Association. Gastroenterology. 2014;146(1):291–304 e1. doi:10.1053/j.gastro.2013.11.004
5. Philip PA, Mooney M, Jaffe D, et al. Consensus report of the national cancer institute clinical trials planning meeting on pancreas cancer treatment. J Clin Oncol. 2009;27(33):5660–5669.
6. Chu LC, Goggins MG, Fishman EK. Diagnosis and Detection of Pancreatic Cancer. Cancer J. 2017;23(6):333–342.
7. Fahrmann JF, Bantis LE, Capello M, et al. A Plasma-Derived Protein-Metabolite Multiplexed Panel for Early-Stage Pancreatic Cancer. J Natl Cancer Inst. 2019;111(4):372–379.
8. Scara S, Bottoni P, Scatena R. CA 19-9: biochemical and Clinical Aspects. Adv Exp Med Biol. 2015;867:247–260.
9. Vila-Navarro E, Duran-Sanchon S, Vila-Casadesus M, et al. Novel Circulating miRNA Signatures for Early Detection of Pancreatic Neoplasia. Clin Transl Gastroenterol. 2019;10(4):e00029.
10. Sun X, Zhou X, Zhang Y, Zhu X, Liu H. Systematic Review and Meta-Analysis of Diagnostic Accuracy of miRNAs in Patients with Pancreatic Cancer. Dis Markers. 2018;2018:6292396. doi:10.1155/2018/6292396
11. Wei L, Yao K, Gan S, Suo Z. Clinical utilization of serum- or plasma-based miRNAs as early detection biomarkers for pancreatic cancer: a meta-analysis up to now. Medicine. 2018;97(35):e12132. doi:10.1097/MD.0000000000012132
12. Wu Y, Tan X, Liu P, et al. ITGA6 and RPSA synergistically promote pancreatic cancer invasion and metastasis via PI3K and MAPK signaling pathways. Exp Cell Res. 2019;379(1):30–47. doi:10.1016/j.yexcr.2019.03.022
13. Yan Y, Fu G, Ming L. Role of exosomes in pancreatic cancer.. Oncol Lett. 2018;15(5):7479–7488. doi:10.3892/ol.2018.8348
14. Kowal J, Tkach M, Thery C. Biogenesis and secretion of exosomes. Curr Opin Cell Biol. 2014;29:116–125. doi:10.1016/j.ceb.2014.05.004
15. Pan BT, Teng K, Wu C, Adam M, Johnstone RM. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes.. J Cell Biol. 1985;101(3):942–948. doi:10.1083/jcb.101.3.942
16. Liu R, Chen X, Du Y, et al. Serum microRNA expression profile as a biomarker in the diagnosis and prognosis of pancreatic cancer. Clin Chem. 2012;58(3):610–618. doi:10.1373/clinchem.2011.172767
17. Pant S, Hilton H, Burczynski ME. The multifaceted exosome: biogenesis, role in normal and aberrant cellular function, and frontiers for pharmacological and biomarker opportunities. Biochem Pharmacol. 2012;83(11):1484–1494. doi:10.1016/j.bcp.2011.12.037
18. Kitagawa T, Taniuchi K, Tsuboi M, et al. Circulating pancreatic cancer exosomal RNA s for detection of pancreatic cancer. Mol Oncol. 2019;13(2):212–227. doi:10.1002/1878-0261.12398
19. Melo SA, Luecke LB, Kahlert C, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523(7559):177–182. doi:10.1038/nature14581
20. Li Z, Tao Y, Wang X, et al. Tumor-Secreted Exosomal miR-222 Promotes Tumor Progression via Regulating P27 Expression and Re-Localization in Pancreatic Cancer. Cell Physiol Biochem. 2018;51(2):610–629. doi:10.1159/000495281
21. Lima LG, Chammas R, Monteiro RQ, Moreira MEC, Barcinski MA. Tumor-derived microvesicles modulate the establishment of metastatic melanoma in a phosphatidylserine-dependent manner. Cancer Lett. 2009;283(2):168–175. doi:10.1016/j.canlet.2009.03.041
22. Huang X, Liang M, Dittmar R, Wang L. Extracellular MicroRNAs in Urologic Malignancies: chances and Challenges. Int J Mol Sci. 2013;14(7):14785–14799. doi:10.3390/ijms140714785
23. Goto T, Fujiya M, Konishi H, et al. An elevated expression of serum exosomal microRNA-191, − 21, −451a of pancreatic neoplasm is considered to be efficient diagnostic marker. BMC Cancer. 2018;18(1):116. doi:10.1186/s12885-018-4006-5
24. Hannafon BN, Trigoso YD, Calloway CL, et al. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res. 2016;18(1):90. doi:10.1186/s13058-016-0753-x
25. Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
26. Wang C, Cui W, Wang J, Chen X, Tong H, Wang Z. Differentiation between solid pseudopapillary neoplasm of the pancreas and hypovascular pancreatic neuroendocrine tumors by using computed tomography. Acta Radiol. 2019;60(10):1216–1223. doi:10.1177/0284185118823343
27. Chen D, Wu X, Xia M, et al. Upregulated exosomic miR-23b-3p plays regulatory roles in the progression of pancreatic cancer. Oncol Rep. 2017;38(4):2182–2188. doi:10.3892/or.2017.5919
28. Madhavan B, Yue S, Galli U, et al. Combined evaluation of a panel of protein and miRNA serum-exosome biomarkers for pancreatic cancer diagnosis increases sensitivity and specificity. Int J Cancer. 2015;136(11):2616–2627. doi:10.1002/ijc.29324
29. Kishikawa T. Circulating RNAs as new biomarkers for detecting pancreatic cancer. World J Gastroenterol. 2015;21(28):8527–8540. doi:10.3748/wjg.v21.i28.8527
30. Deng T, Yuan Y, Zhang C, et al. Identification of Circulating MiR-25 as a Potential Biomarker for Pancreatic Cancer Diagnosis. Cell Physiol Biochem. 2016;39(5):1716–1722. doi:10.1159/000447872
31. Bloomston M, Frankel WL, Petrocca F, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297(17):1901–1908. doi:10.1001/jama.297.17.1901
32. Ikenaga N, Ohuchida K, Mizumoto K, et al. MicroRNA-203 expression as a new prognostic marker of pancreatic adenocarcinoma. Ann Surg Oncol. 2010;17(12):3120–3128. doi:10.1245/s10434-010-1188-8
33. Zhou M, Chen J, Zhou L, Chen W, Ding G, Cao L. Pancreatic cancer derived exosomes regulate the expression of TLR4 in dendritic cells via miR-203. Cell Immunol. 2014;292(1–2):65–69. doi:10.1016/j.cellimm.2014.09.004
34. Bastos N, Ruivo CF, da Silva S, Melo SA. Exosomes in cancer: use them or target them?. Semin Cell Dev Biol. 2018;78:13–21.
35. Cazzoli R, Buttitta F, Di Nicola M, et al. microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J Thorac Oncol. 2013;8(9):1156–1162. doi:10.1097/JTO.0b013e318299ac32
36. Lan B, Zeng S, Grutzmann R, Pilarsky C. The role of exosomes in pancreatic cancer. Int J Mol Sci. 2019;20(18):4332. doi:10.3390/ijms20184332
37. Zhang W, Ni M, Su Y, et al. MicroRNAs in serum exosomes as potential biomarkers in clear-cell renal cell carcinoma. Eur Urol Focus. 2018;4(3):412–419. doi:10.1016/j.euf.2016.09.007
38. Franzen CA, Blackwell RH, Foreman KE, Kuo PC, Flanigan RC, Gupta GN. Urinary exosomes: the potential for biomarker utility, intercellular signaling and therapeutics in urological malignancy. J Urol. 2018;15(5):1331–1339. doi:10.1016/j.juro.2015.08.115
39. Xu Y-F, Hannafon BN, Zhao YD, Postier RG, Ding W-Q. Plasma exosome miR-196a and miR-1246 are potential indicators of localized pancreatic cancer. Oncotarget. 2017;8(44):77028–77040. doi:10.18632/oncotarget.20332
40. Nakamura S, Sadakari Y, Ohtsuka T, et al. Pancreatic juice exosomal micrornas as biomarkers for detection of pancreatic ductal adenocarcinoma. Ann Surg Oncol. 2019;26(7):2104–2111. doi:10.1245/s10434-019-07269-z
41. Jin C, Rajabi H, Kufe D. miR-1226 targets expression of the mucin 1 oncoprotein and induces cell death. Int J Oncol. 2010;37(1):61–69.
42. Dreau D, Moore LJ, Wu M, et al. Combining the Specific Anti-MUC1 Antibody TAB004 and Lip-MSA-IL-2 limits pancreatic cancer progression in immune competent murine models of pancreatic ductal adenocarcinoma. Front Oncol. 2019;9:330.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.