Back to Journals » OncoTargets and Therapy » Volume 9
Screening miRNAs for early diagnosis of colorectal cancer by small RNA deep sequencing and evaluation in a Chinese patient population
Authors wang X, Chen L, Jin H, Wang S, Zhang Y, tang X, Tang G
Received 12 November 2015
Accepted for publication 3 February 2016
Published 7 March 2016 Volume 2016:9 Pages 1159—1166
DOI https://doi.org/10.2147/OTT.S100427
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr William C. Cho
Xiaofeng Wang,1,2,* Li Chen,3,* Heiying Jin,1,4 Shuiming Wang,1 Yajie Zhang,1 Xiaoqing Tang,3 Guiliang Tang3
1National Center of Colorectal Surgery, The Third Affiliated Hospital of Nanjing University of Chinese Medicine, 2Graduate School of Nanjing University of Traditional Chinese Medicine, Nanjing, People’s Republic of China; 3Department of Biological Sciences, Michigan Technological University, Houghton, MI, USA; 4Department of General Surgery, Peking University International Hospital, Beijing, People’s Republic of China
* These authors contributed equally to this work
Purpose: This study aims to screen microRNAs (miRNAs), for an early diagnosis of colorectal cancer, by deep sequencing and evaluation of total miRNAs using clinical samples from a Chinese patient population.
Methods: Total small RNAs from normal colonic mucosa, colonic adenomas, and colorectal cancer tissues were prepared for miRNA analysis by deep sequencing. The sequencing data were then analyzed by bioinformatics for candidate diagnostic miRNAs, which were further validated for their up- or downregulation status.
Results: Comparison of cancer tissues with normal mucosa identified 99 upregulated and 90 downregulated miRNAs. Comparison of adenomas and normal mucosa found 114 upregulated and 107 downregulated miRNAs. Comparison of cancer and adenoma tissues found 70 upregulated and 27 downregulated miRNAs. Selected up- and downregulated miRNAs were validated for their expressions in 12 cases of patients with cancer and polyps. Specifically, for the upregulated miRNAs, miR-18a-5p and miR-21-3p were significantly upregulated in adenomas and cancer tissues, compared with the normal mucosa; miR-135b-5p, miR-17-5p, miR-182-5p, miR-200a-5p, and miR-200c-3p were significantly upregulated in cancer tissues compared to the normal mucosa, but their differential expression was not significant in adenoma tissues when compared with the normal mucosa. miR-183-5p and miR-96-5p were significantly upregulated in adenoma tissues when compared with normal mucosa, but these differences were not significant in cancer tissues when compared to normal mucosa. For the downregulated miRNAs, miR-133a-3p was significantly downregulated in both adenoma and cancer tissues when compared to normal mucosa; miR-204-5p, miR-125b-5p, miR-139-5p, miR-100-5p, and miR-30a-5p were significantly downregulated in cancer tissues compared to the normal mucosa, but their differential expression was not significant in adenoma tissue compared to normal mucosa.
Conclusion: The findings of this study show that a number of miRNAs might be important in the diagnosis and prognosis of colorectal cancer in Chinese patients using the method of small RNA deep sequencing. Upregulation of miR-18a-5p and miR-21-3p or downregulation of miR-133a-3p in adenoma and cancer tissues may serve as an index for early screening of colorectal cancer. Other miRNAs, such as miR-135b-5p, miR-17-5p, miR-182-5p, miR-200a-5p, miR-200c-3p, miR-183-5p, and miR-96-5p, which were either up- or downregulated, in cancer tissues, but not in adenoma tissues, have limited significance in early diagnosis. Further study is needed to determine a screening index with diagnostic value.
Keywords: colorectal cancer, colorectal adenoma, miRNA, deep sequencing
Introduction
Colorectal cancer accounts for one third of cancer-related deaths worldwide and this trend is still increasing. Although application of comprehensive treatments, including surgery, has achieved a significant improvement in the 5-year survival rate, the overall estimated death rate is still ~50%−60%.1 Early diagnosis is thus an effective way to prolong life of patients with colorectal cancer.
The term micro-RNA (miRNA) refers to a type of small RNA of length 19–22 nucleotides in vivo. After a series of reactions, it finally forms an RNA-induced silencing complex. The miRNA-associated RNA-induced silencing complex binds to its target messenger RNA (mRNA) at the 3′-untranslated region, so that it can cleave and degrade the target mRNA or inhibit further translation of the target mRNA, leading to a loss of function of the target gene.
miRNA plays an important role in the occurrence and development of colorectal cancer. miRNA affects many known oncogenes and antioncogenes and is involved in the regulation of the signal transduction pathway related to colorectal cancer. Thus, miRNAs, many of which have differential expression profiles, may be used to serve as biological markers for predicting the development of and prognosis of colorectal cancer.2,3
In recent years, changes of miRNA levels in the human circulation have gained more attention for their role in the early diagnosis of colorectal cancer.3–5 Luo et al3 used TaqMan miRNA slugs in 50 cases of colorectal cancer and 50 cases of adenoma and found nine miRNAs that had differential expression in colorectal cancer when compared to normal mucosa. They hypothesized that assay of these nine markers can contribute to the early diagnosis of colorectal cancer. Ng et al4 assayed 90 known miRNAs in 90 cases of colorectal cancer, 20 cases of gastric cancer, 20 cases of inflammatory bowel disease, and 50 healthy people. Their findings showed that miR-17-3p and miR-92 were increased in patient blood, and further verification showed that, among the 180 cases, miR-92 had a distinct increase in patient blood. In contrast to the report by Ng et al,4 Faltejskova et al,6 by repeating the research performed by Ng et al4 among Czech patients, found that the levels of miR-17-3p (P=0.18), miR-29a (P=0.14), and miR-92a (P=0.60) had no significant difference in colorectal cancer when compared to a control group. The differences in findings might have been due to the distinct sources of samples, including the regional environment distinction of patient samples.
The colorectal cancer screen using miRNAs was claimed to have sensitivity of 89% and specificity of 70%. Research reported by Huang et al5 found that using miR-29a and miR-92a as biomarkers for early diagnosis had 83% sensitivity and 84.7% specificity for colorectal cancer diagnosis and 77% sensitivity and 79.7% of specificity for colorectal polyps.
Although previous studies showed valuable data on miRNAs as potential biomarkers in colorectal cancer, results were still limited and were sometimes contradictory with each other due to different sources of samples.3–5 Cancer-stage-related miRNAs as valuable biomarkers are still lacking. In this study, controlled experiments were performed on colorectal cancer with its distinct developmental stages, including colorectal polyps, and on normal mucosa. miRNA deep sequencing was performed on cancer and precancer tissues of all stages, to determine simultaneous variations in the miRNA index for colorectal cancer and colorectal polyps. This index can be used in further studies to confirm its value in the early diagnosis of colorectal cancer.
Materials and methods
Clinical specimens
The specimens used for the deep sequencing were from four typical patients who had undergone surgery or endoscopy at the National Center of Colorectal Surgery, the Third Affiliated Hospital of Nanjing University of Traditional Chinese Medicine. One case was a 52-year-old female, a patient with colorectal cancer (T2N0M0) who had anterior resection operation; the cancer tissue and normal mucosa were taken for study. Another case was a 50-year-old male with sigmoid cancer with a colon polyp (T3N0M0), who had received a sigmoidectomy operation for sigmoid cancer and endoscopic polypectomy for colon adenoma; the cancer, adenoma, and normal mucosa tissues were taken for study. A third case was a patient with polyps in the cecum and cancer of the horizontal colon (T3N1M0); a right hemicolectomy surgery was performed; his cancer, adenoma, and normal mucosa tissues were obtained for study. A fourth case had a polyp of the sigmoid colon; endoscopic mucosal resection was performed and specimens were taken for study. In total, there were three samples each of cancer, adenoma, and normal mucosa tissues. All samples were preserved in liquid nitrogen. The study was approved by the Ethics Committee of The Third Affiliated Hospital of Nanjing, University of Traditional Chinese Medicine, and all patients provided written informed consent.
miRNA deep sequencing
We implemented miRNA deep sequencing with the method published by Schee et al7 and carried out high-throughput sequencing on the specimens using the 50 bp single-end sequencing model on an Illumina HiSeq2500 (Illumina, San Diego, CA, USA) sequencing platform. Frozen specimens were used with the Sangong kit (Sangong Biotechnology, Shanghai, People’s Republic of China) for total RNA extraction. The total RNA was quantified with NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), and 500 ng of total RNA was visualized on 1% agarose gel after electrophoresis. A miRNA library was established with the TruSeq® miRNA Sample Prep Kit v2 (Illumina Company), with reverse transcriptase polymerase chain reaction (RT-PCR) to enrich the library and with Qubit kit (Thermo Fisher Scientific) to quantify miRNA libraries. Sequencing was performed with the HiSeq2500. The data were transformed into the FASTQ format for expression and differential profile analysis. The results were subjected to cluster analysis with Illumina’s software packages (SCS2.9/RTA1.9 and Off-line Basecaller v1.9). miRNAs that were differentially expressed, either increased or decreased, in colorectal cancer and colorectal adenoma samples were selected for the analysis.
Quantitative RT-PCR test and verification of miRNA deep-sequencing results
Twelve colorectal cancer tissues as well as normal mucosa (>10 cm away from the cancer) and 12 samples of colonic adenoma were used for quantitative RT-PCR (qRT-PCR) validation of the differentially expressed miRNAs revealed by the deep sequencing. The RNA samples were derived from patients with colorectal cancer; seven were males and five were females, with ages ranging from 35 years to 75 years and a median age of 65 years. These included six cases of colorectal cancer, four cases of sigmoid cancer, and two cases of ascending colon cancer. Additional validations were conducted by using RNA samples from 12 patients with colorectal adenomas, eight males and four females with ages ranging from 40 years to 73 years and a median age of 62 years. These included seven cases of rectal polyps, four cases of sigmoid colon polyps, and one case of ascending colon polyps.
We designed reverse transcription stem–loop primer and specificity amplification primers, and the RNU6 qRT-PCR was used as an internal reference for miRNA quantification. The average Ct value of three replicates was calculated for each sample. The Ct values of the internal reference were also obtained three times and used for sample value corrections. The corrected Ct values were directly plotted for differential expression analysis. Please note that the samples were not paired in experiments, and therefore their Ct values were unfit for the calculation of 2−ΔΔCt (cannot appropriately subtract the Ct value of one sample as a control group). Therefore, direct plotting of the Ct value, which was subtracted from the expression of the internal reference of normal, adenoma, and cancer tissues, was used for their expressions. One-way analysis of variance in a normal distribution and the Mann–Whitney U-test equivalent, an inspection method for nonnormal distribution or variance, were used to test all groups studied.
Results
Differential expression of miRNAs in different developmental stages of colorectal cancerous tissues
To analyze the differential expression of miRNAs in different stages of colorectal cancerous tissues, miRNAs with two-fold changes were compared for their expression patterns among adenoma, cancer, and normal mucosa (P≤0.05, expression quantity with two-fold changes). We conducted the following comparisons (Figure 1 and Table 1): 1) between cancerous tissue and normal mucosa tissue; 2) between adenoma and normal mucosa; and 3) between cancer and adenoma. Our data analysis showed that 99 miRNAs were upregulated and 90 miRNAs were downregulated in cancers relative to normal mucosa. Furthermore, 114 miRNAs were found to be upregulated and 107 downregulated in adenoma compared with normal mucosa. Seventy miRNAs were upregulated and 27 downregulated in cancers relative to adenoma miRNAs. Among all the upregulated miRNAs, 32 miRNAs were simultaneously upregulated in both cancer and adenoma, and nine had significantly differential expression between adenoma and cancer. Among all the downregulated miRNAs, 27 miRNAs were simultaneously decreased in both adenoma and cancer, and only two had significant differential expression between adenoma and cancer. Nine miRNAs, increased or decreased in both colorectal cancer and colorectal adenomas, were chosen for further validation.
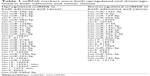  | Table 1 miRNA markers were both upregulated and downregulated in both adenoma and cancer tissues |
Validation of the differentially expressed miRNAs in colorectal cancer tissues of patients
To verify the reliability of the deep-sequencing data, we picked 12 miRNAs that were found to be differentially expressed by deep sequencing. The validation was conducted using more samples of a Chinese patient population, and the data are shown in Figures 2 and 3.
For the upregulated miRNAs (Figure 2), miR-18a-5p and miR-21-3p were statistically significantly increased in both adenoma and cancer tissues, compared to the normal mucosa. miR-135b-5p, miR-17-5p, miR-182-5p, miR-200a-5p, and miR-200c-3p were significantly upregulated in cancer compared with the normal mucosa tissues but not significantly in adenoma relative to normal mucosa tissues. miR-183-5p and miR-96-5p were significantly upregulated in adenoma compared with normal mucosa tissues but showed no differential expression between cancer and normal mucosa.
For the downregulated miRNA (Figure 3), only miR-133a-3p was significantly downregulated in both adenoma and cancer tissues when compared with normal mucosa. miR-204-5p, miR-125b-5p, miR-139-5p, miR-100-5p, and miR-30a-5p were significantly downregulated in cancer relative to normal mucosa but not significantly in adenoma compared with normal mucosa tissues. Among these downregulated miRNAs, no miRNA showed any differential expression either between adenoma and cancer tissues or between cancer and normal mucosa tissues. The statistical results are shown in Table 2.
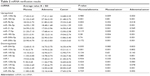  | Table 2 miRNA verification results |
Discussion
The use of molecular biomarkers for the early diagnosis of colorectal cancer is an important topic of current research.2–4 Recent studies have found that miRNA plays an important role in the pathogenesis and development of colorectal cancer.2–4 The use of the characteristics of miRNAs, which can be detected at early stages in the blood circulation system, for early diagnosis of cancers has achieved very promising results.4–7 However, due to miRNA expression variations among different races, environments, diet styles, and so on, the results of the detected miRNA expressions may be different. In this study, small RNA deep-sequencing experiments were specifically conducted on samples of patients with colorectal cancer from Southeast China for studying the differential expression patterns of miRNAs among samples of different cancer developmental stages. The objective was to determine a standard and valuable miRNA diagnostic index for the early detection of colorectal cancers in patients of Southeast China.
In this study, using deep sequencing, we detected two major types of miRNAs that are differentially expressed in different colorectal samples: 99 upregulated miRNAs and 90 downregulated miRNAs in colorectal cancer, as well as 70 upregulated and 27 downregulated miRNAs in adenoma tissues. The results were largely similar to previous reports2–7 and confirmed the reliability of our experiments used for the analysis of distinct samples of Southeast China. Our data were also cross-validated by comparison between the deep-sequencing results and the qRT-PCR validation results. We further identified that miR-18a-5p and miR-21-3p were upregulated in both adenoma and cancer tissues, while miR-133a-3p was downregulated in both adenoma and cancer tissues, which offers potential early miRNA indicators for colorectal cancer screening.
Research reported by Slattery et al8 found that overexpression of hsa-miR-18a-5p can reduce the risk of death from colorectal cancer. A systematic review reported by Clancy et al9 found that patients with colorectal cancers show upregulation of circulating hsa-miR-18a-5p, which may imply that hsa-miR-18a-5p can be used as a diagnostic marker. Calvano Filho et al10 also found that the expression of hsa-miR-18a-5p increases in breast cancer and this suggested a correlation may exist for other types of cancers.
Similarly, upregulation of miR-21-3p was found not only for colorectal cancer but also for a variety of cancers. Rotelli et al11 found that miR-21-3p was increased in the stools of patients with colorectal cancer and that it decreased after surgery, which suggests that it can be used as a prognostic factor for colorectal cancer. Other studies have found that miR-21-3p is associated with gynecological tumors and drug resistance.12 A few studies investigating whether it increases in colorectal cancer and colorectal adenoma have been conducted.12,13 Research reported by Doberstein et al14 found that miR-21-3p increased with tumorigenesis in a variety of tumors. In contrast, few studies have been conducted on the role of miR-133a-3p in the pathogenesis and development of colorectal cancer.15
In this research, differences in expression of miR-135b-5p, miR-17-5p, miR-182-5p, miR-200a-5p, and miR-200c-3p were statistically significant in comparisons between cancer and normal mucosa tissues, while no significance was observed for adenoma and normal mucosa tissues. The differences in miR-183-5p and miR-96-5p expression were statistically significant for adenoma and normal mucosa tissues and showed no significant differences in cancer and normal mucosa tissues.
In our miRNA deep-sequencing study, the expression patterns of miRNAs were similar in both adenoma and cancer tissues. This result might be due to several reasons. One explanation may be the small number of patients in this study, which requires further investigations either using larger sample numbers for the deep sequencing or more validations using larger patient samples. Our next step will be the verification of its value in more colorectal cancer samples. According to the literature, variations in miRNA detection were also associated with varying sources of patients with cancer. There may exist miRNA expression differences between different human races or humans with different environmental or diet habits. One of our research objectives is to identify such potential distinctions of miRNA expressions in different Chinese patient populations for a more precise medication. This study aimed to determine the links between miRNA biomarker variation and the occurrence of colorectal cancer in patients of Southeast China.16–22
In conclusion, this research found a number of important miRNAs that may have diagnostic and prognostic significance in the occurrence and development of colorectal cancer. miR-18a-5p, miR-21-3p, and miR-133a-3p were found to be up- or downregulated in both adenoma and cancer tissues and can be used as early indicators of colorectal cancer during screening, while the increase or decrease of miR-135b-5p, miR-17-5p, miR-182-5p, miR-200a-5p, miR-200c-3p, miR-183-5p, and miR-96-5p in both cancer and adenoma tissues showed no obvious differences. Further research is necessary for determining the appropriate screen index values that can be used for the diagnosis of colorectal cancer.
Acknowledgments
This study was funded by the National Science Foundation of China (numbers 30572447, 30973837, 81273944), the Jiangsu National Science Foundation (BK20151081), the Nanjing Science Fundation (201402041), and the Nanjing Medical Science Foundation (YKK14140).
Disclosure
The authors report no conflicts of interest in this work.
References
Kumar R, Price TJ, Beeke C, et al. Colorectal cancer survival: an analysis of patients with metastatic disease synchronous and metachronous with the primary tumor. Clin Colorectal Cancer. 2014;13(2):87–93. | ||
Slaby O, Svoboda M, Michalek J, Vyzula R. MicroRNAs in colorectal cancer: translation of molecular biology into clinical application. Mol Cancer. 2009;8:102. | ||
Luo X, Burwinkel B, Tao S, Brenner H. MicroRNA signatures: novel biomarker for colorectal cancer? Cancer Epidemiol Biomarkers Prev. 2011;20(7):1272–1286. | ||
Ng EK, Chong WW, Jin H, et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009;58(10):1375–1381. | ||
Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127(1):118–126. | ||
Faltejskova P, Bocanek O, Sachlova M, et al. Circulating miR-17-3p, miR-29a, miR-92a and miR-135b in serum: evidence against their usage as biomarkers in colorectal cancer. Cancer Biomark. 2012;12(4):199–204. | ||
Schee K, Lorenz S, Worren MM, et al. Deep sequencing the microRNA transcriptome in colorectal cancer. PLoS One. 2013;8(6):e66165. | ||
Slattery ML, Herrick JS, Mullany LE, et al. An evaluation and replication of miRNAs with disease stage and colorectal cancer-specific mortality. Int J Cancer. 2015;137(2):428–438. | ||
Clancy C, Joyce MR, Kerin MJ. The use of circulating microRNAs as diagnostic biomarkers in colorectal cancer. Cancer Biomark. 2015;15(2):103–113. | ||
Calvano Filho CM, Calvano-Mendes DC, Carvalho KC, et al. Triple-negative and luminal A breast tumors: differential expression of miR-18a-5p, miR-17-5p, and miR-20a-5p. Tumour Biol. 2014;35(8):7733–7741. | ||
Rotelli MT, Di Lena M, Cavallini A, et al. Fecal microRNA profile in patients with colorectal carcinoma before and after curative surgery. Int J Colorectal Dis. 2015;30(7):891–898. | ||
Karakaya C, Guzeloglu-Kayisli O, Uyar A, et al. Poor ovarian response in women undergoing in vitro fertilization is associated with altered microRNA expression in cumulus cells. Fertil Steril. 2015;103(6):1469–1476. e1461–e1463. | ||
Pink RC, Samuel P, Massa D, Caley DP, Brooks SA, Carter DR. The passenger strand, miR-21-3p, plays a role in mediating cisplatin resistance in ovarian cancer cells. Gynecol Oncol. 2015;137(1):143–151. | ||
Doberstein K, Bretz NP, Schirmer U, et al. miR-21-3p is a positive regulator of L1CAM in several human carcinomas. Cancer Lett. 2014;354(2):455–466. | ||
Chung SH, Gillies M, Sugiyama Y, Zhu L, Lee SR, Shen W. Profiling of microRNAs involved in retinal degeneration caused by selective Muller cell ablation. PLoS One. 2015;10(3):e0118949. | ||
Bertoli G, Cava C, Castiglioni I. MicroRNAs: new biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics. 2015;5(10):1122–1143. | ||
Chang CW, Wu HC, Terry MB, Santella RM. microRNA expression in prospectively collected blood as a potential biomarker of breast cancer risk in the BCFR. Anticancer Res. 2015;35(7):3969–3977. | ||
Della Vittoria Scarpati G, Calura E, Di Marino M, et al. Analysis of differential miRNA expression in primary tumor and stroma of colorectal cancer patients. Biomed Res Int. 2014;2014:840921. | ||
Kara M, Yumrutas O, Ozcan O, et al. Differential expressions of cancer-associated genes and their regulatory miRNAs in colorectal carcinoma. Gene. 2015;567(1):81–86. | ||
Krishnan K, Steptoe AL, Martin HC, et al. MicroRNA-182-5p targets a network of genes involved in DNA repair. RNA. 2013;19(2):230–242. | ||
Tang JF, Yu ZH, Liu T, et al. Five miRNAs as novel diagnostic biomarker candidates for primary nasopharyngeal carcinoma. Asian Pac J Cancer Prev. 2014;15(18):7575–7581. | ||
Yan X, Ding L, Li Y, et al. Identification and profiling of microRNAs from skeletal muscle of the common carp. PLoS One. 2012;7(1):e30925. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.



