Back to Journals » Veterinary Medicine: Research and Reports » Volume 12
Seroprevalence and Molecular Detection of Foot and Mouth Disease Virus in Dairy Cattle Around Addis Ababa, Central Ethiopia
Authors Awel SM, Dilba GM, Abraha B , Zewde D , Wakjira BS, Aliy A
Received 30 April 2021
Accepted for publication 24 June 2021
Published 12 July 2021 Volume 2021:12 Pages 187—197
DOI https://doi.org/10.2147/VMRR.S317103
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Young Lyoo
Shazali Mohammed Awel,1,2 Getachew Mulatu Dilba,3 Bruk Abraha,2 Demeke Zewde,4 Bayeta Senbata Wakjira,4 Abde Aliy4
1Jigjiga University, College of Veterinary Medicine, Jigjiga, Ethiopia; 2Haramaya University, College of Veterinary Medicine, Dire Dawa, Ethiopia; 3Ambo University, College of Agriculture and Veterinary Science, Department of Veterinary Laboratory Technology, Ambo, Oromia, Ethiopia; 4National Animal Health Diagnostic and Investigation Center, Sebeta, Ethiopia
Correspondence: Getachew Mulatu Dilba
Ambo University, College of Agriculture and Veterinary Science, Department of Veterinary Laboratory Technology, P.O. Box:19, Ambo, Oromia, Ethiopia
Tel +251-912717217
Email [email protected]
Background: Foot and mouth disease is a debilitating and highly contagious transboundary disease of cattle that can cause a huge economical loss globally. It is a notifiable disease in Ethiopia, and it is thought to be causing a decrease in cattle productivity and production.
Methods: A cross-sectional study and outbreak investigation were performed to estimate seroprevalence, identify associated factors and serotypes of FMDV in dairy cattle around Addis Ababa. A multi-stage random sampling technique was employed for the selection of sampling units for the seroprevalence study. A total of 383 blood samples were collected using plain vacutainer tubes and the obtained sera were tested by 3ABC-Ab ELISA at the NAHDIC lab. Also, from outbreak cases, 20 epithelial tissue samples were collected purposively for the molecular detection of FMDV serotypes.
Results: The overall seroprevalence of FMD in dairy cattle was 72.1% (95% CI=67.27– 76.50). The seroprevalence in dairy cattle of Ada Berga, Holeta, and Sululta districts was 97.2%, 71.4%, and 57.6%, respectively. Up on Chi-square analysis, age, body condition, and management system were significantly associated with FMD seroprevalence (p< 0.05). Besides, multivariable logistic regression analysis showed that district, age, body condition, and management were significantly associated with FMD seroprevalence (p< 0.05). The odds of being seropositive to FMD were 6.9 (95% CI=1.8– 24.9; p=0.005) and 2.3 (95% CI=1.2– 4.7; p=0.01) times higher in cattle found in Ada Berga and Holeta Woreda. From outbreak cases, 18 (90.0%) were identified positive for FMDV serotype O.
Conclusion: The current study revealed higher seroprevalence was recorded in the study area and associated risk factors identified statically, serotype O of FMDV was identified from outbreak cases. Therefore, it is critical to design and implement feasible control and prevention mechanisms based on the type of circulating virus serotype.
Keywords: central Ethiopia, dairy cattle, FMD virus, molecular detection, seroprevalence
Introduction
The Foot and mouth disease virus (FMDV), the causative agent of foot and mouth disease (FMD), belongs to the genus Aphthovirus and the family Picornaviridae. It has seven serotypes, namely: O, A, C, South African Territories (SAT) SAT1, SAT 2, SAT 3, and Asia 1. By one time or another, one type or the other was widely reported in most parts of the world.1 The highly infectious nature of the virus, the generation of high titers in respiratory secretions, the prolonged survival of the virus in secretions, the fast replication cycle, and the brief incubation time contribute to the virus’s rapid dissemination to fully susceptible populations. In addition to the animal-to-animal transmissions, FMDV is easily transmitted mechanically via fomites such as clothes, shoes, vehicles, and veterinary instruments. Moreover, its spread is aggravated by uncontrolled movements of infected animals across geographic boundaries.3
FMD is the most infectious disease that affects cloven-hoofed livestock, and it can cause significant economic losses in susceptible animals, such as cattle, sheep, horses, pigs, dogs, goats, and water buffalo.2 The disease is a global threat, which through the years has affected most of the countries. It occurs throughout the world, most commonly in Asia, Africa, the Middle East, and parts of South America.3
FMD is considered endemic in most African countries, including Ethiopia. The disease is frequently listed as the most economically important disease of livestock in many developed and developing countries.4 FMD causes annual global losses of 6.5 −21 billion USD in endemic regions, resulting from visible production losses, vaccination costs, and treatment costs, while an additional annual loss of >1.5 billion USD was reported from areas with FMD outbreaks (ie, non-endemic areas).5
In Ethiopia, the disease has been affecting mainly cattle, while also causing problems in small ruminants at infrequent intervals. Historically, the disease was first identified in 1957, but it had certainly been present in the country before that, since most livestock keepers were familiar with it and some were using traditional methods of immunization against it, such as “mouthing.”6 According to the report of Martel7 FMD serotypes O, A, and C were responsible for FMD outbreaks from 1957 to 1979. A separate report on the genetic characterization of FMDV from 1981 to 2007 disclosed additional serotypes such as SAT 1 and SAT 2.8 Furthermore, a study from 2011 to 2012 showed that serotype O was identified from bovine and swine FMD outbreaks in different regions of Ethiopia. Meanwhile, the study showed that all serotype O isolates belong to a single topotype, ie, East Africa-3 (EA-3).9 A study conducted by Gizaw et al10 has shown the molecular characteristics of FMDV circulating in Ethiopia between 2008 −2019.
Due to its high contagiousness, wide geographical distribution, broad host range, ability to establish carrier status, poor cross-immunity, and relatively short duration of immunity, FMD is difficult to control and ranks the disease as the most feared viral disease.11,12 In FMD endemic areas, control of the disease is complicated due to poor disease surveillance, lack of FMDV molecular characterization, and lack of proper study on the origin of infection. This is particularly true in central Ethiopia, where there is repeated FMD outbreaks among milk-producing dairy animals, constituting one of the largest milk shedding areas in the country.11 Thus, in FMD endemic countries like Ethiopia continuous surveillance and an understanding of the serotype and genotype of the virus are critical for the implementation of control schemes. Therefore, the objectives of the current study were to estimate the seroprevalence of FMD, to assess factors associated with FMD seropositivity, and to identify FMD virus serotypes circulating in outbreak cases from dairy cattle around Addis Ababa, central Ethiopia.
Materials and Methods
General Description of the Study Area
The current study was conducted in Ada Berga, Holeta, and Sululta districts around Addis Ababa, central Ethiopia from January 2019–2021 to estimate FMD seroprevalence and to identify the FMDV serotype from FMD outbreaks. In most of the study areas, animals were reared under an extensive management system, while semi-intensive and intensive management systems were also practiced in farms located in Ada Berga (Ada Berga dairy farm) and Wolmera (Holeta Agricultural Research Center (HARC) dairy farm). The specific districts, kebeles, and farms were selected purposively based on the presence of suspected FMD outbreak cases. The average annual rainfall and average maximum and minimum temperature for the area were 1180 mm and 22.8°C and 10.6°C, respectively. Ada Berga is located between 9°14′60″N latitude and 38°24′59.99″E longitude, while Wolmera is located between 9°03′24″N latitude and 38°30′72″E longitude. Central Ethiopia has a humid subtropical climate with moderate seasonality, which is characterized by mild dry winters and mild rainy summers.13
Study Population
Dairy cattle of greater than six months old and all breeds under different husbandry and management system were the study population for seroprevalence. Besides, during an outbreak of the diseases, cattle with symptoms of FMD in the vicinity were included as a study population for molecular detection of FMDV. An animal was suspected for FMD case if it exhibits profuse salivation and/or lameness. Moreover, the oral cavity and hooves of animals have been examined for the presence of intact/rupture vesicles, erosions, and ulcers.
Study Design
A cross-sectional study was performed to estimate the seroprevalence of FMD; assess putative risk factors; and detect FMDV and its serotype in dairy farms of Ada Berga, Holeta, and Sululta districts.
Sample Size Determination
For estimation of FMD seroprevalence, the sample size was estimated/calculated according to the formula given by Thrusfield14 for a theoretically infinite population. Thus, based on 50% expected prevalence (P), 95% confidence interval (Z), and 5% desired precision (d) 384 dairy cattle sera were sampled.
Thus, with missed information from one sample, the total number of samples processed for serological examination was 383. In addition, epithelial tissues were collected from 20 FMD suspected cattle for molecular detection.
For seroprevalence study, the farms were considered as clusters and a simple random technique was used to select sampling clusters, and animals with clusters were selected by simple random sampling strategy after assigning an identification number to each animal. When the active outbreak of FMD was encountered or reported, the field-level investigation was conducted in areas where FMD outbreaks have happened. Cattle with evident clinical signs and symptoms of FMD, including a history of oral lesion, history of infection but having healing lesion, and any other asymptomatic cattle (with no evident clinical signs for FMD) in the same farm or grazing with the symptomatic cattle (with evident clinical signs of FMD) were sampled as per the extent of the outbreak.
Sample and Data Collections
About 10mL blood sample was aseptically collected from the jugular vein of apparently healthy animals using plain vacutainer tubes. From clinical cases, at least one gram of tongue epithelial tissues, feet epithelial tissue, and vesicular fluids was collected from non-ruptured or freshly ruptured vesicles then placed in the bottle with transport medium (ie, an equal amount of buffer saline solution). The samples were labeled with identification numbers and types of tissue. Both samples were transported using an icebox to the NAHDIC (Sebeta) for serological and molecular laboratory diagnosis. Besides information on the potential risk factors, such as age, breed, sex, and body condition score were recorded. Also, the cattle owner or manager was interviewed with semi-structured questionnaires regarding the season of outbreaks, the occurrence of the disease, and history of infected cattle which can help to trace cases of outbreaks.
Laboratory Analysis
Serological Test
A serum sample was examined for antibody to 3ABC non-structural proteins of FMD viruses using commercially available ELISA test for identifying the infected animal from non-infected one according to Bergmann et al15 3ABC-Ab ELISA test provides a rapid, simple, sensitive, and specific method for detecting antibodies against non-structural proteins (NSP) of the FMD virus in serum bovine origin. The test allows differentiation between samples from infected (presence of antibodies against NSP of FMD virus) and vaccinated (no antibodies against NSP of FMD virus) animals. The test was performed as per the manufacturer’s instructions (IDEXX Laboratories Inc). (e optical density (OD) reading was recorded using a spectrophotometer at a wavelength of 450nm.16
The validation of the test result was validated if
- The mean value of the negative control (NC) is less than or equal to 0.3 (NC
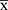 ≤ 0.3)
≤ 0.3) - The mean value of the positive control (PC) is less than or equal to 2 (PC
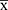 ≤ 2)
≤ 2) - PC
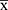 - NC
- NC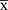 ≥ 0.3
≥ 0.3
The interpretation for each serum sample, the competition percentage was calculated (S/P%) as follows:
(S/P) % less than 35 was recorded as negative, while S/P% greater than or equal to 35 was considered as positive.
Molecular Characterization
Extraction of Viral RNA
Total RNA was extracted from supernatants of the homogenized epithelial tissue suspensions by QIAgen RNA extraction kit (Qiagen, Crawley, West Sussex, UK) following the manufacturer’s recommendation. First, 140 μL epithelial tissue suspensions samples were separately added with 560 μL of prepared buffer AVL containing carrier RNA into a 1.5 mL microcentrifuge Eppendorf tube, mixed by pulse vertexing for 15 seconds then incubated at 25°C for 10 minutes (min) for lysing purpose. The tubes were then briefly centrifuged to remove drops from the inside of the lid. Binding was done by adding 560μL of ethanol (96%), mixed by pulse vortexing for 15sec then briefly centrifuged the tube to remove drops from inside the lid. A 630 μL of the solution was transferred into the QIAamp Mini spin (silica) column, then centrifuged at 8000rpm for 1min through placing the QIAamp Mini column and collected into a clean 2 mL collection tube. Any left solution was processed again similarly to get more quantity of RNA material. Washing was first conducted by 500μL of buffer AW1 and centrifuged at 8000rpm for 1min. The filtrate was then discarded and the column was placed in a fresh 2mL collection tube. Then, 500μL of buffer AW2 was added to the column then centrifuged at 14,000 rpm for 3 min. Then, 60μL of buffer AVE was added to the column and incubated at room temperature for 1 min before centrifuged at 8000rpm 1min. Finally, the RNA of the FMDV was obtained. Extracted RNA was kept at +4°C for immediate use or stored at −80 °C until used for real-time reverse transcription-polymerase chain reaction (rtRT-PCR).17
Detection of FMDV by Real-Time RT-PCR
The one-step rtRT-PCR assay was applied for primary detection of the FMDV. Real-time RT-PCR has a sensitivity comparable to that of virus isolation and automated procedures enhanced sample throughput.18 Reverse transcription of FMDV RNA and PCR amplification of reverse-transcribed RNA was conducted using automated one-step real-time RT-PCR as reported before by Callahan et al19 that detects the 3D RNA polymerase encoding gene. The 3D non-structural protein in viral RNA-dependent RNA polymerase, which is responsible for RNA replication and is highly conserved (94–99% similarity).20 The nucleotide sequence of forwarding primer (FMDV 3DF) 5ʹ - ACT GGG TTT TAC AAA CCT GTG A-3′, the reverse primer (FMDV 3DR) 5ʹ - GCG AGT CCT GCC ACG GA-3′ and the probe (FMDV Probe 3DP) 5ʹ- [6FAM] TCC TTT GCA CGC CGT GGG AC [TAM]-3′ were used.19 The probe was labeled with 5ʹ- reporter dye, 6-carboxyfluorescein, and 3ʹ- quencher, tetramethylrhodamine in real-time RT-PCR reaction to detect the 3Dpol gene sequence in all the FMDV serotypes. Real-time RT-PCR was carried out by using a superscript III/platinum Taq one-step rRT-PCR kit. The master mix reaction components for one-step real-time RT-PCR were prepared using, 12.5μL of 2x - reaction mix, 1.5μL of RNAse-free water, 2μL of forward primer, 2μL of reverse primer, 1.5μL of TaqMan probe, and 0.5μL of superscript ®III reverse transcription (RT) to make a total of 20μL per sample for each reaction of PCR per well per plate including positive and negative control master mix. It is then thoroughly mixed by pulse vertexing.
A 5μL extracted RNA template was added to each PCR plate and the total volume of the PCR plate was 25μL. The PCR plate was sealed by adhesive film then inserted into the thermal cycler machine slots and adjusted according to QIAgen one-step RT-PCR kit protocols. The one-step rRT-PCR amplification was started with RT cDNA synthesis at 50°C for 30 min; followed by initial denaturation at 95°C for 10 min; denaturation at 95°C for 15 sec, annealing at 60°C for 1min and extension at 72°C for 30 seconds. 50 cycles were taken to finish amplifications.21 Negative control (nuclease-free water) and positive control (field isolate) were included in each run.
Interpretation of Real-Time PCR
The PCR amplification was carried out in the thermal cycler Rotor-Gene Q (Qiagen®, Germany). The successfully amplified target gave an amplification curve and the cycle threshold (Ct) at which the target amplicon was initially detected above the background fluorescent levels as determined by SDS software. Then, FMDV was detected through threshold cycle (Ct) values based on baseline and graphs. The Ct values <32.0 were considered as positive and amplification with Ct values of undetermined were considered as negative. Samples that had Ct values ≥32 and <50 were “inconclusive” and the test was repeated.21 Positive reactions were defined as those which gave a detectable Ct.22 Strong positive FMD samples have a Ct value below 20.0.23
Serotyping of FMDV by Taq One-Step qRT-PCR
All positive 3D rRT-PCR results (n=18) were brought to serotyping of FMDV through platinum Taq one-step qRT-PCR kit protocol (Invitrogen) assay. Master mix reagents such as 12.5μL of 2x - reaction mix, 2μL of RNAse-free water, 2μL of forwarding primer (FP), 2μL of reverse primer (RP), 1μL of TaqMan probe (P), 5μL RNA, and 0.5μL of superscript ®III reverse transcription (RT) to make a total of 25μL per sample for each reaction of PCR per well per plate including positive and negative control master mix. Also, the expected FMDV serotype in Ethiopia oligonucleotides was custom synthesized according to the following sequences (Pirbright, UK).18,19,24–30
Oligo name Nucleotide sequence (5ʹ 3ʹ)
FMDV/A/FP: GCCACRACCATCCACGA
FMDV/A/RP: GAAGGGCCCAGGGTTGGACTC
FMDV/A/P: FAM-CTCGTGCGMATGAARCGGC-BHQ1
FMDV/O/FP: CCTCCTTCAAYTACGGTG
FMDV/O/RP: GCCACAATCTTYTGTTTGTG
FMDV/O/P: FAM-CCCTCTTCATGCGGTARAGCAG-BHQ1
FMDV/SAT1/FP: CTYGACCGGTTCACYCTG
FMDV/SAT1/RP: CCGAGAAGTAGTACGTRGC
FMDV/SAT1/P: FAM-CAGGAYTGCGCCCACCA-BHQ1
FMDV/SAT2/FP: CRATCCGCGGTGAYCG
FMDV/SAT2/RP: CGCTTCATYCTGTARACGTC
FMDV/SAT2/P: FAM-TTTGGACAYGTGACCGCCG-BHQ1
Total mixed reagent (25μL) again mixed with oligonucleotides for each sample (n=18) and amplified by using real-time PCR.
Interpretations:
In positive results, the fluorescent signal accumulated during amplification crosses the threshold value. A Ct value was calculated at end of the amplification.
In negative results assay that did not reach the threshold were reported as no Ct.
Data Management and Analysis
The data collected from the questionnaire survey and laboratory investigations were entered into a Microsoft Excel spreadsheet (Microsoft Corporation) and analyzed using STATA version 11.0 for Windows (Stata Corp. College Station, TX, USA). The sum of seropositive samples divided by the overall number of samples tested was used to measure the seroprevalence. The association of seropositivity with the potential risk factors was computed by Pearson’s Chi-square test with a 95% confidence interval and a significance level at P≤0.05. The degree of association between FMD seroprevalence and categorical independent variables was assessed using odds ratio (OR) with multivariable logistic regression analyses. Before regression analysis, the data was checked for fulfillments of assumptions, such as the correlation of each variable (not more than 0.7), correlation of independent variables with the dependent variable (minimum of 0.3), and multi-collinearity tests (VIF (>10) and tolerance (>0.1)). And all tested variables did not show multi-collinearity. However, sex, breed, and herd size had a low correlation with the outcome variable (<0.3), thus decided to be omitted from the model. Statistical outputs were considered significant at p≤ 0.05.
Results
Overall Seroprevalence of FMD
From the total 383 sera tested by 3ABC-Ab ELISA, 72.1% (276/383) were found positive for FMD. A higher prevalence was observed in the Ada Berga district compared to other districts (Table 1) and the difference was statistically significant (p<0.05).
 |
Table 1 Overall FMD Seroprevalence and Associated Putative Risk Factors |
Animal-Related Risk Factors Associated with FMD Seropositivity
Comparison of FMD seroprevalence was done in two age groups, young (<3years) and adult (>3years) animals. An increased seroprevalence was observed with an increase in the age of animals (Table 1) and the age-wise difference was statistically significant (χ2 = 34.5; p = 0.000). The seroprevalence estimate for male and female cattle was 72.5% and 72.0%, respectively (Table 1). However, the difference was not statistically significant (p > 0.05). Breed-specific prevalence of 83.3% for local and 71.7% for the cross was observed (Table 1). The difference between breeds was not statistically significant (p>0.05). Moreover, the prevalence of 47.4%, 71.2%, and 77.0% was recorded in poor, moderate, and good body conditioned animals, respectively. Statistically significant variation (χ2 = 14.1; p = 0.001) in FMD seroprevalence was recorded among different body condition groups of animals.
Environment Related Risk Factors for FMD Seroprevalence
Animals managed semi-intensively have greater FMD prevalence than intensively managed animals (Table 1), and the variation was statistically significant (χ2 = 89.02; p = 0.000). Seropositivity was assessed in herd size and a higher seroprevalence of FMD was observed in small herd size (Table 1). But, the difference was not statistically significant for herds of cattle (p > 0.05).
Logistic Regression Analysis of FMD Seroprevalence and Associated Risk Factors
The multivariable logistic regression analysis showed that the odds of animals with FMD was 6.9 times (OR=6.9; 95% CI=1.8–24.9; p-value=0.005) higher in dairy animals who are found in Ada Berga district as compared to Sululta (reference category) (Table 2). Regarding odds of the management system, FMD was 5.3 times (OR=5.3; 95% Cl=2.9–9.9; p-value=0.000) higher in dairy cattle managed under an intensive management system compare to the semi-intensive system (Table 2). All tested variables did not show multi-collinearity.
 |
Table 2 Results of Multivariable Logistic Regression Analysis and Associated Risk Factors |
Molecular Detection of FMD Virus from Outbreak Cases
The successfully amplified target gave an amplification curve and the cycle threshold (Ct), the target amplicon was initially detected above the background fluorescent levels determined by the embedded software. The Ct value (cycle threshold or crossing point) corresponds to the number of cycles required for a given sample to reach the threshold above is considered as positive. For detection of FMDV, real-time RT-PCR (using universal primers and probe of FMDV) was performed on all 20 collected samples from the Ada Berga district. Out of 20 samples, 18 (90%) samples were found positive having a Ct value ranging from 19.01 to 32.01 and the fluorescence of samples rises above the background fluorescence. A total of 18 (90%) samples that were collected from Ada Berge districts were found positive with serotype O (Table 3).
 |
Table 3 Molecular Detection of FMD Virus and Serotyping from Outbreak Cases |
Discussion
In the current study, based on serological tests the prevalence of FMD was found to be 72.1%. The estimated prevalence obtained is higher than the previous reports from different parts of the country. Thus, prevalence of 12.5–30% in Addis Ababa;31,32 24–41.5% in districts of Tigray region;31,33 4.9–13.5% in areas of southern nation and nationalities;34,35 38.7–52.5% in Amhara region;36 12.5–55.6% in Oromia region,3,37–39 and 14.2–15.1% in Somali regional state by Mohamoud et al40 were reported from Ethiopia. According to previous reports, the eastern Tigray has 41.5% FMD seroprevalence, followed by the selected districts of West Shewa,41 Guji region of Oromia, and the Yeka district of Addis Ababa city, which have 40.4%, 32.7%, and 30% FMD seroprevalence, respectively.31 On the other hand, the current finding is in line with 79% overall seroprevalence of FMD in cattle reported from Sudan.42 In addition, Namatovu et al43 from Uganda reported a seroprevalence of 77%. Because FMD has a short duration of immunity and in the absence of recent outbreaks among the study farms, the higher seroprevalence report in the current study indicates that cattle are serving as FMDV carriers for a prolonged duration. This carrier state has been previously reported by Alexandersen et al,44 who stated that cattle can act as FMDV carriers for more than 3 years.
In the present study, statistically significant variation (X2 = 34.5; P<0.001) in FMD seropositivity was observed among age categories with higher prevalence in older (80.3%) than young animals (<3 years old) (50%). This is in agreement with the report of Thrusfield,14 which states that young cattle with less prior exposure did not show a measurable response against FMD. In contrast to previous reports by Rufael et al39 in Borena pastoral area; Megersa et al34 in Gamo Gofa and Sidama Zones; and Ahmed et al41 in West Shewa zone Oromia of Regional State; revealed significantly higher FMD seroprevalence in young as compared with adult and old cattle. On the other hand, Gelaye et al45 in the Bench Maji Zone of southern Ethiopia documented no significant association between seropositivity of FMD between groups of different ages of cattle. The variation in reports might be due to the unproportionally allocate sample size among age categories and variation in handling young animals, ie, separate housing of young animals or keeping young animals apart from adult animals around the homestead and camps.11,46,47
The FMD seroprevalence was almost the same in both sex categories, with 72.0% and 72.5% in females and males, respectively. This finding is consistent with previous reports from different parts of Ethiopia, where sex appeared not to have a significant effect on FMD seropositivity.34,45 On the contrary, Mazengia et al48 in the study on FMDV occurrence among dairy cattle in the Northwest part of Ethiopia, found that female cattle had a higher rate of infection than male cattle. The observed variation in FMD seroprevalence between the sexes of cattle may be related to the unproportionally allocated sample size, in which the sample size of female was higher than male animals.
Regarding the breed of cattle, the prevalence was slightly higher in local (83.3%) than crossbreds (71.7%). This is in agreement with the report of Misgana et al,49 which showed a non-significance difference in seroprevalence of FMD between local and crossbreeds. However, the higher prevalence in local breeds might be attributed to uncontrolled movement and poor management, unlike the relatively controlled movement in the crossbreed animals.
The current study revealed that the body condition status of cattle has a significant association with seroprevalence of FMD, in that it was lower in poor body condition (47.4%) followed by moderate and good scores with the proportion of 71.2% and 77.0%, respectively. It was also observed that animals with good body conditions had 4.0 times the odds of being positive to FMD than poor body conditions (Table 2). The higher prevalence in animals with good conditions is in line with the report of Radostits et al,50 who stated that animals in good conditions are relatively more susceptible. This might be due to the favorable environment for easy replication of the FMDV, virus-receptor interactions as well as unique aspects of virus translation and shutoff of host macromolecular synthesis in epithelial tissue.51
Management system had shown significant association with FMD seroprevalence, in that it was higher in animals managed under intensive (89.6%) than semi-intensive (45.4%) farming systems. Moreover, the odds of animals with FMD were 5.3 times higher in intensively managed dairy animals than semi-intensively managed. In line with this result, Vosloo et al52 reported that intensive livestock production is highly vulnerable to the effect of FMD. This could be attributed to the crowding of animals that can facilitate the frequency of direct contact, thus enhances the chances of FMD transmission. Further, this could be attributed to the fact that the FMD virus can stay for several months in shaded premises, which are not exposed to sunlight.50
In the present study, out of 20 bovine epithelial tissue samples tested by real-time PCR, 18 were found positive for 3D regions of FMDV (Table 3). Previously, Paixão et al53 suggested that real-time RT-PCR that targets the 3D region of the viral genome is a powerful technique for reliable detection of FMDV which currently is becoming a key diagnostic test used to confirm FMDV presence in field samples. The negative findings for those two samples may be due to the small amount of viral RNA in the initial samples, which may have caused sample dissemination during lab work. In support of this observation, OIE42 reported that the preferred sample for virus detection is the epithelial tissue that was previously confirmed by Urge et al54 and Getachew55 who reported the presence of higher levels of viral RNA in the epithelial tissues. In agreement with this, Reid et al18 indicated epithelial tissue samples from the vesicular lesions could be used as the sample of choice for FMDV detection.
Moreover, the current study revealed that the FMDV detected in all of (18) the epithelial tissue samples belong to serotype O, which is an indication of predominant serotype circulating in sites of outbreaks from Ada Berga district of central Ethiopia. In agreement with this finding, previous studies showed that serotype O was the most prevalent and dominant serotype causing outbreaks in the different parts of the country.8,9,36,54–56 Moreover, serotype O was the main serotype reported from FMD outbreaks in neighboring countries to Ethiopia, such as Sudan and Kenya.57,58
Conclusion
In the current study, serological examination confirmed that foot and mouth disease is highly prevalent among cattle managed under different production systems of central Ethiopia. Associate risk variables like district, age, body condition score, and management system showed a strong association with FMD seropositivity. From outbreak cases, FMDV serotype O was identified using molecular tools. So, further study should be conducted at a wider range on virus isolation, serotyping, and molecular characterization of the FMD virus.
Data Sharing Statement
The data used to validate the results of this analysis are available from the first and correspondent authors upon reasonable request.
Ethical Approval and Consent to Participate
The best practice guidelines for veterinary care were applied in our study, dairy cattle owners were informed and aware about the purpose of the study, and verbal informed consent was allowed and approved by the Animal Research Scientific and Ethics Review Committee of the National Animal Health Diagnostic and Investigation Center, Sebeta, Ethiopia (National referral and reference laboratory). Regarding the cattle owners and managers, we don’t include personal data, images, videos, etc that can violate individual participants. Samples (blood, tissue) collection was carried out under aseptic conditions by experienced veterinary laboratory technicians without affecting an animal’s life.
Acknowledgments
This study was financed by Haramaya University and National Animal Health Diagnostic and Investigation Center (NAHDIC). So, the authors deeply acknowledge both organizations for various occasions during the duration of the study.
Author Contributions
All authors contributed significantly to the overall research activities, whether in the study’s creation, implementation, data collection, analysis, and interpretation or all of these fields. Moreover, all contributors contributed to the revision or critical review of the paper; gave final approval to the version to be published; decided on the journal to which the article was submitted; and agreed to be responsible for all facets of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Quinn PJ, Markey BK, Leonard FC, Hartigan P, Fanning S, Fitzpatrick E. Veterinary Microbiology and Microbial Disease. John Wiley & Sons; 2011.
2. Larska M, Wernery U, Kinne J, Schuster R, Alexandersen G, Alexandersen S. Differences in the susceptibility of dromedary and Bactrian camels to foot-and-mouth disease virus. Epidemiol Infect. 2009;137(4):549–554.
3. Rufael T, Sahle M, Asseged B Participatory appraisal and seroprevalence study of foot and mouth disease in Borana pastoral system, South Ethiopia. MSc thesis. Addis Ababa University, Faculty of veterinary medicine; 2006.
4. Grace D, Songe M, Knight-Jones TJ. Impact of Neglected Diseases on Animal Productivity and Public Health in Africa. 2015.
5. Knight‐Jones TJ, McLaws M, Rushton J. Foot‐and‐mouth disease impact on smallholders‐What do we know, what don’t we know and how can we find out more? Transbound Emerg Dis. 2017;64(4):1079–1094.
6. Gulima D. Disease reporting presentation on VACNADA Project closes out workshop. CBPP Ethiopia. 2011;27.
7. Martel J. Foot and mouth disease in Ethiopia: distribution of foot and mouth disease virus serotypes. Revue d’elevage et de medecine veterinaire des pays tropicaux. 1974;0035-1865.
8. Ayelet G, Mahapatra M, Gelaye E, et al. Genetic characterization of foot-and-mouth disease viruses, Ethiopia, 1981–2007. Emerg Infect Dis. 2009;15(9):1409.
9. Menda S, Jenberie S, Negussie H, Ayelet G, Amasalu K. Molecular epidemiology of foot and mouth disease Virus Outbreaks in Ethiopia in 2011/2012. AJAD. 2014;3:8–16.
10. Gizaw D, Tesfaye Y, Wood BA, et al. Molecular characterization of foot-and-mouth disease viruses circulating in Ethiopia between 2008 and 2019. Transbound Emerg Dis. 2020;67(6):2983–2992. doi:10.1111/tbed.13675
11. Maclachlan NJ, Dubovi EJ. Fenner’s Veterinary Virology. Academic press; 2010.
12. Rweyemamu M, Roeder P, Mackay D, et al. Epidemiological patterns of foot‐and‐mouth disease worldwide. Transbound Emerg Dis. 2008;55(1):57–72.
13. Authority(CSA) CS. Federal Democratic Republic of Ethiopia; Central Statistical Agency Agricultural Sample Survey: Report on Livestock and Livestock Characteristics 2014/15 [2007 E.C]. Ethiopia, Addis Ababa 2015.
14. Thrusfield M. Veterinary Epidemiology. John Wiley & Sons; 2018.
15. Bergmann I, Malirat V, Neitzert E, et al. Improvement of a serodiagnostic strategy for foot-and-mouth disease virus surveillance in cattle under systematic vaccination: a combined system of an indirect ELISA-3ABC with an enzyme-linked immunoelectrotransfer blot assay. Arch Virol. 2000;145(3):473–489.
16. FMD. Foot and Mouth Disease, Multispecies Antibody Test- IDEXX US. Westbrook M, ed. USA;2019:1–21.
17. Goodwin S, Tuthill TJ, Arias A, Killington RA, Rowlands DJ. Foot-and-mouth disease virus assembly: processing of recombinant capsid precursor by exogenous protease induces self-assembly of pentamers in vitro in a myristoylation-dependent manner. J Virol. 2009;83(21):11275–11282.
18. Reid SM, Grierson SS, Ferris NP, Hutchings GH, Alexandersen S. Evaluation of automated RT-PCR to accelerate the laboratory diagnosis of foot-and-mouth disease virus. J Virol Methods. 2003;107(2):129–139.
19. Callahan JD, Brown F, Osorio FA, et al. Use of a portable real-time reverse transcriptase polymerase chain reaction assay for rapid detection of foot-and-mouth disease virus. J Am Vet Med Assoc. 2002;220(11):1636–1642.
20. Feng Q, Yu H, Liu Y, et al. Genome comparison of a novel foot-and-mouth disease virus with other FMDV strains. Biochem Biophys Res Commun. 2004;323(1):254–263.
21. Mukasa HK, Mwiine FN, Atuhaire DK, Ochwo S, Nanteza A. Comparative detection of foot-and-mouth disease virus by reverse transcription loop-mediated isothermal amplification assay and real time polymerase chain reaction in Uganda. Int J Biotechnol Food Sci. 2016;4:22–33.
22. Howson E, Armson B, Madi M, et al. Evaluation of two lyophilized molecular assays to rapidly detect foot‐and‐mouth disease virus directly from clinical samples in field settings. Transbound Emerg Dis. 2017;64(3):861–871.
23. Reid SM, Ferris NP, Hutchings GH, Samuel AR, Knowles NJ. Primary diagnosis of foot-and-mouth disease by reverse transcription polymerase chain reaction. J Virol Methods. 2000;89(1–2):167–176.
24. Saduakassova MA, Sultanov AA, Kutumbetov LB, et al. Development and evaluation of a novel real-time RT-PCR to detect foot-and-mouth disease viruses from the emerging A/ASIA/G-VII lineage. J Virol Methods. 2018;252:37–41. doi:10.1016/j.jviromet.2017.10.023
25. Kasanga CJ, Yamazaki W, Mioulet V, et al. Rapid, sensitive and effective diagnostic tools for foot-and-mouth disease virus in Africa. 2014;81(2):2219–2635. doi:10.4102/ojvr.v81i2.727
26. Shaw AE, Reid SM, Ebert K, Hutchings GH, Ferris NP, King DP. Implementation of a one-step real-time RT-PCR protocol for diagnosis of foot-and-mouth disease. J Virol Methods. 2007;143(1):81–85.
27. Bachanek-Bankowska K, Mero HR, Wadsworth J, et al. Development and evaluation of tailored specific real-time RT-PCR assays for detection of foot-and-mouth disease virus serotypes circulating in East Africa. J Virol Methods. 2016;237:114–120.
28. Reid SM, Mioulet V, Knowles NJ, Shirazi N, Belsham GJ, King DP. Development of tailored real-time RT-PCR assays for the detection and differentiation of serotype O, A and Asia-1 foot-and-mouth disease virus lineages circulating in the Middle East. J Virol Methods. 2014;207:146–153.
29. Ahmed H, Salem S, Habashi A, et al. Emergence of foot‐and‐mouth disease virus SAT 2 in E gypt During 2012. Transbound Emerg Dis. 2012;59(6):476–481.
30. Knowles N, Bachanek‐Bankowska K, Wadsworth J, et al. Outbreaks of foot‐and‐mouth disease in Libya and Saudi Arabia during 2013 due to an exotic O/ME‐SA/Ind‐2001 lineage virus. Transbound Emerg Dis. 2016;63(5):e431–e435.
31. Ayelet G, Gelaye E, Negussie H, Asmare K. Study on the epidemiology of foot and mouth disease in Ethiopia. Rev Sci Tech. 2012;31(3):789–798.
32. Abunna F, Fikru S, Rufael T. Seroprevalence of Foot and Mouth Disease (FMD) at Dire Dawa and Its Surroundings, Eastern Ethiopia. Global Veterinaria. 2013;11(5):575–578.
33. Zerabruk G, Romha G, Rufael T. Sero-epidemiological investigation of foot and mouth disease in cattle managed under extensive husbandry system in Tigray, northern Ethiopia. Global Vet. 2014;13:112–116.
34. Megersa B, Beyene B, Abunna F, Regassa A, Amenu K, Rufael T. Risk factors for foot and mouth disease seroprevalence in indigenous cattle in Southern Ethiopia: the effect of production system. Trop Anim Health Prod. 2009;41(6):891–898.
35. Molla B, Ayelet G, Asfaw Y, Jibril Y, Ganga G, Gelaye E. Epidemiological study on foot‐and‐mouth disease in cattle: seroprevalence and risk factor assessment in South Omo zone, south‐western Ethiopia. Transbound Emerg Dis. 2010;57(5):
36. Negusssie H, Kyule MN, Yami M, Ayelet G. Outbreak investigations and genetic characterization of foot-and-mouth disease virus in Ethiopia in 2008/2009. Trop Anim Health Prod. 2011;43(1):
37. Birhanu T, Nekemte E. Prevalence of the major infectious animal diseases affecting livestock trade industry in Ethiopia. Prevalence. 2014;4:17.
38. Tesfaye A, Mengistu A, Rufael T. Sero-prevalence status of foot and mouth disease in the North Western Amhara Regional State, Ethiopia. Ethiopian Vet J. 2016;20(2):43–53.
39. Rufael T, Catley A, Bogale A, Sahle M, Shiferaw Y. Foot and mouth disease in the Borana pastoral system, southern Ethiopia and implications for livelihoods and international trade. Trop Anim Health Prod. 2008;40(1):
40. Mohamoud A, Tessema E, Degefu H. Seroprevalence of bovine foot and mouth disease (FMD) in Awbere and Babille districts of Jijiga zone, Somalia Regional State, Eastern Ethiopia. Afr J Microbiol Res. 2011;5(21):3559–3563.
41. Ahmed B, Megersa L, Mulatu G, Siraj M, Boneya G. Seroprevalence and associated risk factors of foot and mouth disease in cattle in West Shewa Zone, Ethiopia. Vet Med Int. 2020;2020:2090–8113.
42. OIE. Foot and Mouth Disease, Manual of Diagnostic Tests and Vaccines for Terrestrial Animals (Mammals, Birds, and Bees). Paris; 2012.
43. Namatovu A, Tjørnehøj K, Belsham GJ, et al. Characterization of foot-and-mouth disease viruses (FMDVs) from Ugandan cattle outbreaks during 2012-2013: evidence for circulation of multiple serotypes. PLoS One. 2015;10(2):e0114811.
44. Alexandersen S, Zhang Z, Donaldson AI. Aspects of the persistence of foot-and-mouth disease virus in animals—the carrier problem. Microbes Infect. 2002;4(10):1099–1110.
45. Gelaye E, Ayelet G, Abera T, Asmare K. Seroprevalence of foot and mouth disease in Bench Maji zone, Southwestern Ethiopia. J Vet Med Animal Health. 2009;1(1):005–010.
46. Alexandersen S, Mowat N. Foot-and-mouth disease: host range and pathogenesis. Foot Mouth Dis Virus. 2005;9–42.
47. Murphy FA, Gibbs EPJ, Horzinek MC, Studdert MJ. Veterinary Virology. Elsevier; 1999.
48. Mazengia H, Taye M, Negussie H, Alemu S, Tassew A. Incidence of foot and mouth disease and its effect on milk yield in dairy cattle at Andassa dairy farm, Northwest Ethiopia. Agri Biol J North Am. 2010;1(5):969–973.
49. Misgana D, Yasmin J, Ahmed I, Addisalem H. Seroprevalence of foot and mouth disease of cattle in Bale Zone, Oromiya regional state, Ethiopia. Global Veterinaria. 2013;11(1):59–64.
50. Radostits OM, Gay C, Hinchcliff KW, Constable PD. A textbook of the diseases of cattle, horses, sheep, pigs, and goats. Vet Med. 2007;10:2045–2050.
51. Grubman MJ, Baxt B. Foot-and-mouth disease. Clin Microbiol Rev. 2004;17(2):465–493.
52. Vosloo W, Bastos A, Sangare O, Hargreaves S, Thomson G. Review of the status and control of foot and mouth disease in sub-Saharan Africa. Revue Scientifique Et Technique-Office International Des Épizooties. 2002;21(3):437–445.
53. Paixão TA, Neta AVC, Paiva NO, et al. Diagnosis of foot-and-mouth disease by real-time reverse transcription-polymerase chain reaction under field conditions in Brazil. BMC Vet Res. 2008;4(1):
54. Urge B, Dawo F, Aklilu H, et al. Detection and molecular characterization of foot and mouth disease virus serotypes from outbreaks in Ada Berga District. Livestock Res Results. 40.
55. Getachew M, Sissay M, Delesa D Serotyping and molecular characterization of foot and mouth disease of cattle in central Ethiopia. MSc in Biotechnology. Harmaya University; 2017.
56. Sulayeman M, Dawo F, Mammo B, Gizaw D, Shegu D. Isolation, molecular characterization and sero-prevalence study of foot-and-mouth disease virus circulating in central Ethiopia. BMC Vet Res. 2018;14(1):1–10.
57. Habiela M, Ferris N, Hutchings G, et al. Molecular characterization of foot‐and‐mouth disease viruses collected from Sudan. Transbound Emerg Dis. 2010;57(5):305–314.
58. Chepkwony EC. An Epidemiological Survey of the Serotypes of Foot and Mouth Disease Virus in Circulation in the Somali-Ecosystem in Kenya. Applied Veterinary Microbiology Msc, University of Nairobi; 2011.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.