Back to Journals » Journal of Hepatocellular Carcinoma » Volume 8
SERPINA11 Inhibits Metastasis in Hepatocellular Carcinoma by Suppressing MEK/ERK Signaling Pathway
Authors Song Y, Li Z, Li L, Zhou H, Zeng TT, Jin C, Lin JR, Gao S, Li Y, Guan XY, Zhu YH
Received 15 April 2021
Accepted for publication 11 June 2021
Published 6 July 2021 Volume 2021:8 Pages 759—771
DOI https://doi.org/10.2147/JHC.S315634
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Ahmed Kaseb
Ye Song,1,2,* Zhuo Li,1,* Lei Li,3,* Houming Zhou,4 Ting-Ting Zeng,1 Chuan Jin,2 Jin-Rong Lin,2 Sha Gao,2 Yan Li,1 Xin-Yuan Guan,1,3 Ying-Hui Zhu1
1Sun Yat-sen University Cancer Center, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangzhou, 510060, People’s Republic of China; 2Department of Medical Oncology, Affiliated Cancer Hospital and Institute of Guangzhou Medical University, Guangzhou, 510000, People’s Republic of China; 3Department of Clinical Oncology, The University of Hong Kong, Hong Kong, People’s Republic of China; 4Department of Chinese Medicine, The First Affiliated Hospital of Sun Yat-Sen University, Guangzhou, 510080, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Ying-Hui Zhu
State Key Laboratory of Oncology in Southern China, Sun Yat-sen University Cancer Center, 651 Dongfeng East Road, Guangzhou, 510060, People’s Republic of China
Tel + 86-20-87342283
Email [email protected]
Xin-Yuan Guan
Department of Clinical Oncology, The University of Hong Kong, Room L10-56, 10/F, Laboratory Block, 21 Sassoon Road, Hong Kong, People’s Republic of China
Tel +86 852-39179782
Fax + 852-28169126
Email [email protected]
Purpose: By using integrative RNA sequencing analysis, we identified a novel tumor suppressor, serpin family A member 11 (SERPINA11), which is a serine proteinase inhibitor that belongs to the serpin superfamily. However, the function of SERPINA11 in hepatocellular carcinoma (HCC) remains unclear. The aim of this study was to investigate the role and molecular mechanism of SERPINA11 in HCC.
Methods: Gene expression patterns of SERPINA11 were analyzed in tissue samples of HCC patients by qRT-PCR. In vitro and in vivo experiments were performed to characterize the function and molecular mechanism of SERPINA11 in the tumor metastasis capacity.
Results: SERPINA11 was downregulated in approximately 50% of HCC and significantly associated with metastasis and poor outcome of patients. Functional study demonstrated that SERPINA11 could inhibit cell growth, cell migration and tumor metastasis. Mechanistic investigations suggested that SERPINA11 accelerated urokinase-type plasminogen activator (uPA) degradation to suppress extracellular signal-regulated kinase (ERK1/2) phosphorylation, and thereby subdued metastatic capabilities of HCC cells.
Conclusion: SERPINA11 plays an important tumor suppressive role in HCC, with possible use as a biomarker and an intervention point for new therapeutic strategies.
Keywords: SERPINA11, HCC, metastasis, uPA, ERK
Introduction
Hepatocellular carcinoma (HCC), the fifth most commonly malignancies, ranks as the second leading cause of cancer-related deaths across the globe.1 Patients are commonly diagnosed at the advanced stage and lose the curative surgery opportunity. The majority of patients have an extremely poor prognosis with a median survival of only 6 months, primarily because of high metastatic and recurrent rate.2 Similar to other solid tumors, it is believed that HCC development is a multi-step process which involves the accumulation of multi-genetic and epigenetic alterations.3,4 In order to identify new sensitive and specific molecular prognostic markers and therapeutic targets, we performed RNA sequencing (RNA-Seq) on three pairs of matched HCC and their nontumorous counterparts.5,6 Downregulation of serpin family A member 11 (SERPINA11) was detected in all three HCC tumor tissues compared with adjacent non-tumor tissues.
SERPINA11 is a serine proteinase inhibitor belonging to the serpin superfamily, which share a highly conserved tertiary structure with three beta sheets, eight or nine α-helices, and a reactive center loop (RCL) in the C-terminal segment.7–9 Although some non-inhibitory serpins act as chaperones involved in transport, storage and other roles, 10−12 the majority of the serpins are inhibitors of serine proteases through a suicide substrate-like mechanism.13 The RCL of inhibitory-type serpins contains a unique primary sequence that acts as lure for serine proteases to inhibit its enzymatic activity.13 Serpins, distributed in tissues throughout the body, participate in intracellular and extracellular physiological processes, such as inflammation, blood clotting, immune function, tumor suppression, cancer metastasis, remodeling of the extracellular matrix (ECM).14–17 Recently, downregulation of SERPINA11 was found to be correlated with later stages in HCC.18 However, the molecular function of SERPINA11 in HCC remains obscure.
In the present study, in vitro and in vivo functional assays demonstrated the tumor-suppressive effects of SERPINA11 in HCC. Its molecular mechanism in HCC progression was also investigated.
Materials and Methods
Tissue Specimens and Cell Lines
A total of 155 paired HCC tumor and nontumorous surgical specimens were harvested at Sun Yat-sen University Cancer Center (Guangzhou, China). All patients were diagnosed with primary HCC and had not received preoperative treatments. All clinical samples used in this study were authorized by the Institute Research Ethics Committee at Sun Yat-sen University Cancer Center. The study was conducted in accordance with the Declaration of Helsinki. The HCC cell lines QGY-7703 and PLC8024 were obtained from the Institute of Virology, Chinese Academy of Medical Sciences (Beijing, China). The HCC cell lines HepG2, Hep3B, SMMC7721, QGY-7701, BEL-7402 and Huh7 were purchased from National Collection of Authenticated Cell Cultures (Beijing, China). The cells were cultured in high-glucose DMEM supplemented with 10% fetal bovine serum (FBS).
Quantitative Real-Time Reverse Transcription Polymerase Chain Reaction
Total RNA was extracted from cultured cells or clinical tissues using TRIzol (Invitrogen, Carlsbad, CA, USA), and was reverse transcribed with random primers using a PrimeScriptRT reagent Kit (Promega, Madison, WI, USA) according to the manufacturer’s instructions. Quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) was performed with SYBR Green SuperMix (Roche, Basel, Switzerland) and detected by ABI 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The expression level was normalized against endogenous glyceraldehyde-3-phosphate dehydrogenase (GAPDH) for tested genes. Supplementary Table S1 provides a list of the primer sequence used in this study.
Lentiviral-Mediated SERPINA11 Overexpression and Knockdown
Lentiviral construct containing the full length SERPINA11 (GeneCopoeia, Germantown, MD, USA) was packaged into 293FT cells using the ViraPower Mix (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. QGY7701 and BEL7402 cells were stably transfected with SERPINA11-expressing lentivirus or lentiviral vector plus 10 mg/mL polybrene (Sigma, St Louis, Missouri, USA). To establish SERPINA11 knockdown cells, short hairpin RNAs (shRNA) in lentivirus against SERPINA11 (Genepharma, Shanghai, China) were stably transduced into Huh7 cells. The sequences of the two shRNAs against SERPINA11 are as follows: shRNA1: 5ʹ-CACCGGACACGTTCATGGTTCTTGCCGAAGCAAGAACCATGAACGTGTCC-3ʹ; shRNA2: 5ʹ-CACCGGACAGCATCAAGGAGCTTTACGAATAAAGCTCCTTGATGCTGTCC-3ʹ. The scramble shRNA (Scr)-transfected cells were used as negative controls.
Cell Growth and Foci Formation Assays
For cell growth assay, cells were seeded at a density of 1×103 cells per well in 96-well plate. The cell growth rate was measured by Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). For foci formation assay, 1×103 cells were seeded in each well of a six-well plate. Surviving colonies (>50 cells per colony) were counted with crystal violet staining after 2 weeks of culture. Experiments were performed in triplicate and repeated three times.
Tumor Formation Assay in Nude Mice
Animal care and experiments were performed according to the institutional standard guidelines approved by the Committee of the Use of Live Animals at Sun Yat-sen University Cancer Center. For 7701 lines group, SERPINA11- and vector-transfected cells (4 × 106) were subcutaneously injected into the right and left dorsal flank of mice, respectively. For 7402 lines group, SERPINA11- and vector-transfected cells (3.5 × 106) were subcutaneously injected into the right and left dorsal flank of mice, respectively. Three weeks after the injection, mice were killed with 80 mg/kg pentobarbital, followed by tumor dissection.
Cell Motility and Invasion Assay
For cell motility assay, cells were added to the upper chambers (Corning Costar, Corning, NY, USA) in serum-free medium. Medium containing 10% FBS served as the attractant in the lower chamber. The migrated cells were fixed, stained with crystal violet and counted under microscope. Invasion assay was performed with Matrigel Invasion Chamber (BD Biosciences, Franklin Lakes, NJ) and the procedures were similar to the migration assay. The experiments were repeated thrice.
In vivo Metastasis Assay
Female BALB/c nude mice at 4–6 weeks were randomly divided into several groups (six mice for 7402 lines group and five mice for 7701 lines group) and inoculated with SERPINA11- or vector-transfected cells (1 × 106) via the lateral tail vein. After 8 weeks, mice were killed with 80 mg/kg pentobarbital. Following euthanasia, livers and lungs were harvested, fixed and embedded in paraffin for further H.E. staining. The intrahepatic and pulmonary metastatic nodules were carefully counted.
Antibodies and Western Blot Analysis
Western blot analysis was performed according to the standard protocol with antibodies against SERPINA11 (Sigma, St Louis, Missouri, USA), uPA (Abcam, Cambridge, UK), Phospho-MEK, Phospho-p42/44MAPK, Phospho-p90RSK, Phospho-Elk, p42/44MAPK (Cell Signaling Technology, Danvers, MA), and GAPDH (Santa Cruz Biotechnology, Santa Cruz, CA, USA).
ELISA Assay for SERPINA11
Cells were cultured in serum-free medium for 24 hr, then the medium was collected and centrifuged for 5 minutes at 1000 RPM at 4°C. The supernatant was concentrated by centrifugation using an Amicon® Ultra filter (10K MWCO). Then, 100 μL ultrafiltrate was added to each well to test the concentration of SERPINA11 using the ELISA kits (CUSABIO, Wuhan, China) according to the manufacturer’s instructions. The optical densities were measured at 450 nm. Experiments were performed in triplicate and repeated three times.
Immunohistochemistry (IHC)
The tissue slides were first deparaffinized and rehydrated, and then blocked with 3% H2O2 for 10 min at room temperature. Antigen retrieval was performed by high-pressure-cooking the samples in a 10 mM citrate buffer (pH 6.0) for 4 min. Slides were blocked with 5% normal goat serum for 30 min at room temperature and then incubated in monoclonal antibody against SERPINA11 at 4°C overnight. The Envision detection system (Dako, Agilent Technologies, Inc., Santa Clara, CA, USA) was used for color development followed by Mayer’s hematoxylin counterstaining.
Octet Red System
Recombinant protein of SERPINA11 and uPA were purchased from Novoprotein Scientific Inc. (Shanghai, China) and Sino Biological Inc. (Beijing, China), respectively. The purified human protein of SERPINA11 contained a Fc tag while uPA contained a His tag. The Octet Red System (ForteBio, Menlo Park, CA, USA) was carried out following the manufacturer’s instructions. SERPINA11 was immobilized to the SA sensor and increasing concentrations (0, 13.6, 27.2, 54.4, 108.7 and 217.4 nM) of uPA were run through the protein-coated biosensors. Octet Red System Data Analysis software was used to analyze the association rate, dissociation rate and affinity constant.
Cycloheximide Chase Assay
HCC cells were pretreated with 10 μM cycloheximide (Sigma-Aldrich, St. Louis, MO, USA) for 10 min, followed by an incubation with or without recombinant SERPINA11 (10 ng/mL). The cell extracts from 0, 2, 6, 12 hr treatment groups were assessed by Western blotting to determine the expression of uPA.
Statistical Analysis
SPSS standard V.16.0 (SPSS, Inc., IL, USA) was used for data analysis. Results were presented as means ± SD of three independent experiments. The Student’s t-test was used for statistical comparisons of two independent groups. The disease-specific survival time of HCC patients with different SERPINA11 expression levels was analyzed by Kaplan–Meier analysis with Log rank tests. The correlation between SERPINA11 expression and clinicopathological characteristics of HCC patients was explored by Pearson χ2 test. Spearman correlation was used to assess the relationship between SERPINA11 and uPA expression. Univariable and multivariable Cox proportional hazard model with a forward stepwise procedure was used to estimate the survival hazard. P values of <0.05 were considered statistically significant.
Results
Downregulation of SERPINA11 is Frequently Detected in HCC
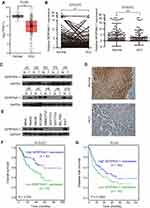
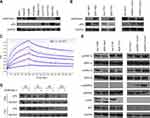
Our previous RNA-seq data showed that SERPINA11 was downregulated in all three tested HCC tissues.19 We first validated this finding in TCGA datasheet. The results demonstrated that SERPINA11 expression was significantly lower in tumor tissues than that in normal liver tissues (Figure 1A). The expression pattern of SERPINA11 was subsequently tested in 155 pairs of primary HCCs samples collected at Sun Yat-sen University Cancer Center (SYSUCC) by qRT-PCR. The results showed that SERPINA11 was significantly downregulated in 75 of 155 (48.4%) of HCC tissues compared with their nontumor counterparts (defined as >2-fold change; P < 0.001, paired Student’s t test; Figure 1B, left panel). The average level of SERPINA11 expression in tumor specimens was dramatically lower than that in nontumor specimens (2.62 versus 5.21, P < 0.001, paired Student’s t-test; Figure 1B, right panel). Consistently, downregulation of SERPINA11 protein was detected in 6 of 10 randomly selected HCC tissues by Western blotting and IHC (Figure 1C and D). The expression levels of SERPINA11 in eight HCC cell lines and one immortalized liver cell line MIHA were also tested by Western blotting. Downregulation of SERPINA11 was detected in 4 of 8 HCC cell lines (SMMC7721, QGY7701, QGY7703, BEL7402) compared with MIHA (Figure 1E).
SERPINA11 Downregulation is Associated with HCC Metastasis and Poor Prognosis
To determine clinical significance of SERPINA11 in HCC, the association of SERPINA11 with clinical-pathological features from 155 HCC patients was statistically analyzed. The results showed that the low expression of SERPINA11 was significantly correlated with tumor size (P = 0.037), the completeness of the tumor encapsulation (P = 0.035) and metastasis and recurrence (P = 0.024, Table 1). To address the prognostic significance of SERPINA11 in HCC, Kaplan-Meier analysis and Log rank test were performed. As shown in Figure 1F, resected HCC patients with low SERPINA11 expression had poorer overall survival (OS) than those with high SERPINA11 expression (P = 0.020, Log rank test). TCGA database also suggested that disease-free survival rate of HCCs with low SERPINA11 expression were significantly lower than those with high SERPINA11 expression (P = 0.0054, Log rank test, Figure 1G). Moreover, multivariate Cox regression analysis showed that SERPINA11 expression was an independent prognostic factor for resected HCC patients (P = 0.043, Table 2). These results imply that SERPINA11 plays an important role in the pathogenesis of HCC. Further, we evaluated the clinical significance of SERPINA11 in HCC patients with or without Hepatitis B virus (HBV) infection. The results showed that the low expression of SERPINA11 was significantly correlated with metastasis and recurrence in HBsAg positive cases (P = 0.013, Supplementary Table S2), while tumor size in HBsAg negative cases (P = 0.032, Supplementary Table S3). In a stratified survival analysis according to the HBV infection, no significant difference in OS was observed in patients with or without HBV infection (Supplementary Figure 1A).
 |
Table 1 Association of SERPINA11expression with Clinicopathological Features in HCCs |
 |
Table 2 Cox Proportional Hazard Regression Analyses for Overall Survival |
SERPINA11 has Tumor-Suppressive Ability
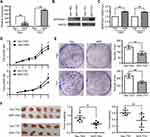
To determine the tumor-suppressive potential of SERPINA11 in HCC, SERPINA11 was cloned into a lentiviral vector and stably transfected into two HCC cell lines, BEL-7402 and QGY-7701. Empty vector-transfected cells were used as controls. The enhanced expression of SERPINA11 in stable transfectants was confirmed by qRT-PCR and Western blotting (Figure 2A and B). Ectopic secretion of SERPINA11 in conditional medium was determined by ELISA assay (Figure 2C). Compared with control cells, the ectopic expression of SERPINA11 effectively suppressed cell growth rate (P < 0.05, Student’s t test, Figure 2D) and foci formation frequency (P < 0.01, Student’s t-test, Figure 2E). To explore the in vivo tumor suppressive capacity of SERPINA11, SERPINA11 stably expressing BEL-7402 (SER11-7402) and QGY-7701 (SER11-7701) cells were subcutaneously injected into nude mice, while Vec-7402 or Vec-7701 cells were used as controls. Consistent with in vitro results, tumor formation in nude mice was markedly dampened in SERPINA11-expressing cells (P < 0.05, Student’s t-test, Figure 2F).
SERPINA11 Suppresses HCC Metastasis
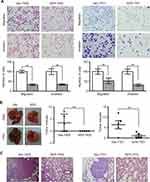
Since downregulation of SERPINA11 has been significantly associated with HCC metastasis, its potential effects on cell migration and invasion were further investigated by both in vitro and in vivo assays. Cell migration and invasion assays revealed that SERPINA11 significantly hampered cell motility and invasiveness, compared with the control cells, respectively (P < 0.01, Figure 3A). To further validate the in vivo suppressive effect of SERPINA11 on tumor metastasis, SERPINA11-expressing or empty vector-transfected cells were injected into the tail vein of nude mice. Eight weeks after injection, tested mice were sacrificed, and the metastatic nodules formed in the lungs and livers were counted. Less pulmonary metastatic nodules were observed in mice injected with SER11-7402 or SER11-7701 cells, compared with the mice injected with control cells (3.4±1.60 vs 0 ± 0, P < 0.01 for 7402; 6.4± 1.97 vs 1.0±0.63, P < 0.01 for 7701, Student’s t-test, Figure 3B). However, no visible metastatic nodule was observed in the livers of mice (data not shown). Histologic study confirmed that the nodules on the surfaces of mice lungs were metastatic tumors (Figure 3C). These results suggest that SERPINA11 effectively impedes HCC metastasis in vitro and in vivo.
Knockdown of SERPINA11 Promotes HCC Growth and Metastasis
To further confirm the tumor-suppressive ability of endogenous SERPINA11, RNA interference (RNAi) was used to knock down SERPINA11 expression in Huh7 cells with high endogenous SERPINA11 expression. qRT-PCR and Western blot analysis demonstrated that the introduction of two specific short hairpin RNAs (shRNA) against SERPINA11 could dramatically reduce SERPINA11 expression (Figure 4A). Knockdown of SERPINA11 enhanced the cell growth rate, foci formation efficiency, and the abilities of migration and invasion (Figure 4B–D).
SERPINA11 Prohibits Cell Metastasis Through Accelerating Urokinase-Type Plasminogen Activator (uPA) Degradation
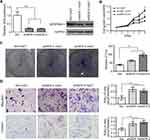
SERPINA11 is a member of the serpin superfamily and its conserved structure suggests that SERPINA11 may have the serine proteinase inhibitor activity.8 uPA, a well-known mediator of tumor cell invasion,17 could be inhibited by several members of serpin family: SERPINE1, SERPINB2, SERPINA5, SERPINE2 and SERPINI1.20–24 We first explored the potential relationship between SERPINA11 and uPA in HCC cell lines. Interestingly, protein expression of uPA was strongly negatively correlated with SERPINA11 (Figure 5A). Moreover, the protein levels of uPA were significantly decreased in SERPINA11-transfected cells compared with vector-transfected cells, while the RNA expression levels were not obviously affected (Figure 5B; Supplementary Figure 1B). Conversely, knockdown of SERPINA11 in Huh7 cells yielded opposing effects without impacting uPA RNA expression (Figure 5B; Supplementary Figure 1C). In addition, we verified the interaction between SERPINA11 and uPA by Octet Red System (Figure 5C). The KD (affinity constant) value for the binding of SERPINA11 to uPA was 1.13×10−8 Mol. The high affinity of uPA for SERPINA11 implies that SERPINA11 may act as a new serine proteinase inhibitor to uPA.
It has been reported that serine proteinase inhibitor can bind uPA to form a stable serpin-protease complex, which not only inhibits the proteolytic activity of uPA, but also promotes the endocytosis and degradation of uPA.16,25,26 In light of a strong negative correlation between SERPINA11 and uPA in HCC cell lines and the decreased protein level of uPA in SERPINA11-expressing cells, we examined the decay rate of uPA in HCCs by cycloheximide (CHX) chase assay. Incubation of 7402 and 7701 cells with recombinant SERPINA11 (rSERPINA11) at 10 ng/mL notably decreased the half-life of uPA (Figure 5D). Our results indicate that SERPINA11 suppresses cell metastasis through accelerating uPA degradation in HCC cells.
SERPINA11 Inhibits Cell Metastasis by Suppressing MEK/ERK Signaling
The uPA system promotes tumor metastasis not only by breaking down the ECM,10 but also by activating divers signal pathways.27 It has been reported that binding of uPA to uPAR can activate MEK/ERK, p38 MAPK and PI3K/Akt signaling pathways.27–30 To further gain insight into detail mechanism by which SERPINA11 inhibits cell metastasis in HCC, we analyzed the effects of SERPINA11 expressing or knockdown on the activation of uPA downstream signaling pathways. The results showed that the phosphorylation levels of MEK1/2, ERK1/2, Elk1 were decreased in SERPINA11-expressing cells but were increased in SERPINA11-knockdown cells (Figure 5E). Increased expression of phosphorylated p90RSK was detected in SERPINA11-knockdown Huh7 cells compared with their control cells (Figure 5E). However, phosphorylated p90RSK could not be detected in neither 7402 nor 7701 cells. These results suggest that SERPINA11 influence cell migration and invasion via suppressing uPA downstream MEK/ERK signaling pathway.
Discussion
SERPINA11 belongs to the serpins superfamily and is predicted to own a serine proteinase inhibitor activity for its conserved protein structure.7,8 SERPINA11 is highly expressed in the liver, implying it may have some important roles in maintaining the normal physiological function of liver.31 Our RNA-Seq results demonstrated that SERPINA11 was downregulated in HCC tumor tissues compared with their corresponding non-tumor tissues. Consistent with the RNA-Seq profiling data, downregulation of SERPINA11 in RNA and protein level was detected in HCC tissues. Moreover, correlation of clinicopathological data showed that downregulation of SERPINA11 was significantly associated with HCC metastasis and poor outcome for resected HCC patients. Further investigation may be needed to determine the clinical significance of SERPINA11 in unresectable cases. All these data suggest that SERPINA11 is an important tumor suppressor gene in HCC development and progression.
Functional studies in vitro and in vivo depicted that SERPINA11 dampened HCC cell growth, motility, and invasiveness. Serine proteases are the second largest family of proteases and have long been associated with tumor invasion, angiogenesis, and metastases.16,17,20 uPA-mediated plasmin generation through extracellular matrix is an important proteolytic event in cancer cell invasion.27,30,32 Moreover, recent evidence indicates that the uPA system is involved in cancer cell invasion through plasmin independent mechanisms.27,29 Serine proteinase inhibitors can bind uPA to form a serpin-protease covalent complex, and subsequently associate with low-density lipoprotein receptor (LDLR) proteins, leading the uPA-uPAR complex to be endocytosed and degraded.25,26,33 It has been found that the degradation of uPA is in parallel with increased expression of SERPINE1 (the serine proteinase inhibitor E1) in endometrial stromal cells.34 In the present study, we elucidated that SERPINA11 could bind uPA and act as a serine proteinase inhibitor to accelerate uPA degradation.
HCC has high rates of intrahepatic metastasis. It is well known that the uPA system is involved in cancer cell invasion through degrading the extracellular matrix.35,36 uPA is overexpressed in HCC, which leads to invasiveness and metastasis.37–39 uPA and uPAR convert plasminogen into the active plasmin to activate metalloproteinases (MMPs), then degrade the ECM.40 Here, we analyzed the activity of MMP-2 and MMP-9 in SERPINA11-expressing cells and found that there was no difference between SERPINA11 expression cells and control cells (data not shown). It reminds us that there might be an alternative mechanism to explain the effect of SERPINA11-mediated uPA deficiency on HCC metastasis. Moreover, recent evidence suggests that the uPA system effects migration, adhesion and metastasis in plasmin-independent ways, which activate various intracellular signaling molecules such as the serine kinase Raf, the tyrosine kinase Src and MEK/ERK.28,29,41 Consistently, our results showed that the phosphorylation levels of MEK1/2, ERK1/2, Elk1 were decreased in SERPINA11-expressing cells, indicating that SERPINA11 metastasis-suppression function was executed through accelerating uPA degradation to impair the downstream MEK/ERK signaling pathway.
HBV is one of the most prominent risk factors for HCC genesis and development. Our study showed that the low expression of SERPINA11 was significantly correlated with recurrence and metastasis in HBsAg positive HCCs, but not in HBsAg negative HCCs. Interestingly, Hepatitis B spliced protein (HBSP), a protein involved in HBV persistence and pathogenesis, was found to interact with cathepsin B (CTSB) to enhance the migration and invasion of hepatoma cells via activation of MMP-9 and uPA.42 Thus, we speculated that SERPINA11 might has more important metastasis-suppression role in HBsAg positive HCCs.
Conclusions
Collectively, our findings reveal that SERPINA11 is a novel tumor suppressor gene, which inhibits tumor metastasis by accelerating the degradation of uPA and thereby hindering MEK/ERK activity in HCCs. Further characterization of SERPINA11 may lead to identify a biomarker and intervention point for new therapeutic strategies to patients with HCCs.
Abbreviations
SERPINA11, serpin family A member 11; HCC, hepatic cell carcinoma; RNA-Seq, RNA sequencing; RCL, reactive center loop; ECM, extracellular matrix; qRT-PCR, quantitative real-time polymerase chain reaction; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; shRNA, short hairpin RNAs; Scr, scramble shRNA; IHC, immunohistochemistry; CHX, Cycloheximide; RNAi, RNA interference; uPA, urokinase-type plasminogen activator; H&E, hematoxylin and eosin.
Data Sharing Statement
The data described in this manuscript are contained in published articles or available from the corresponding author upon reasonable request.
Ethics Statement
This study was approved by the Institute Research Ethics Committee at Sun Yat-sen University Cancer Center with written informed consent obtained from all patients. The animal study was approved by the Committee of the Use of Live Animals at Sun Yat-sen University Cancer Center.
Consent for Publication
Not applicable.
Funding
This work was supported by National Natural Science Foundation of China (81802335, 81872007, 81772554); Natural Science Foundation of Guangdong Province (2018A030310175, 2018A030313034, 2021A1515010589); Young Doctoral Plan of Guangzhou Medical University (2016C42); Young Doctoral Plan of Guangdong Health Commission (B2018059); General Guidance Plan of Guangzhou Health commission (20181A010062); Young Talent Teachers Plan of Sun Yat-sen University (15ykpy33); National Basic Research Program of China (2012CB967001); China National Key Sci-Tech Special Project of Infectious Diseases (2018ZX10723204-006-005); Shenzhen Peacock team Project (KQDT2015033117210153).
Disclosure
The authors declare no potential conflicts of interest.
References
1. El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365:1118–1127. doi:10.1056/NEJMra1001683
2. Parkin DM, Bray F, Ferlay J, et al. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi:10.3322/canjclin.55.2.74
3. Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339–346. doi:10.1038/ng0802-339
4. Santoni-Rugiu E, Jensen MR, Thorgeirsson SS. Disruption of the pRb/E2F pathway and inhibition of apoptosis are major oncogenic events in liver constitutively expressing c-myc and transforming growth factor alpha. Cancer Res. 1998;58:123–134.
5. Morozova O, Marra MA. Applications of next-generation sequencing technologies in functional genomics. Genomics. 2008;92:255–264. doi:10.1016/j.ygeno.2008.07.001
6. Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi:10.1038/nrg2484
7. Heit C, Jackson BC, McAndrews M, et al. Update of the human and mouse SERPIN gene superfamily. Hum Genomics. 2013;7:22. doi:10.1186/1479-7364-7-22
8. Law RH, Zhang Q, McGowan S, et al. An overview of the serpin superfamily. Genome Biol. 2006;7:216. doi:10.1186/gb-2006-7-5-216
9. Silverman GA, Bird PI, Carrell RW, et al. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, Mechanism of Inhibition, Novel Functions, and a Revised Nomenclature. J Biol Chem. 2001;276:33293–33296.
10. Potempa J, Korzus E, Travis J. The serpin superfamily of proteinase inhibitors: structure, function, and regulation. J Biol Chem. 1994;269:15957–15960. doi:10.1016/S0021-9258(17)33954-6
11. Nagata K. Hsp47: a collagen-specific molecular chaperone. Trends Biochem Sci. 1996;21:22–26. doi:10.1016/S0968-0004(06)80023-X
12. Zou Z, Anisowicz A, Hendrix MJ, et al. Maspin, a serpin with tumor-suppressing activity in human mammary epithelial cells. Science. 1994;263:526–529. doi:10.1126/science.8290962
13. Huntington JA, Read RJ, Carrell RW. Structure of a serpin-protease complex shows inhibition by deformation. Nature. 2000;407:923–926. doi:10.1038/35038119
14. Carrell RW, Aulak KS, Owen MC. The molecular pathology of the serpins. Mol Biol Med. 1989;6:35–42.
15. Andreasen PA, Georg B, Lund LR, et al. Plasminogen activator inhibitors: hormonally regulated serpins. Mol Cell Endocrinol. 1990;68:1–19. doi:10.1016/0303-7207(90)90164-4
16. Andreasen PA, Kjoller L, Christensen L, et al. The urokinase-type plasminogen activator system in cancer metastasis: a review. Int J Cancer. 1997;72:1–22.
17. Andreasen PA, Egelund R, Petersen HH. The plasminogen activation system in tumor growth, invasion, and metastasis. Cell Mol Life Sci. 2000;57:25–40. doi:10.1007/s000180050497
18. Lin KT, Shann YJ, Chau GY, et al. Identification of latent biomarkers in hepatocellular carcinoma by ultra-deep whole-transcriptome sequencing. Oncogene. 2014;33:4786–4794. doi:10.1038/onc.2013.424
19. Chen L, Li Y, Lin CH, et al. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat Med. 2013;19:209–216. doi:10.1038/nm.3043
20. Fortenberry Y. The role of serpins in tumor cell migration. Biol Chem. 2015;396:205–213. doi:10.1515/hsz-2014-0254
21. Van De Craen B, Declerck PJ, Gils A. The biochemistry, physiology and pathological roles of PAI-1 and the requirements for PAI-1 inhibition in vivo. Thromb Res. 2012;130:576–585. doi:10.1016/j.thromres.2012.06.023
22. Al-Ejeh F, Croucher D, Ranson M. Kinetic analysis of plasminogen activator inhibitor type-2: urokinase complex formation and subsequent internalisation by carcinoma cell lines. Exp Cell Res. 2004;297:259–271. doi:10.1016/j.yexcr.2004.03.030
23. Suzuki K. The multi-functional serpin, protein C inhibitor: beyond thrombosis and hemostasis. J Thromb Haemost. 2008;6:2017–2026. doi:10.1111/j.1538-7836.2008.03181.x
24. Yepes M, Sandkvist M, Wong MK, et al. Neuroserpin reduces cerebral infarct volume and protects neurons from ischemia-induced apoptosis. Blood. 2000;96:569–576. doi:10.1182/blood.V96.2.569
25. Cubellis MV, Wun TC, Blasi F. Receptor-mediated internalization and degradation of urokinase is caused by its specific inhibitor PAI-1. EMBO J. 1990;9:1079–1085. doi:10.1002/j.1460-2075.1990.tb08213.x
26. Dano K, Andreasen PA, Grondahl-Hansen J, et al. Plasminogen activators, tissue degradation, and cancer. Adv Cancer Res. 1985;44:139–266.
27. Tang L, Han X. The urokinase plasminogen activator system in breast cancer invasion and metastasis. Biomed Pharmacother. 2013;67:179–182. doi:10.1016/j.biopha.2012.10.003
28. Luo J, Sun X, Gao F, et al. Effects of ulinastatin and docetaxel on breast cancer invasion and expression of uPA, uPAR and ERK. J Exp Clin Cancer Res. 2011;30:71. doi:10.1186/1756-9966-30-71
29. Chandrasekar N, Mohanam S, Gujrati M, et al. Downregulation of uPA inhibits migration and PI3k/Akt signaling in glioblastoma cells. Oncogene. 2003;22(3):392–400. doi:10.1038/sj.onc.1206164
30. Nguyen DH, Webb DJ, Catling AD, et al. Urokinase-type plasminogen activator stimulates the Ras/Extracellular signal-regulated kinase (ERK) signaling pathway and MCF-7 cell migration by a mechanism that requires focal adhesion kinase, Src, and Shc. Rapid dissociation of GRB2/Sps-Shc complex is associated with the transient phosphorylation of ERK in urokinase-treated cells. J Biol Chem. 2000;275:19382–19388. doi:10.1074/jbc.M909575199
31. UniProt Consortium T. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2018;46:2699. doi:10.1093/nar/gky092
32. Holst-Hansen C, Johannessen B, Hoyer-Hansen G, et al. Urokinase-type plasminogen activation in three human breast cancer cell lines correlates with their in vitro invasiveness. Clin Exp Metastasis. 1996;14:297–307. doi:10.1007/BF00053903
33. Croucher DR, Saunders DN, Lobov S, et al. Revisiting the biological roles of PAI2 (SERPINB2) in cancer. Nat Rev Cancer. 2008;8:535–545. doi:10.1038/nrc2400
34. Casslen B, Nordengren J, Gustavsson B, et al. Progesterone stimulates degradation of urokinase plasminogen activator (u-PA) in endometrial stromal cells by increasing its inhibitor and surface expression of the u-PA receptor. J Clin Endocrinol Metab. 1995;80(9):2776–2784. doi:10.1210/jcem.80.9.7673423
35. Stillfried GE, Saunders DN, Ranson M. Plasminogen binding and activation at the breast cancer cell surface: the integral role of urokinase activity. Breast Cancer Res. 2007;9:R14. doi:10.1186/bcr1647
36. De Cremoux P, Grandin L, Dieras V, et al. Urokinase-type plasminogen activator and plasminogen-activator-inhibitor type 1 predict metastases in good prognosis breast cancer patients. Anticancer Res. 2009;29(5):1475–1482.
37. De Petro G, Tavian D, Copeta A, et al. Expression of urokinase-type plasminogen activator (u-PA), u-PA receptor, and tissue-type PA messenger RNAs in human hepatocellular carcinoma. Cancer Res. 1998;58:2234–2239.
38. Zhou L, Hayashi Y, Itoh T, et al. Expression of urokinase-type plasminogen activator, urokinase-type plasminogen activator receptor, and plasminogen activator inhibitor-1 and −2 in hepatocellular carcinoma. Pathol Int. 2000;50:392–397. doi:10.1046/j.1440-1827.2000.01059.x
39. Chan CF, Yau TO, Jin DY, et al. Evaluation of nuclear factor-kappaB, urokinase-type plasminogen activator, and HBx and their clinicopathological significance in hepatocellular carcinoma. Clin Cancer Res. 2004;10:4140–4149. doi:10.1158/1078-0432.CCR-03-0574
40. Matrisian LM. The matrix-degrading metalloproteinases. Bioessays. 1992;14:455–463. doi:10.1002/bies.950140705
41. Blasi F, Sidenius N. The urokinase receptor: focused cell surface proteolysis, cell adhesion and signaling. FEBS Lett. 2010;584:1923–1930. doi:10.1016/j.febslet.2009.12.039
42. Chen WN, Chen JY, Jiao BY, et al. Interaction of the hepatitis B spliced protein with cathepsin B promotes hepatoma cell migration and invasion. J Virol. 2012;86:13533–13541. doi:10.1128/JVI.02095-12
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.