Back to Journals » Drug Design, Development and Therapy » Volume 15
Selection of DNA Aptamers Recognizing EpCAM-Positive Prostate Cancer by Cell-SELEX for in vitro and in vivo MR Imaging
Authors Zhong J , Ding J, Deng L, Xiang Y, Liu D, Zhang Y, Chen X, Yang Q
Received 4 June 2021
Accepted for publication 6 September 2021
Published 21 September 2021 Volume 2021:15 Pages 3985—3996
DOI https://doi.org/10.2147/DDDT.S322854
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Georgios Panos
Jinman Zhong,1,* Jianke Ding,2,* Lei Deng,1 Ying Xiang,1 Duoduo Liu,1 Yanyan Zhang,1 Xin Chen,1 Quanxin Yang1
1Department of Radiology, The Second Affiliated Hospital, Xi’ an Jiaotong University, Xi’an, Shaanxi Province, 710004, People’s Republic of China; 2Department of Plastic and Reconstructive Surgery, Xijing Hospital, Fourth Military Medical University, Xi’an, Shaanxi Province, 710032, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Quanxin Yang; Xin Chen
Department of Radiology, The Second Affiliated Hospital, Xi’ an Jiaotong University, 157 Xiwu Road, Xi’an, Shaanxi Province, 710004, People’s Republic of China
Email [email protected]; [email protected]
Background: The sensitive and specific detection of pathogenic cells is important in tumor diagnosis at an early stage. Aptamers are short single-stranded oligonucleotides evolved from systematic evolution of ligands by exponential enrichment (SELEX). It has been proved that aptamers can interact with cognate target molecules with high affinity and specificity and have great potential in the development of medical imaging at molecular level.
Purpose: To select epithelial cell adhesion molecule (EpCAM) specific aptamers targeting prostate cancer and further to conjugate aptamers with GoldMag nanoparticles (a typical iron oxide core/gold shell structure) to construct magnetic molecular probes for medical imaging.
Methods: EpCAM-specific aptamers were selected by Cell-SELEX. The enrichment of specific aptamer candidates was monitored by flow cytometric analysis. Aptamers were further conjugated with GoldMag nanoparticles to construct magnetic molecular probes. The affinity and specificity of aptamer candidates and aptamer-conjugated GoldMag nanoparticles were evaluated. The MR imaging of aptamer-conjugated GoldMag nanoparticles to prostate cancer was further explored in vitro and in vivo.
Results: After 12 rounds of selection, aptamer candidates Eppc6 and Eppc14 could specifically target three types of prostate cancer cells, revealing a high affinity of Eppc6 and Eppc14. Moreover, aptamer-conjugated GoldMag nanoparticles not only exhibited good affinity to different prostate cancer cells but also produced strong T2WI signal intensity reduction distinguished from peritumoral tissue in MRI, indicating that the molecular probes possess both the affinity properties of EpCAM-specific aptamer and the superparamagnetic features of iron oxide.
Conclusion: Our study indicates that aptamer Eppc6 and Eppc14 can recognize prostate cancer cells and tissues. The aptamer-conjugated GoldMag nanoparticles constructed in the study can be used as a molecular imaging agent for detection of PCa in MRI.
Keywords: EpCAM, aptamer, Cell-SELEX, prostate cancer
Introduction
Prostate cancer (PCa) is one of the most common genitourinary system malignant tumor in men worldwide. In Asian countries, many patients with PCa are often diagnosed at an advanced stage maybe mostly because of a large population base with relatively backward economic development and imperfect cancer screening system of PCa, which leads to a poor prognosis. Epithelial cell adhesion molecule (EpCAM, also known as CD326 or ESA) is a type I glycosylated transmembrane protein with a molecular weight of 37kDa that mediates epithelial-specific cell–cell adhesion, cell signalling, cell proliferation, migration and invasion.1,2 EpCAM is overexpressed in most carcinomas and cancer stem cells, but exhibits low expression levels in normal tissues and cells.3–5 Went et al analysed by immunohistochemical staining tissue microarrays with 4046 primary human carcinoma samples from colon, stomach, lung cancers and PCa for both frequency and intensity of EpCAM expression under highly standardised conditions. The results showed that high-level EpCAM expression was observed in 87.2% of PCa, and no detectable EpCAM staining was found with only 1.9% of PCa.6 It has been found that EpCAM is stably and highly expressed in primary and metastatic PCa tissues and EpCAM overexpression is considered as an independent marker for poor survival.7 During cancer progression, the expression pattern of EpCAM changes from basal and basolateral membrane in normal epithelial to the surface in tumor epithelial cells.7 The differential expression makes EpCAM as an ideal candidate of tumor-related marker to detect PCa in early at the cellular and molecular level.8
Aptamers are single-stranded DNA or RNA oligonucleotides derived from systematic evolution of ligands by exponential enrichment (SELEX) technology.9 Currently, aptamers have been selected against varieties of targets such as proteins, nucleic acids, cells among others, with high affinity and specificity.10 Compared with antibody, aptamers are much smaller and thus have better tissue penetration properties. They are also thermally stable, and easy to synthesize with low batch-to-batch variability. The easy functionalization makes it possible to introduce various imaging labels or functional groups to aptamers. In addition, aptamers are non-immunogenic in vivo.10–12 Therefore, aptamers have been served as valuable alternatives in tumor diagnosis and treatment. Previously, there have been several reports of EpCAM-specific aptamers used in colorectal cancer,13 ovarian cancer,14 breast cancer15 and brain metastasis16 for drug delivery and therapy, indicating that EpCAM may be a useful marker for developing antitumor therapeutic and diagnostic strategies. Currently, EpCAM-specific aptamers targeting PCa have not been reported in previous studies.
Recent pre-clinical research efforts to develop multi-modality molecular imaging methods for diagnosis of tumors have the potential for noninvasive PCa diagnosis in early. Superparamagnetic iron oxide (SPIO) nanoparticles can be readily bound to various molecular markers including ligands, proteins and nucleic acid for magnetic resonance imaging (MRI), which allows for the characterization of structural and functional cellular changes. Currently, SPIO nanoparticles have attracted broad attention due to their potential applications in MRI.17–19 In the study, we used GoldMag nanoparticles, essentially a kind of SPIO, as MRI contrast agents. The nanoparticles are a typical core/shell structure with iron oxide (Fe3O4) as the core and a layer of gold deposited on the core surface as the shell.20–22 The Fe3O4 core provides a particle of a small size with significant magnetic moment, and a gold coating on the Fe3O4 core introduces a good platform for further conjugation with biomolecules. In this study, we screened anti-EpCAM DNA aptamers Eppc6 and Eppc14 by Cell-SELEX, and coupled aptamers with GoldMag nanoparticles to construct the molecular probe Eppc6-GoldMag for MR imaging of PCa xenografts. Our findings indicate that the aptamer Eppc6 and Eppc14 may be a valuable agent for diagnosis of PCa.
Materials and Methods
Cell Lines and Cell Culture
PC-3, DU145, LNCaP (PCa cells) and HEK293T (human embryonic kidney cells) cell lines were purchased from Typical Culture Preservation Commission Cell Bank, Chinese Academy of Sciences (Shanghai, China) and identified by short tandem repeat (STR) methods before the experiments. DU145, LNCaP and HEK293T were cultured in the basic medium of Dulbecco’s Modified Eagle Medium (DMEM, Gibco, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, USA); PC-3 cells were cultured in the medium of RPMI-1640 (Gibco, USA) with 10% FBS. All cell lines were cultured at 37°C under a mixture of 95% air and 5% CO2.
Construction of Recombinant Vector and Transfection
EpCAM cDNA was cloned into a pCDH-CMV-MCS-EF1-Puro lentiviral expression vector (Invitrogen), and the resulting expression plasmid pCDH-EpCAM was confirmed by DNA sequencing. HEK293T cells were transfected with pCDH-EpCAM plasmid (HEK293T-EpCAM) using Lipofectamine 3000 according to manufacturer’s instructions. In addition, HEK293T cells were mock-transfected with the empty pCDH-CMV-MCS-EF1-Puro plasmid (mock cells) by the same method.
SELEX Library and Primers
All oligonucleotides, including single-stranded DNA (ssDNA) library and primers, were synthesized by Sangon Biotech Co. Ltd. (Shanghai, China) and purified by HPLC. The library contained a central randomized sequence of 40 nucleotides flanked by 20-nucleotide sequences for primer annealing (5′-TTCTTCAAGTCCGCCATGCC-(40N)-CGACAAGCAGAAGAACGGCA-3′). The forward primer sequence was 5′-FAM-TTCTTCAAGTCCGCCATGCC-3′, and the reverse primer sequence was 5′-Biotin- TGCCGTTCTTCTGCTTGTCG −3′, which were synthesized for polymerase chain reaction (PCR) amplification.
Cell-SELEX
HEK293T-EpCAM cells were used as the target cell line, and mock cells as the control cell line. The initial ssDNA library (2 OD) was dissolved in 1 mL of binding buffer (0.1 M Hepes, 0.75 M NaCl, 0.025 M KCl, 0.01 M CaCl2, 0.01 M MgCl2·6H2O in double distilled water, pH 7.4). The library was denatured at 95°C for 5 min and immediately cooled on ice for 10 min. For the first cycle of selection, the cells were incubated with the initial library for 2 h at 37°C. After incubation, cells were rinsed with PBS three times for 5 min to remove unbound sequences. The bound ssDNAs were then extracted by phenol-chloroform-isoamyl alcohol (125:24:1) extraction method before centrifuging at 12,000 g for 5 min at 4°C, after which the DNAs in the liquid phase were precipitated with sodium acetate (3 M) and cooled isopropanol, and incubated at −20°C for 3 h. The mixture was centrifuged at 12,000 g for 10 min at 4°C, following which the supernatant was removed and the DNA pellet was rinsed with 1 mL of 75% cooled ethanol and then resuspended in 200 µL of double distilled water. The DNA was amplified by PCR using FAM-labeled forward primer and Biotin-labeled reverse primer under the following conditions: 95°C for 5 min, 15–30 cycles of 10 s at 95°C, 10 s annealing at 57°C, and 10 s extension at 72°C, followed by 72°C for 5 min. The PCR product was precipitated with sodium acetate and cooled isopropanol at −20°C for 3 h, and then centrifuged at 12,000 g for 10 min at 4°C, after which the supernatant was removed and the DNA pellet was washed with 1 mL of 75% cooled ethanol and then resuspended in 200 µL of Buffer I (10 mM Tris-HCl, 1 mM EDTA, 1 M NaCl, 0.01–0.1% Tween-20 in double distilled water). The DNA was incubated with streptavidin-coated sepharose beads for 30 min at room temperature, then denatured by incubation with 0.15 M NaOH for 20 min. The selected ssDNA was lyophilized and resuspended in binding buffer for the subsequent round of selection or cytometric analysis. From the sixth round, the ssDNA pool was incubated with negative control cells and the unbound sequences were collected and incubated with the target cells. After 12 rounds of selection, the enriched ssDNA pool was cloned and sequenced by Sangon Biotech Co. Ltd. (Shanghai, China). Two effective ssDNA sequences Epp6 and Eppc14 with high affinity for recognizing target cells were selected and synthesized. The secondary structure of Epp6 and Eppc14 was predicted by using the RNA structure (http://rna.urmc.rochester.edu/RNAstructureWeb/).
Flow Cytometric Analysis
To evaluate the enrichment of specific aptamer candidates and the binding affinity of the aptamer candidates to target cells, cells were incubated with FAM-labeled ssDNA at 37°C for 45 min after detached by treatment with 0.1% trypsin‑0.02% EDTA solution. Cells were rinsed with PBS binding buffer containing 10% FCS and 0.02% NaN3 three times for 5 min and resuspended in 200 µL of binding buffer. The fluorescence signal was analyzed with a FACSCalibur flow cytometer (BD Biosciences, USA).
The binding affinity of the molecular probe aptamer-GoldMag to target cells was also evaluated by flow cytometric analysis. After cells were detached by treatment with 0.1% trypsin‑0.02% EDTA solution, cells were placed in fetal calf serum (FCS)-supplemented medium to inactivate trypsin and EDTA. Then the cells were incubated with biotin-labeled aptamer-GoldMag at 37°C for 45 min in PBS binding buffer containing 10% FCS and 0.02% NaN3. After washing, cells were incubated with FAM-labeled streptavidin in binding buffer at 4°C for 30 min. Cells were then rinsed with washing buffer three times and resuspended in 200 µL of binding buffer. The fluorescence signal was analyzed with a FACSCalibur flow cytometer.
To measure the equilibrium dissociation constant (Kd) of aptamers, HEK293T-EpCAM cells were incubated with a series of concentrations of FAM-labeled aptamers in 200 µL of binding buffer at 37°C for 45 min. The FAM-labeled random DNA library was used as a negative control. Cells were rinsed with washing buffer three times for 5 min and resuspended in 200 µL of binding buffer for flow cytometric analysis. The Kds of aptamers were determined by fitting the dependence of fluorescence intensity on aptamer concentration with the equation Y=Bmax × X/(Kd+X), where Y is fluorescence intensity of cells, X is concentration of the aptamer and Bmax is maximum binding potential. All the experiments for the binding assay were repeated in triplicate.
Confocal Microscopy Imaging
Confocal microscopy imaging was performed to monitor the binding affinity of the aptamer candidates and their molecular probes to target cells. 1×105 cells were seeded in a 20 mm glass bottom dish and cultured overnight. Then the cells were fixed in 4% paraformaldehyde in PBS for 15 min and washed three times with PBS, following which the cells were blocked with 5% BSA in PBS at room temperature for 1 h and stained with 200 pmol of FAM-labeled aptamers/biotin-labeled molecular probe at 4°C overnight. After washing, cells incubated with the latter were incubated with Cy3-labeled streptavidin at room temperature for 2 h. Nuclei were counterstained with 10 µg/mL DAPI (Invitrogen, Carlsbad, CA, USA). After washing, the stained cells were imaged by confocal laser scanning microscope (Olympus, Japan).
Immobilization of Aptamer Onto GoldMag Surface
GoldMag nanoparticles were purchased from GoldMag Biotechnology Co., Ltd (Shanxi, China). The nanoparticles require only a single step for nucleic acid immobilization. 20 to 160 µg aptamer Eppc6/Eppc14 were dissolved in a 500 µL of binding buffer (0.01 M PBS, pH 7.4) containing 1mg GoldMag nanoparticles. The mixture was incubated at 37°C for 30 min. The GoldMag nanoparticles coupled with aptamers were magnetically separated from free aptamers. The coupled GoldMag was then rinsed with washing buffer three times and resuspended in PBS. Immobilization efficiency of aptamers onto the GoldMag surface was calculated by using the equation: (OD260(pre) × D1 - OD260(post) × D2)/(OD260(pre) × D1) × 100%, where OD260(pre) and OD260(post) are the absorbance values at 260 nm of pre- and post-coupling aptamer solution, respectively; D1 and D2 are the dilution factors of pre- and post-coupling aptamer solution, respectively. ssDNA-GoldMag was used for negative control and synthesized in the same way.
In vitro MR Imaging of Magnetic Molecular Probe Eppc6-GoldMag
To evaluate the ability of aptamer-conjugated GoldMag to target EpCAM-positive PCa cells for MRI in vitro, aptamer Eppc6 that has higher Kd compared with Eppc14 was primarily selected for detection. Three types of PCa cells PC-3, DU145 and LNCaP were, respectively, incubated with Eppc6-GoldMag at 37°C for 30 min. Subsequently, the cells were rinsed with washing buffer three times and resuspended in 3% agarose gel in eppendorf tubes, respectively. After the agarose solidification, the cells in eppendorf tubes were put in the test-tube stand and scanned by using a 3.0T MRI scanner (Siemens Magnetom Tim Trio MRI). The T2-weighted imaging (T2WI) of three types of PCa cell groups were, respectively, acquired with a repetition time (TR) of 3500 ms, an echo time (TE) of 90 ms, a field-of-view (FOV) of 109 mm, a slice thickness of 2.0 mm, and an average of 8. ssDNA-GoldMag served as a negative control. After MRI examination, the data were sent to a dedicated MRI image processing workstation (Leonardo; Siemens) for further analysis. Region of interest (ROI) was drawn in the longitudinal section of respective tubes to measure the T2WI signal intensity of PCa cells. At least three ROIs were drawn in each tube.
MRI of Tumor-Bearing Mice Injected with Eppc6-GoldMag
Male nude mice (Bab/c, 5–6 weeks) under specific pathogen-free conditions were used for the study. According to the power analysis, at least 6 samples (mice) of each group for T2WI signal intensity statistic analysis were needed; thus, we used 12 nude mice (experimental group for 6 and control group for 6, respectively) in the animal experiments and repeated the experiments three times independently. Experiments on animals were performed in accordance with the Animal Care and Use Committee of Xi’ an Jiaotong University (Xi’an, China) and followed the guidelines of “Laboratory Animal-General requirements for animal experiments” (GB/T, 35823-2018, China). The Animal Care and Use Committee of Xi’ an Jiaotong University has approved the animal experiments conducted in our study. The prostate tumor-bearing nude mice model was established by subcutaneously injecting 5×106 PC-3 cells into the right armpit of nude mice. Experiment was performed when tumors reached 0.8–1.0 cm in average diameter. The tumor-bearing nude mice underwent MRI using small-animal coils before the injection of Eppc6-GoldMag. The mice were anesthetized with an intraperitoneal injection of 1% sodium pentobarbital (0.5 mL/100 g). Subsequently, tumor-bearing nude mice (experimental group) were injected with 200 µL (1 mg GoldMag nanoparticles mixed with 40 µg aptamer in each mL) Eppc6-GoldMag via tail vein and underwent MRI examination 1 h, 6 h and 12 h after the injection, respectively. The tumor-bearing nude mice (control group) intravenously injected with 200 µL ssDNA-GoldMag served as negative control. T2-weighted coronal images were obtained with the following parameters: Repetition time, 7400 ms; echo time, 66 ms; field of view, 200 mm; slice thickness, 2 mm; flip angle, 150°; and number of signal averaging, 4. After MRI examination, all mice were euthanized by cervical dislocation. The MR imaging data were sent to a dedicated MRI image processing workstation (Leonardo; Siemens) for further analysis. ROI was drawn in the coronal section of respective mouse tumors to measure the T2WI signal intensity of tumors. At least three ROIs were drawn in each mouse tumor at one time point.
Statistical Analysis
The experiments were repeated three times independently. The data were presented as median with interquartile range (IQR). Kruskal–Wallis H-test followed by Nemenyi test were used to compare T2WI signal intensity measurements from in vitro sample vials containing different cell lines, and to compare in vivo T2WI signal intensity measurements in tumors at pre-injection and three post-injection time intervals. A P value less than 0.05 was considered statistically significant. The statistical analysis was performed by GraphPad Prism (Version 6.0 for Windows, GraphPad Software Inc., San Diego, CA, USA).
Results
Expression of EpCAM in Transfected HEK293T Cells and PCa Cells
To effectively select aptamers targeting EpCAM, we sought to express EpCAM protein on the surface of HEK293T cells in order to maintain its native conformation. HEK293T cells were transfected with pCDH-EpCAM. After transfection, we confirmed the overexpression and location of EpCAM by Flow cytometry. As shown in Figure 1, the fluorescence intensity of HEK293T-EpCAM cells as well as PCa cells PC-3, DU145 and LNCaP were increased. In contrast, almost no increase in fluorescence signal was observed for negative control mock cells.
Selection of DNA Aptamers Against EpCAM-Expressing HEK293T Cells
To isolate aptamers specifically targeting EpCAM-positive cells, we selected HEK293T-EpCAM cells to be the target cells, and mock cells to be control cells. For evaluating the process of enrichment, we analyzed the ssDNA libraries derived from the 5th, 9th, 11th and 12th round of selection by flow cytometry. As shown in Figure 2, the fluorescence intensity curve reflects the amount of FAM-conjugated aptamer bound to cells. When the number of turns increased, the fluorescence intensity of HEK293T-EpCAM cells gradually increased (Figure 2A). In contrast, mock cells exhibited only weak fluorescence strength, similar to the negative control group (Figure 2B). The results suggest that the target cell-binding DNA sequences were gradually enriched during the selection process.
Identification of DNA Aptamer Candidates
After sequencing, two representative sequences, termed Eppc6 and Eppc14 were selected (Table 1) and chemically synthesized for further characterization. The predicted secondary structure of Eppc6 and Eppc14 are shown in Figure 3A and B. To quantitatively evaluate the binding affinity of Eppc6 and Eppc14 to HEK293T-EpCAM cells, the Kds of the two aptamers were measured by flow cytometry. The Kd of Eppc6 and Eppc14 for HEK293T-EpCAM cells were calculated, respectively, by using the equation Y=Bmax × X/(Kd+X). As shown in Figure 3C and D, aptamer Eppc6 was determined to possess a Kd of 41.7 ± 5.9 nM, aptamer Eppc14 was determined to possess a Kd of 80.8 ± 7.4 nM, indicating that the two selected aptamers could specifically recognize target HEK293T-EpCAM cells with high affinity.
 |
Table 1 Sequences of Aptamer Eppc6 and Eppc14 |
 |
Figure 3 Predicted secondary structures of aptamer candidates Eppc6 (A) and Eppc14 (B); and binding curves of Eppc6 (C) and Eppc14 (D). |
Aptamer Eppc6 and Eppc14 Could Specifically Target EpCAM-Positive HEK293T-EpCAM Cells and PCa Cells
To investigate the binding affinity of aptamer Eppc6 and Eppc14 to HEK293T-EpCAM cells and PCa cells, flow cytometry and confocal microscopy imaging were performed. The unselected initial ssDNA library was used as control. As shown in Figure 4A, the fluorescence intensity of HEK293T-EpCAM cells as well as PCa cells PC-3, DU145 and LNCaP were increased after incubation with Eppc6 and Eppc14, respectively. There was no increase in fluorescence signal in cells incubated with ssDNA library, neither in mock cells. In addition, the fluorescence signal was observed mainly on the surface of three types of PCa cells after incubation with Eppc6 and Eppc14, respectively, but not on cells incubated with ssDNA library (Figure 4B).
Immobilization of Aptamers on GoldMag to Construct Molecular Probes
The rate of aptamer-immobilized coupling was gradually reduced with increasing concentrations of aptamer Eppc6/Eppc14. 40 µg/ 50 µg of aptamer Eppc6 mixed with 1mg GoldMag nanoparticles yielded immobilization efficiency close to 92% and 86%, respectively (Supplementary Materials Figure S1A); 40 µg/50 µg of aptamer Eppc14 mixed with 1mg GoldMag nanoparticles yielded immobilization efficiency close to 89% and 83%, respectively (Supplementary Materials Figure S1B). Therefore, for 1mg of sample, about 40 µg of Eppc6 or 30 µg of Eppc14 were surface-coupled to the GoldMag nanoparticles. To avoid biases during later in vitro and in vivo comparison studies, immobilization efficiency ssDNA to GoldMag nanoparticles was also determined under the same conditions.
Aptamers-Conjugated GoldMag Nanoparticles Could Bind to PCa Cells
Binding of aptamer Eppc6 and Eppc14, respectively, conjugated with magnetic GoldMag to PCa cells were investigated by flow cytometry and confocal microscopy imaging. As shown in Figure 5A, the fluorescence intensity of HEK293T-EpCAM cells as well as three types of PCa cells were increased after incubation with the molecular probe Eppc6-GoldMag/Eppc14-GoldMag, while no increase in fluorescence signal in cells incubated with ssDNA-GoldMag. With ssDNA-GoldMag serving as a negative control, specific staining with Eppc6-GoldMag/Eppc14-GoldMag was detected on three types of PCa cells (Figure 5B), suggesting that Eppc6-GoldMag could specifically target EpCAM-positive PCa cells.
In vitro MR Imaging for PCa Cells Incubated with Eppc6-GoldMag
To evaluate the ability of Eppc6-GoldMag to recognize PCa cells for MRI in vitro, three types of EpCAM-positive PCa cells incubated with Eppc6-GoldMag (experimental group) were, respectively, fixed by 3% agarose gel and underwent MRI. PCa cells incubated with ssDNA-GoldMag served as negative control (control group). As shown in Figure 6, MRI of three types of PCa cells PC-3, DU145 and LNCaP treated with Eppc6-GoldMag, respectively, showed significantly greater T2WI signal intensity reductions compared to the corresponding three types of PCa cells treated with ssDNA-GoldMag (All P < 0.0001 for each comparison).
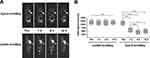
In vivo MR Imaging for Prostate Tumor-Bearing Nude Mice
To investigate that Eppc6-GoldMag had the ability to recognize and target EpCAM-positive PCa cells with high specificity in vivo, PC-3 cell prostate tumor-bearing nude mice were used. As shown in Figure 7, T2WI of tumor-bearing nude mice were acquired and showed that the signal intensity of tumors slightly decreased after 1 h following injection, and continued to decrease after 6 h following injection. The T2WI signal intensity changes of tumors after injection of Eppc6-GoldMag nanoparticles were statistically significant when compared to the baseline pre-injection T2WI signal intensity levels of tumors (P < 0.0001 for each comparison). Significant reductions in T2WI signal intensity of tumor were observed between the 1 and 6 hour post-injection time points (P = 0.0009). The T2WI signal intensity of tumors gradually increased 12 h later. Accumulation of Eppc6-GoldMag was greatest in tumors with relatively little in normal tissue distribution. There was no significant difference of tumor T2WI signal intensity between the 6 and 12 hours post-injection time points (P = 0.4376). Significant difference was found between the 1 and 12 hours post-injection time points (P = 0.0081). In addition, there were no statistically significant T2WI signal intensity changes after injection of negative control ssDNA-GoldMag (All P > 0.05). The results showed that aptamer Eppc6 could accumulate and efficiently penetrate into tumors, and aptamer Eppc6-conjugated GoldMag had the ability for PCa MR imaging.
Discussion
In the study, we successfully selected EpCAM-specific DNA aptamers Eppc6 and Eppc14 via Cell-SELEX, and further coupled aptamers with GoldMag, a kind of SPIO nanoparticles, to construct the molecular probe Eppc6-GoldMag for MR imaging of PCa xenografts. The aptamers Eppc6 and Eppc14 as well as their corresponding molecular probes were identified to have high affinity and specificity for EpCAM. Furthermore, the molecular probe Eppc6-GoldMag showed a strong effect that enhanced the visual contrast between tumors and surrounding tissues. Our findings indicate that aptamer Eppc6 and Eppc14 may be a valuable agent for early recognition and diagnosis of PCa.
EpCAM is a pleiotropic molecule that potentially offers diagnostic and therapeutic applications in tumor. Recent data show that EpCAM is expressed exclusively in epithelia and epithelial-derived neoplasms, and involved in cell signaling, migration, proliferation and differentiation.23,24 In PCa, high expression of EpCAM is identified in about 90% of prostate acinar adenocarcinoma, while in hormone-refractory adenocarcinomas of the prostate, significantly higher EpCAM expression is found than in untreated adenocarcinomas.25,26 In the current study, we examined the expression of EpCAM in three types of PCa cells, as well as in HEK293T cells transfected with pCDH-EpCAM plasmid or mock-transfected with empty pCDH-CMV-MCS-EF1-Puro plasmid to further select EpCAM-specific aptamers via Cell-SELEX. The results confirmed that EpCAM is highly expressed in three types of PCa cells including androgen-dependent prostate cancer (ADPC) LNCap cells, and castration-resistant prostate cancer (CRPC) PC-3 and DU145 cells. Furthermore, EpCAM expression level of the latter was higher than that of the former, which is in accordance with previous studies.5–7
Aptamers are short RNA or single-stranded DNA oligonucleotides. In the early stage, researchers preferred to develop RNA aptamers for their higher affinity and diversity of 3D structures.27,28 However, RNA aptamers are easily degraded by RNase, which increases the difficulty of its screening and application.28 On the other hand, DNA aptamers are much more stable in structures, thus attract more attention than RNA aptamers.29 On that basis, we screened and isolated EpCAM-specific aptamers Eppc6 and Eppc14, which were DNA aptamers. Both aptamers could specifically recognize target cell lines with structural stability, thus be further stably conjugated with SPIO nanoparticles to construct molecular probes for in vitro and in vivo MR imaging.
Aptamers as ssDNA can fold into various structures (such as hairpin, stem loop, pseudoknot) through intramolecular interactions under certain conditions.27 Similar to conformational recognition that mediates antibody-antigen recognition and complex formation, aptamers bind to their cognate targets with high specificity and affinity via van der Waals forces, hydrogen bonding, electrostatic interactions, stacking of flat moieties, and shape complementarity with Kd usually ranging from pico- to nanomolar.30 Aptamers selected in the study have different nucleic acid sequences compared with other previous sequences of EpCAM aptamers. However, most of them have similar loop-stem or hairpin structures according to their predicted secondary structures which help them bind to the targets.1,31,32 The EpCAM aptamers selected in our study have advantages over those reported in previous studies. Firstly, the Cell-SELEX was applied in place of traditional SELEX in vitro selection of target aptamers. Traditional SELEX are usually based on knowledge of the target for aptamer selection, while the Cell-SELEX can be used to generate highly specific aptamers by using whole cells without prior knowledge of target molecules,33 which greatly improves the efficiency of aptamer selection. In addition, our study used HEK293T cells transfected with pCDH-EpCAM plasmid as the positive cell line and HEK293T cells mock-transfected as the negative cell line for EpCAM-specific aptamers selection, which ensures elimination of common molecules on positive and negative cells and reduces non-specific sequences. Furthermore, aptamer Eppc6 possesses a Kd of 41.7 ± 5.9 nM and aptamer Eppc14 possesses a Kd of 80.8 ± 7.4 nM. The Kd of the two aptamers is in agreement with previously reported RNA and DNA aptamers against EpCAM (approximately 30–70 nM).1,5
The GoldMag used in the study to conjugate EpCAM aptamers for in vitro and in vivo MR imaging are essentially SPIO nanoparticles. In the presence of SPIO, a change in relaxation rate of water protons is detectable by MRI which is responsible for the image contrast. When cellular uptake of the nanoparticles occurs, the SPIO causes local magnetic field inhomogeneities which result in signal decrease on T2WI.34 GoldMag nanoparticles are a typical core/shell structure with Fe3O4 as the core and a layer of gold deposited on the core surface as the shell. Magnetic Fe3O4 nanoparticles as the core can considerably disturb the magnetic field, leading to localized signal loss in MR images. A layer of gold as the shell can provide a platform for conjugation with biomolecules. The gold-coated Fe3O4 nanoparticles were reported to exhibit good biocompatibility and affinity.35,36 On that basis, the aptamer-conjugated GoldMag molecular probes constructed in the study possess both the affinity properties of EpCAM-specific aptamer and the superparamagnetic features of iron oxide. The former properties could lead molecular probes to target tumor cells and tissues specifically, and the latter features could produce strong T2WI signal intensity reduction distinguished from peritumoral tissue. In vivo studies, the molecular probe Eppc6-GoldMag was intravenously injected into prostate tumor-bearing nude mice, and T2WI of tumors showed significantly decreased signal intensity compared to pre-injection levels. The evidence indicates the potential to use Eppc6-GoldMag as in vivo MRI probe targeted to EpCAM-positive PCa cells.
In conclusion, aptamer Eppc6 and Eppc14 successfully obtained by Cell-SELEX after 12 rounds of selection can specifically recognize EpCAM-positive PCa cell lines. The aptamer-conjugated GoldMag nanoparticles constructed in this study also display specific binding to EpCAM-positive PCa cell lines. Furthermore, Eppc6-GoldMag could be used as a molecular imaging agent for detection of PCa cells and tissues in MRI. Our study is of significance for early recognition and diagnosis of PCa, which has the potential for diagnosis and targeting therapy of other EpCAM-relative tumors.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Grant number: 82102347) and the Shanxi Provincial key research and development project (Grant number: 2021SF-267).
Disclosure
The authors declare that there are no conflicts of interest in this work.
References
1. Shigdar S, Lin J, Yu Y, Pastuovic M, Wei M, Duan W. RNA aptamer against a cancer stem cell marker epithelial cell adhesion molecule. Cancer Sci. 2011;102(5):991–998. doi:10.1111/j.1349-7006.2011.01897.x
2. Zhang J, Qi YP, Ma N, et al. Overexpression of Epcam and CD133 correlates with poor prognosis in dual-phenotype hepatocellular carcinoma. J Cancer. 2020;11(11):3400–3406. doi:10.7150/jca.41090
3. Ababneh N, Alshaer W, Allozi O, et al. In vitro selection of modified RNA aptamers against CD44 cancer stem cell marker. Nucleic Acid Ther. 2013;23(6):401–407. doi:10.1089/nat.2013.0423
4. Subramanian N, Kanwar JR, Kanwar RK, et al. EpCAM aptamer-siRNA chimera targets and regress epithelial cancer. PLoS One. 2015;10(7):e132407. doi:10.1371/journal.pone.0132407
5. Song Y, Zhu Z, An Y, et al. Selection of DNA aptamers against epithelial cell adhesion molecule for cancer cell imaging and circulating tumor cell capture. Anal Chem. 2013;85(8):4141–4149. doi:10.1021/ac400366b
6. Went P, Vasei M, Bubendorf L, et al. Frequent high-level expression of the immunotherapeutic target Ep-CAM in colon, stomach, prostate and lung cancers. Br J Cancer. 2006;94(1):128–135. doi:10.1038/sj.bjc.6602924
7. Benko G, Spajic B, Kruslin B, Tomas D. Impact of the EpCAM expression on biochemical recurrence-free survival in clinically localized prostate cancer. Urol Oncol. 2013;31(4):468–474. doi:10.1016/j.urolonc.2011.03.007
8. Patriarca C, Macchi RM, Marschner AK, Mellstedt H. Epithelial cell adhesion molecule expression (CD326) in cancer: a short review. Cancer Treat Rev. 2012;38(1):68–75. doi:10.1016/j.ctrv.2011.04.002
9. Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249(4968):505–510. doi:10.1126/science.2200121
10. Zhang Y, Hong H, Cai W. Tumor-targeted drug delivery with aptamers. Curr Med Chem. 2011;18(27):4185–4194. doi:10.2174/092986711797189547
11. Kim JW, Kim EY, Kim SY, et al. Identification of DNA aptamers toward epithelial cell adhesion molecule via cell-SELEX. Mol Cells. 2014;37(10):742–746. doi:10.14348/molcells.2014.0208
12. Szeitner Z, Andras J, Gyurcsanyi RE, Meszaros T. Is less more? Lessons from aptamer selection strategies. J Pharm Biomed Anal. 2014;101:58–65. doi:10.1016/j.jpba.2014.04.018
13. Xie X, Li F, Zhang H, et al. EpCAM aptamer-functionalized mesoporous silica nanoparticles for efficient colon cancer cell-targeted drug delivery. Eur J Pharm Sci. 2016;83:28–35. doi:10.1016/j.ejps.2015.12.014
14. Zheng J, Zhao S, Yu X, Huang S, Liu HY. Simultaneous targeting of CD44 and EpCAM with a bispecific aptamer effectively inhibits intraperitoneal ovarian cancer growth. Theranostics. 2017;7(5):1373–1388. doi:10.7150/thno.17826
15. Alibolandi M, Ramezani M, Sadeghi F, Abnous K, Hadizadeh F. Epithelial cell adhesion molecule aptamer conjugated PEG-PLGA nanopolymersomes for targeted delivery of doxorubicin to human breast adenocarcinoma cell line in vitro. Int J Pharm. 2015;479(1):241–251. doi:10.1016/j.ijpharm.2014.12.035
16. Macdonald J, Denoyer D, Henri J, et al. Bifunctional aptamer-Doxorubicin conjugate crosses the blood-brain barrier and selectively delivers its payload to EpCAM-positive tumor cells. Nucleic Acid Ther. 2020;30(2):117–128. doi:10.1089/nat.2019.0807
17. Yigit MV, Mazumdar D, Lu Y. MRI detection of thrombin with aptamer functionalized superparamagnetic iron oxide nanoparticles. Bioconjug Chem. 2008;19(2):412–417. doi:10.1021/bc7003928
18. Liang G, Cai S, Zhang P, et al. Magnetic relaxation switch and colorimetric detection of thrombin using aptamer-functionalized gold-coated iron oxide nanoparticles. Anal Chim Acta. 2011;689(2):243–249. doi:10.1016/j.aca.2011.01.046
19. Islam T, Josephson L. Current state and future applications of active targeting in malignancies using superparamagnetic iron oxide nanoparticles. Cancer Biomark. 2009;5(2):99–107. doi:10.3233/CBM-2009-0615
20. Cui YL, Hu DD, Fang Y, Ma JB. Preparation and mechanism of Fe3O4/Au core/shell super-paramagnetic microsphere. Sci China Series B. 2001;44(6):404–410. doi:10.1007/BF02879815
21. Park HY, Schadt MJ, Wang L, et al. Fabrication of magnetic core@shell Fe oxide@Au nanoparticles for interfacial bioactivity and bio-separation. Langmuir. 2007;23(17):9050–9056. doi:10.1021/la701305f
22. Lin J, Zhou WL, Kumbhar A, et al. Gold-coated iron (Fe@Au) nanoparticles: synthesis, characterization, and magnetic field-induced self-assembly. J Solid State Chem. 2001;159(1):26–31. doi:10.1006/jssc.2001.9117
23. Bellone S, Siegel ER, Cocco E, et al. Overexpression of epithelial cell adhesion molecule in primary, metastatic, and recurrent/ chemotherapy-resistant epithelial ovarian cancer: implications for epithelial cell adhesion molecule-specific immunotherapy. Int J Gynecol Cancer. 2009;19(5):860–866. doi:10.1111/IGC.0b013e3181a8331f
24. Passebosc-Faure K, Li G, Lambert C, et al. Evaluation of a panel of molecular markers for the diagnosis of malignant serous effusions. Clin Cancer Res. 2005;11(19 Pt 1):6862–6867. doi:10.1158/1078-0432.CCR-05-0043
25. Went PT, Lugli A, Meier S, et al. Frequent EpCam protein expression in human carcinomas. Hum Pathol. 2004;35(1):122–128. doi:10.1016/j.humpath.2003.08.026
26. Rao CG, Chianese D, Doyle GV, et al. Expression of epithelial cell adhesion molecule in carcinoma cells present in blood and primary and metastatic tumors. Int J Oncol. 2005;27(1):49–57.
27. Zhang Y, Lai BS, Juhas M. Recent advances in aptamer discovery and applications. Molecules. 2019;24(5):941. doi:10.3390/molecules24050941
28. Gotrik MR, Feagin TA, Csordas AT, Nakamoto MA, Soh HT. Advancements in aptamer discovery technologies. Acc Chem Res. 2016;49(9):1903–1910. doi:10.1021/acs.accounts.6b00283
29. Wang T, Chen C, Larcher LM, Barrero RA, Veedu RN. Three decades of nucleic acid aptamer technologies: lessons learned, progress and opportunities on aptamer development. Biotechnol Adv. 2019;37(1):28–50. doi:10.1016/j.biotechadv.2018.11.001
30. Cai S, Yan J, Xiong H, Liu Y, Peng D, Liu Z. Investigations on the interface of nucleic acid aptamers and binding targets. Analyst. 2018;143(22):5317–5338. doi:10.1039/C8AN01467A
31. Alshaer W, Ababneh N, Hatmal M, et al. Selection and targeting of EpCAM protein by ssDNA aptamer. PLoS One. 2017;12(12):e189558. doi:10.1371/journal.pone.0189558
32. Bavi R, Liu Z, Han Z, Zhang H, Gu Y. In silico designed RNA aptamer against epithelial cell adhesion molecule for cancer cell imaging. Biochem Biophys Res Commun. 2019;509(4):937–942. doi:10.1016/j.bbrc.2019.01.028
33. Darmostuk M, Rimpelova S, Gbelcova H, Ruml T. Current approaches in SELEX: an update to aptamer selection technology. Biotechnol Adv. 2015;33(6 Pt 2):1141–1161. doi:10.1016/j.biotechadv.2015.02.008
34. Dulinska-Litewka J, Lazarczyk A, Halubiec P, Szafranski O, Karnas K, Karewicz A. Superparamagnetic iron oxide nanoparticles-current and prospective medical applications. Materials (Basel). 2019;12(4):617. doi:10.3390/ma12040617
35. Mandal M, Kundu S, Ghosh SK, et al. Magnetite nanoparticles with tunable gold or silver shell. J Colloid Interface Sci. 2005;286(1):187–194. doi:10.1016/j.jcis.2005.01.013
36. Meldrum FC, Heywood BR, Mann S. Magnetoferritin: in vitro synthesis of a novel magnetic protein. Science. 1992;257(5069):522–523. doi:10.1126/science.1636086
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.