Back to Journals » International Journal of General Medicine » Volume 15
Screening for Regulatory Network of miRNA–Inflammation, Oxidative Stress and Prognosis-Related mRNA in Acute Myocardial Infarction: An in silico and Validation Study
Authors Yin X, Wang X, Wang S, Xia Y, Chen H, Yin L, Hu K
Received 16 December 2021
Accepted for publication 24 January 2022
Published 18 February 2022 Volume 2022:15 Pages 1715—1731
DOI https://doi.org/10.2147/IJGM.S354359
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Xunli Yin,1 Xuebing Wang,1 Shiai Wang,1 Youwei Xia,2 Huihui Chen,1 Ling Yin,3 Keqing Hu4
1Department of Cardiovascular Medicine, The Seventh People’s Hospital of Jinan, Jinan, 250100, People’s Republic of China; 2Department of Critical Care Medicine, The Seventh People’s Hospital of Jinan, Jinan, 250100, People’s Republic of China; 3Department of Conduit Room, The Seventh People’s Hospital of Jinan, Jinan, 250100, People’s Republic of China; 4Cardiovascular Department, Central Hospital Affiliated to Shandong First Medical University, Jinan, 250013, People’s Republic of China
Correspondence: Keqing Hu, Central Hospital Affiliated to Shandong First Medical University, Cardiovascular Department,105#, Jiefang Road, Jinan 250013, Shandong, China, Tel +86 0531-85695114, Fax +86 0531-86942457 Email [email protected]
Background: Acute myocardial infarction (AMI), which commonly leads to heart failure, is among the leading causes of mortality worldwide. The aim of this study was to find potential regulatory network for miRNA-inflammation, oxidative stress and prognosis-related mRNA to uncover molecular mechanisms of AMI.
Methods: The expression profiles of miRNA and mRNA in the blood samples from AMI patients were downloaded from the Gene Expression Omnibus (GEO) dataset for differential expression analysis. Weighted gene co-expression network analysis (WGCNA) was used to further identify important mRNAs. The negatively regulatory network construction of miRNA–inflammation, oxidative stress and prognosis-related mRNAs was performed, followed by protein–protein interaction (PPI) and functional analysis of mRNAs.
Results: A total of three pairs of negatively regulatory network of miRNA–inflammation and prognosis-related mRNAs (hsa-miR-636/hsa-miR-491-3p/hsa-miR-188-5p/hsa-miR-188-3p-AQP9, hsa-miR-518a-3p-C5AR1 and hsa-miR-509-3-5p/hsa-miR-127-5p-PLAUR), two pairs of negatively regulatory network of miRNA–oxidative stress and prognosis-related mRNAs (hsa-miR-604-TLR4 and hsa-miR-139-5p-CXCL1) and three pairs of negatively regulatory network of miRNA-inflammation, oxidative stress and prognosis-related mRNA (hsa-miR-634/hsa-miR-591-TLR2, hsa-miR-938-NFKBIA and hsa-miR-520h/hsa-miR-450b-3p-ADM) were identified. In the KEGG analysis, some signaling pathways were identified, such as complement and coagulation cascades, pathogenic Escherichia coli infection, chemokine signaling pathway and cytokine–cytokine receptor interaction and Toll-like receptor signaling pathway.
Conclusion: Identified negatively regulatory network of miRNA-inflammation/oxidative stress and prognosis-related mRNA may be involved in the process of AMI. Those inflammation/oxidative stress and prognosis-related mRNAs may be diagnostic and prognostic biomarkers for AMI.
Keywords: acute myocardial infarction, miRNAs, mRNAs, prognosis, inflammation, oxidative stress
Introduction
Acute myocardial infarction (AMI), one of the major cardiovascular diseases that lead to a high morbidity and mortality, is commonly defined as a cardiomyocyte death due to the prolonged ischaemia.1 In addition, other risk factors including age, size and location of infarct, and haemodynamic status are associated with AMI.2 The improved survival rates have been observed in AMI patients.3,4 However, recurrent patients with AMI, a higher risk subgroup of those with AMI, have worse outcomes.5–7 Therefore, it is needed to identify potential prognostic indicators to prevent AMI patients from relapsing. Previous reports showed that the age, creatinine, ejection fraction score and SYNTAX Score II had a prognostic value for patients with ST-elevation myocardial infarction-related cardiogenic shock.8,9
MI is a primarily local event, which results in the activation of the acute systemic inflammatory response. Reactive oxygen species (ROS) play a crucial role in MI.10 In heart cells, the ROS causes mitochondrial dysfunction and result in serious complications, such as remodeling of the left ventricle and infarct expansion after MI.11 It is noted that oxidative stress also up-regulated synthesis of pro-inflammatory cytokine.12 Therefore, novel therapeutic strategy against AMI intends to control both the inflammatory response and the oxidative stress.
MiRNAs regulate mRNA expression at the post-transcriptional level.13,14 MiRNAs have emerged as a kind of promising tool associated with some pathophysiological processes, including cardiovascular diseases. In addition, weighted gene co-expression network analysis (WGCNA) is used to elucidate changes in transcriptome expression patterns in complex diseases.15–17 In contrast to differential expression analysis, WGCNA aims to identify high-order correlations between mRNA products.18 In the present study, we tried to find differentially expressed miRNAs and inflammation, oxidative stress and prognosis-related mRNAs through WGCNA to uncover the molecular regulatory mechanism of AMI.
Materials and Methods
Dataset Retrieval
The expression profiles of mRNA and miRNA were downloaded from Gene Expression Omnibus (GEO) datasets by searching keywords (“acute myocardial infarction” [All Fields]) AND “Homo sapiens”[porgn] AND “gse”[Filter]). Firstly, the following mRNA datasets were selected for differential expression analysis: 1) dataset must be genome-wide transcriptome data of mRNA; 2) data were obtained from blood of AMI group and normal control group; 3) both standardized and raw datasets were considered. Finally, two mRNA expression datasets (GSE34198 and GSE48060) were selected. Secondly, GSE66360 dataset was downloaded and used for WGCNA analysis. Thirdly, three datasets (GSE123342, GSE59867 and GSE62646), including follow-up data of AMI patients after cure and discharge, were downloaded and used to identify mRNAs associated with prognosis. The screening criteria of prognostic mRNAs were as follows: 1) the mRNA expression trend was consistent in the three data sets; 2) the mRNA expression after discharge was significantly different from that the onset of disease. Lastly, GSE31568 dataset was downloaded and used for identification of differentially expressed miRNAs. Detail information of above enrolled datasets is shown in Table 1.
 |
Table 1 Detail Information of Enrolled Datasets of AMI |
Screening of Differentially Expressed miRNAs and mRNAs
After scale standardization of the data set, MetaMA and Limma packages were used for differential expression analysis. The p value was combined by the inverse normal method. The false discovery rate (FDR) was calculated by Benjamini hochberg threshold. Differentially expressed miRNA and mRNA was identified under the threshold value of FDR < 0.05 and |log FC (fold change)| >1, and p value <0.05, respectively.
Construction of the WGCNA Co-Expression Network
Data in the GSE66360 dataset were preprocessed and annotated using the annotation file of the microarray platform. The average value of multiple probes corresponding to the same mRNA was taken. After pretreatment, the mRNA expression matrix was obtained for scale standardization. The WGCNA package in R was utilized for analysis of the top 25% mRNAs in the normalized mRNA expression matrix file to identify meaningful modules and mRNAs. Since gene co-expression analysis is extremely sensitive to the presence of abnormal samples, strict quality control procedures are needed to ensure the highest quality levels. The average linkage method in WGCNA was used to cluster the samples. To ensure the scale-free network, the scale-free fitting index and average connectivity were calculated (the β value was set to 7). A one-step method was adopted to build the network that covers modules. Interaction between modules was analyzed, followed by drawing of heat map. In addition, modules were clustered and the correlation between modules was calculated to explore the common expression similarity of all modules. It is worth mentioning that the common mRNAs in both key module in the WGCNA analysis and differential expression analysis were considered as hub mRNAs.
Construction of miRNA–Inflammation and Oxidative Stress-Related mRNA Network
More and more evidence indicated that inflammation plays an important role in the pathophysiology of MI. Oxidative stress also plays a role in ventricular remodeling after MI. In order to investigate the relationship between inflammatory and oxidative stress and AMI, molecular characteristic database (1) and GeneCards (https://www.genecards.org) database (2) was respectively used to identify mRNAs associated with inflammation and oxidative stress among all differentially expressed mRNAs. MiRWalk (http://mirwalk.umm.uni-heidelberg.de/interactions/) was used to construct the negatively regulatory network between miRNAs and mRNAs associated with inflammation and oxidative stress.
PPI Analysis and Functional Enrichment of Hub mRNAs
To investigate the biological function of hub mRNAs, string database (https://string-db.org/) was firstly used to construct the PPI network, which was visualized by Cytoscape 3.6.1. In addition, DAVID 6.8 dataset (https://david.ncifcrf.gov/) was applied for functional analysis, including Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG). P value <0.05 was considered as statistical significance.
In vitro Validation of Inflammation and Oxidative Stress-Related mRNAs
In vitro validation QRT-PCR was performed. The inclusion criteria of AMI patients was as follows: 1) time of chest pain or distress >30 min within 24 hours and levels of cardiac enzymes creatine phosphokinase-MB type (CK-MB) and cardiac troponin T (cTnT) were higher than normal range; 2) the patient had AMI for the first time; 3) the patient did not receive medication or surgery prior to admission; 4) patient had blood samples at three time points, including before hospitalization, at discharge, and 6 months after AMI; 5) patient had complete clinical data. The exclusion criteria of AMI patients were as follows: 1) patient had myocarditis and other diseases caused by chest pain or distress; 2) patient with a history of kidney failure, malignant tumors, advanced liver disease, and other inflammatory diseases (psoriasis and rheumatoid arthritis); 3) recurrent patient; 4) patients with incomplete clinical data; 5) patients with missing blood samples at three time points of before hospitalization, at discharge, and 6 months after AMI. According to the above criteria, nine AMI patients and nine normal controls were enrolled. Blood samples from these patients were collected. All participating individuals provided informed consent with the approval of the ethics committee of the Seventh People’s Hospital of Jinan. In addition, this study was conducted in accordance with the Declaration of Helsinki.
Total RNA of the blood sample was extracted and synthesized DNA by FastQuant cDNA first strand synthesis kit. Real-time PCR was performed in the SuperReal PreMix Plus (SYBR Green analysis method). Relative mRNAs expression was analyzed by log2 (fold change) method and represented as fold change. FC > 1 and FC < 1 represented up-regulation and down-regulation, respectively.
Results
Identification of Differentially Expressed miRNAs and mRNAs
A total of 250 differentially expressed (103 up-regulated and 147 down-regulated) miRNAs were identified in AMI. In addition, a total of 1981 differentially expressed (999 up-regulated and 982 down-regulated) mRNAs were identified in AMI. The heat map of all differentially expressed miRNAs and top 100 differentially expressed mRNAs is shown in Figure 1A and B, respectively.
 |
Figure 1 The heat map of all miRNAs (A) and top 100 mRNAs (B) in AMI. |
WGCNA Co-Expression Network Analysis
Based on the WGCNA co-expression network analysis, a total of nine modules were identified (Figure 2). Interaction analysis between nine modules showed that each module had high independence and relative independence of mRNA expression. The heat map of the first 400 mRNA in nine modules is shown in Figure 3A. These modules were also clustered and the correlation between modules was calculated to explore the common expression similarity of all modules (Figure 3B). Compared with other modules, green module (involving 357 mRNAs) was found to be highly correlated with disease status, suggesting that mRNAs in green module may play a key role in the occurrence and development of AMI (Figure 3C). The relationship between the number of module membership and AMI in the green module is shown in Figure 3D. It is noted that there were 116 common mRNAs (hub mRNAs) between all 1981 differentially expressed mRNAs and 357 mRNAs in the green module, including 104 up-regulated and 12 down-regulated mRNAs in AMI.
 |
Figure 2 Modules identified by the WGCNA analysis. MRNAs in modules are marked with different colors. |
Network of miRNA-Inflammation and Oxidative Stress Related mRNA
A total of 200 mRNAs were associated with inflammatory response in the molecular characteristic database. Among which, 16 common mRNAs were identified between 200 inflammation-related mRNAs and 116 hub mRNAs, such as NFKB inhibitor alpha (NFKB1A, up-regulation), adrenomedullin (ADM, up-regulation) and Toll-like receptor 2 (TLR2, up-regulation). MiRWalk analysis showed that a total of 11,427 miRNAs targeted the above 16 common mRNAs. Among which, 372 miRNAs were differentially expressed miRNAs in AMI. By analyzing the negative regulation of 372 miRNA and target mRNAs, 214 pairs of negatively regulatory network of miRNA-inflammation related mRNA were obtained, including 3 pairs of up-regulated miRNA-down-regulated mRNA and 211 pairs of down-regulated miRNA-up-regulated mRNA (Figure 4), such as down-regulated hsa-miR-636/hsa-miR-491-3p/hsa-miR-188-5p/hsa-miR-188-3p-up-regulated aquaporin 9 (AQP9), down-regulated hsa-miR-518a-3p-up-regulated complement C5a receptor 1 (C5AR1) and down-regulated hsa-miR-509-3-5p/hsa-miR-127-5p-up-regulated plasminogen activator, urokinase receptor (PLAUR). In addition, a total of 835 mRNAs were associated with oxidative stress in the GeneCards database. Among which, 16 common mRNAs were identified between 835 oxidative stress-related mRNAs and 116 hub mRNAs, such as NFKB1A, ADM and TLR2. MiRWalk analysis showed that a total of 9576 miRNAs targeted the above 16 common mRNAs. Among which, 284 miRNAs were differentially expressed miRNAs in AMI. By analyzing the negative regulation of 284 miRNA and target mRNAs, 155 pairs of negatively regulatory network of miRNA-oxidative stress-related mRNA were obtained, including 2 pairs of up-regulated miRNA-down-regulated mRNA and 153 pairs of down-regulated miRNA-up-regulated mRNA (Figure 5), such as down-regulated hsa-miR-604-up-regulated Toll-like receptor 4 (TLR4) and down-regulated hsa-miR-139-5p-up-regulated C-X-C motif chemokine ligand 1 (CXCL1). It is worth mentioning that three pairs of negatively regulatory network of miRNA-inflammation and oxidative stress-related mRNA were obtained, including down-regulated hsa-miR-634/hsa-miR-591-up-regulated TLR2, down-regulated hsa-miR-938-up-regulated NFKBIA and down-regulated hsa-miR-520h/hsa-miR-450b-3p-up-regulated ADM.
 |
Figure 4 A total of 214 pairs of negatively regulatory network of miRNA-inflammation-related mRNA. Triangle and circle represents miRNA and mRNA, respectively. |
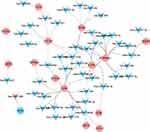 |
Figure 5 A total of 155 pairs of negatively regulatory network of miRNA-oxidative stress-related mRNA. Triangle and circle represents miRNA and mRNA, respectively. |
PPI Analysis of Hub mRNAs
In order to investigate the interactions between 116 hub mRNAs, string database was used for construction of the PPI network. In the PPI network, there were 82 mRNAs (Figure 6). Some mRNAs with a high degree were identified, including TLR2, TLR4 and CXCL1. Top 15 mRNAs with a high degree are listed in Table 2. After further screening of the above 82 mRNAs in datasets of GSE123342, GSE59867 and GSE62646, a total of 40 mRNAs were associated with prognosis of AMI, such as inflammation/oxidative stress-related mRNAs of AQP9, TLR2, TLR4, CXCL1, C5AR1, PLAUR, NFKBIA and ADM (Table 3). The expression box diagram of AQP9, TLR2, TLR4, CXCL1, C5AR1, PLAUR, NFKBIA and ADM in the GSE59867, GSE62646 and GSE123342 datasets is shown in Figure 7. It can be seen that these mRNAs were up-regulated in AMI, while down-regulated 4–6 days, 30 days, 1 month, 6 months and 1 year after AMI. It is indicated that these mRNAs could not only be used as biomarkers to detect AMI incidence but also as prognostic indicators of AMI patients.
 |
Table 2 Top 15 mRNAs with High Degree in the PPI Network in AMI |
 |
Table 3 Forty mRNAs Associated with Prognosis of AMI |
 |
Figure 6 PPI network of 116 hub mRNAs. Red and green represents up-regulation and down-regulation, respectively. The mRNA with black borders represents top 10 up-regulation/down-regulation mRNA. |
Functional Enrichment of Hub mRNAs
To further study the biological function of 116 hub mRNAs, DAVID 6.8 dataset was used for functional analysis. According to the KEGG analysis, these 116 hub mRNAs were involved in only 7 signaling pathways (Figure 8, Table 4). Among which, inflammation-related C5AR1 and PLAUR were involved in complement and coagulation cascades; oxidative stress-related TLR4 and CXCL1 was respectively involved in pathogenic Escherichia coli infection, and chemokine signaling pathway and cytokine–cytokine receptor interaction; inflammation and oxidative stress-related TLR2 and NFKBIA were involved in Toll-like receptor signaling pathway.
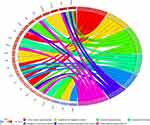 |
Figure 8 KEGG analysis of 116 hub mRNAs in AMI. |
In vitro Validation of Inflammation, Oxidative Stress and Prognosis-Related mRNAs
To validate the expression of inflammation, oxidative stress and prognosis-related mRNAs (AQP9, TLR2, TLR4, CXCL1, C5AR1 and PLAUR), blood samples from nine AMI patients and nine normal controls were collected for QRT-PCR (Figure 9). The clinical information of these individuals is listed in Table 5. The QRT-PCR result showed that AQP9, TLR2 and TLR4 were significantly up-regulated in AMI. The expression trend of CXCL1, C5AR1 and PLAUR was up-regulated without statistical significance. Larger numbers of samples are further needed.
 |
Table 4 Enriched Seven Signaling Pathways of 116 Hub mRNAs in AMI |
 |
Table 5 Clinical Information of Enrolled Individuals in QRT-PCR |
 |
Figure 9 In vitro validation of inflammation and oxidative stress-related AQP9, TLR2, TLR4, CXCL1, C5AR1 and PLAUR. *p value < 0.05; **p value < 0.01. |
Discussion
Hsa-miR-636 is down-regulated and has a potential diagnostic value in the plasma of AMI patients.19 Hsa-miR-491-3p is involved in regulation of cardiac regeneration after AMI.20,21 Hsa-miR-188-5p is down-regulated in hyperhomocysteinemia cardiomyocytes.22,23 The expression levels of hsa-miR-188-3p are decreased upon MI.24 Hsa-miR-518a-3p is down-regulated in the development of coronary artery disease.25 Hsa-miR-509-3-5p is involved in human atrial aging.26 The expression levels of hsa-miR-127-5p are associated with cardiac score and cardiac function in AMI patients.27 In addition, increased expression levels of AQP9 are related to atherosclerotic lesions.28 AQP9 is up-regulated in AMI.29 AQP9 is associated with recovery from MI.30 Huang et al found that silencing of the AQP9 gene improved cardiac function following MI.31 C5AR1 is involved in complement and coagulation cascades in AMI.32 PLAUR, a conserved gene, is up-regulated in AMI.32,33 In this study, we found regulatory networks of miRNA–inflammation and prognosis-related mRNAs (hsa-miR-636/hsa-miR-491-3p/hsa-miR-188-5p/hsa-miR-188-3p-AQP9, hsa-miR-518a-3p-C5AR1 and hsa-miR-509-3-5p/hsa-miR-127-5p-PLAUR) in AMI, which suggested that these molecules may play an important role in myocardial function of AMI. In addition, inflammation-related AQP9, C5AR1 and PLAUR can be considered as potential diagnostic and prognostic biomarkers for AMI.
Hsa-miR-604 is over expressed in the serum of paracoccidioidomycosis patients.34 Hsa-miR-139-5p is affected by ischemic pre- or post-conditioning in the heart.35 Yuan et al found that hsa-miR-139-5p was a potential biomarker of atherosclerosis.36 The expression levels of hsa-miR-139-5p are decreased earlier, within just 7 days following MI.37 In AMI, TLR4 mediates the synthesis of cytokines in circulating monocytes and is associated with the bad disease outcome.38–40 Plasma levels of CXCL1 are significantly increased in patients with AMI.41 In the present study, we found negatively regulatory networks of miRNA–oxidative stress and prognosis-related mRNAs (hsa-miR-604-TLR4 and hsa-miR-139-5p-CXCL1) in AMI. This indicated that these miRNAs and targets play a crucial role in angiogenesis of AMI. Oxidative stress-related TLR4 and CXCL1 may be regarded as potential prognostic biomarkers for AMI.
Hsa-miR-634 is up-regulated in atrial fibrillation and coronary artery disease.13,42 In patients with idiopathic dilated cardiomyopathy, hsa-miR-591 is used to predict time-dependent heart remodeling.43 The expression of hsa-miR-520h is found in human heart disease.44 Hsa-miR-450b-3p is regulated by ischemia.45 TLR2 has been reported as a critical regulator in AMI and identified as a potential biomarker for AMI detection.46–48 Blocking NFKBIA-mediated nuclear factor of kappa B (NF-κB) signaling pathway protects against MI.49 In patients with AMI, high ADM levels are related to impaired left ventricular function and death.50–52 Herein, we found that inflammation and oxidative related TLR2, NFKBIA and ADM could be involved in the pathological process of AMI under the regulation of miRNAs.
The above inflammation/oxidative stress and prognosis-related mRNAs were not only involved in miRNA regulatory network but also some signaling pathways. For example, inflammation-related C5AR1 and PLAUR were involved in complement and coagulation cascades; oxidative stress-related TLR4 and CXCL1 was respectively involved in pathogenic Escherichia coli infection, and chemokine signaling pathway and cytokine–cytokine receptor interaction; inflammation and oxidative stress-related TLR2 and NFKBIA were involved in Toll-like receptor signaling pathway. In blood circulation, the complement and coagulation cascades, evolutionarily related enzymatic cascades, are involved in post-MI responses.53 Gram-negative Escherichia coli has an activating or aggregating effect on platelets.54,55 In the infarcted myocardium, production of chemokines provides exit cues for circulating leukocytes expressing cognate receptors.56–58 Cytokine-cytokine receptor interaction is involved in stable coronary artery disease.59 In the infarcted heart, Toll-like receptor signaling pathway prompts NF-κB triggering and the up-regulation of cytokine.60
Conclusion
Our study found several negatively regulatory networks of miRNA–inflammation/oxidative stress and prognosis-related mRNAs, including hsa-miR-636/hsa-miR-491-3p/hsa-miR-188-5p/hsa-miR-188-3p-AQP9, hsa-miR-518a-3p-C5AR1, hsa-miR-509-3-5p/hsa-miR-127-5p-PLAUR, hsa-miR-604-TLR4, hsa-miR-139-5p-CXCL1, hsa-miR-634/hsa-miR-591-TLR2, hsa-miR-938-NFKBIA and hsa-miR-520h/hsa-miR-450b-3p-ADM) in AMI. In addition, some signaling pathways were also identified. Our study may provide a new field in understanding pathological mechanisms of AMI of the inflammation and oxidative stress levels. Nonetheless, our study has some limitations. In vitro validation of miRNA-mRNA regulatory network is needed. In addition, in vivo animal model is further needed to investigate the potential biological function of identified miRNAs and mRNAs.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Montecucco F, Carbone F, Schindler TH. Pathophysiology of ST-segment elevation myocardial infarction: novel mechanisms and treatments. Eur Heart J. 2016;37(16):1268–1283. doi:10.1093/eurheartj/ehv592
2. Margaritis M, Sanna F, Lazaros G, et al. Predictive value of telomere length on outcome following acute myocardial infarction: evidence for contrasting effects of vascular vs. blood oxidative stress. Eur Heart J. 2017;38(41):3094–3104. doi:10.1093/eurheartj/ehx177
3. Yeh RW, Sidney S, Chandra M, Sorel M, Selby JV, Go AS. Population trends in the incidence and outcomes of acute myocardial infarction. N Engl J Med. 2010;362(23):2155–2165. doi:10.1056/NEJMoa0908610
4. Roe MT, Parsons LS, Pollack CV
5. Motivala AA, Tamhane U, Ramanath VS, et al. A prior myocardial infarction: how does it affect management and outcomes in recurrent acute coronary syndromes? Clin Cardiol. 2008;31(12):590–596. doi:10.1002/clc.20356
6. Shiraishi J, Kohno Y, Sawada T, et al. Predictors of in-hospital outcome after primary percutaneous coronary intervention for recurrent myocardial infarction. Circ J. 2008;72(8):1225–1229. doi:10.1253/circj.72.1225
7. Shotan A, Blondheim DS, Gottlieb S, et al. Comparison of outcome of recurrent versus first ST-segment elevation myocardial infarction (from national Israel surveys 1998 to 2006). Am J Cardiol. 2011;107(12):1730–1737. doi:10.1016/j.amjcard.2011.02.332
8. Hayıroğlu M, Keskin M, Uzun AO, et al. Predictive value of SYNTAX score II for clinical outcomes in cardiogenic shock underwent primary percutaneous coronary intervention; a pilot study. Int J Cardiovasc Imaging. 2018;34(3):329–336. doi:10.1007/s10554-017-1241-9
9. Çinar T, Hayiroğlu M, Şeker M, et al. The predictive value of age, creatinine, ejection fraction score for in-hospital mortality in patients with cardiogenic shock. Coron Artery Dis. 2019;30(8):569–574. doi:10.1097/MCA.0000000000000776
10. Loeper J, Goy J, Rozensztajn L, Bedu O, Moisson P. Lipid peroxidation and protective enzymes during myocardial infarction. Clin Chim Acta. 1991;196(2–3):119–125. doi:10.1016/0009-8981(91)90064-J
11. Ohsawa M, Tsuru R, Hojo Y, et al. [Relationship between redox state of whole arterial blood glutathione and left ventricular function after acute myocardial infarction]. J Cardiol. 2004;44(4):141–146. Japanese
12. Abdelzaher WY, Ahmed SM, Welson NN, Alsharif KF, Batiha GE, Labib DAA. Dapsone ameliorates isoproterenol-induced myocardial infarction via Nrf2/ HO-1; TLR4/ TNF-α signaling pathways and the suppression of oxidative stress, inflammation, and apoptosis in rats. Front Pharmacol. 2021;12:669679. doi:10.3389/fphar.2021.669679
13. Schulte C, Zeller T. microRNA-based diagnostics and therapy in cardiovascular disease-summing up the facts. Cardiovasc Diagn Ther. 2015;5(1):17–36. doi:10.3978/j.issn.2223-3652.2014.12.03
14. Wu H, Ye C, Ramirez D, Manjunath N. Alternative processing of primary microRNA transcripts by Drosha generates 5’ end variation of mature microRNA. PLoS One. 2009;4(10):e7566. doi:10.1371/journal.pone.0007566
15. Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4(1):Article17. doi:10.2202/1544-6115.1128
16. Chen Y, Zhu J, Lum PY, et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452(7186):429–435. doi:10.1038/nature06757
17. Voineagu I, Wang X, Johnston P, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474(7351):380–384. doi:10.1038/nature10110
18. Oldham MC, Konopka G, Iwamoto K, et al. Functional organization of the transcriptome in human brain. Nat Neurosci. 2008;11(11):1271–1282. doi:10.1038/nn.2207
19. Chen L, Bai J, Liu J, Lu H, Zheng K. A four-microRNA panel in peripheral blood identified as an early biomarker to diagnose acute myocardial infarction. Front Physiol. 2021;12:669590. doi:10.3389/fphys.2021.669590
20. Li Y, Hiroi Y, Liao JK. Notch signaling as an important mediator of cardiac repair and regeneration after myocardial infarction. Trends Cardiovasc Med. 2010;20(7):228–231. doi:10.1016/j.tcm.2011.11.006
21. Wu F, Yu B, Zhang X, Zhang Y. Cardioprotective effect of Notch signaling on the development of myocardial infarction complicated by diabetes mellitus. Exp Ther Med. 2017;14(4):3447–3454. doi:10.3892/etm.2017.4932
22. Kemp JR, Unal H, Desnoyer R, Yue H, Bhatnagar A, Karnik SS. Angiotensin II-regulated microRNA 483-3p directly targets multiple components of the renin-angiotensin system. J Mol Cell Cardiol. 2014;75:25–39. doi:10.1016/j.yjmcc.2014.06.008
23. Mishra PK, Tyagi N, Kundu S, Tyagi SC. MicroRNAs are involved in homocysteine-induced cardiac remodeling. Cell Biochem Biophys. 2009;55(3):153–162. doi:10.1007/s12013-009-9063-6
24. Wang K, Liu CY, Zhou LY, et al. APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nat Commun. 2015;6(1):6779. doi:10.1038/ncomms7779
25. He J, Li X. Comprehensive analysis of ceRNA regulation network involved in the development of coronary artery disease. Biomed Res Int. 2021;2021:6658115. doi:10.1155/2021/6658115
26. Yao Y, Jiang C, Wang F, et al. Integrative analysis of miRNA and mRNA expression profiles associated with human atrial aging. Front Physiol. 2019;10:1226. doi:10.3389/fphys.2019.01226
27. Akodad M, Mericskay M, Roubille F. Micro-RNAs as promising biomarkers in cardiac diseases. Ann Transl Med. 2016;4(24):551. doi:10.21037/atm.2016.12.38
28. Inouye M, Ripatti S, Kettunen J, et al. Novel Loci for metabolic networks and multi-tissue expression studies reveal genes for atherosclerosis. PLoS Genet. 2012;8(8):e1002907. doi:10.1371/journal.pgen.1002907
29. Chen J, Yu L, Zhang S, Chen X. Network analysis-based approach for exploring the potential diagnostic biomarkers of acute myocardial infarction. Front Physiol. 2016;7:615. doi:10.3389/fphys.2016.00615
30. Wu K, Zhao Q, Li Z, et al. Bioinformatic screening for key miRNAs and genes associated with myocardial infarction. FEBS Open Bio. 2018;8:897–913. doi:10.1002/2211-5463.12423
31. Huang X, Yu X, Li H, Han L, Yang X. Regulation mechanism of aquaporin 9 gene on inflammatory response and cardiac function in rats with myocardial infarction through extracellular signal-regulated kinase1/2 pathway. Heart Vessels. 2019;34(12):2041–2051. doi:10.1007/s00380-019-01452-8
32. Zhuo LA, Wen YT, Wang Y, et al. LncRNA SNHG8 is identified as a key regulator of acute myocardial infarction by RNA-seq analysis. Lipids Health Dis. 2019;18(1):201. doi:10.1186/s12944-019-1142-0
33. Qiu L, Liu X. Identification of key genes involved in myocardial infarction. Eur J Med Res. 2019;24(1):22. doi:10.1186/s40001-019-0381-x
34. De Lacorte Singulani J, De Fátima Da Silva J, Gullo FP, et al. Preliminary evaluation of circulating microRNAs as potential biomarkers in paracoccidioidomycosis. Biomed Rep. 2017;6(3):353–357. doi:10.3892/br.2017.849
35. Varga ZV, Zvara A, Faragó N, et al. MicroRNAs associated with ischemia-reperfusion injury and cardioprotection by ischemic pre- and postconditioning: protectomiRs. Am J Physiol Heart Circ Physiol. 2014;307(2):H216–227. doi:10.1152/ajpheart.00812.2013
36. Yuan N, Scherzer R, Tanriverdi K, Martin J, Rahalkar S, Hsue P. MicroRNA biomarkers associated with type 1 myocardial infarction in HIV-positive individuals. AIDS. 2019;33(15):2351–2361. doi:10.1097/QAD.0000000000002368
37. Bostjancic E, Zidar N, Glavac D. MicroRNA microarray expression profiling in human myocardial infarction. Dis Markers. 2009;27(6):255–268. doi:10.1155/2009/641082
38. Ishikawa Y, Satoh M, Itoh T, Minami Y, Takahashi Y, Akamura M. Local expression of Toll-like receptor 4 at the site of ruptured plaques in patients with acute myocardial infarction. Clin Sci. 2008;115(4):133–140. doi:10.1042/CS20070379
39. Satoh M, Shimoda Y, Maesawa C, et al. Activated toll-like receptor 4 in monocytes is associated with heart failure after acute myocardial infarction. Int J Cardiol. 2006;109(2):226–234. doi:10.1016/j.ijcard.2005.06.023
40. Ma H, Du J, Feng X, et al. Rosiglitazone alleviates myocardial apoptosis in rats with acute myocardial infarction via inhibiting TLR4/NF-κB signaling pathway. Exp Ther Med. 2020;19(4):2491–2496. doi:10.3892/etm.2020.8479
41. Pordel S, Sajedi Khanian M, Karimi MH, Nikoo H, Doroudchi M. Plasma CXCL1 levels and TRAF3IP2 variants in patients with myocardial infarction. J Clin Lab Anal. 2018;32(6):e22402. doi:10.1002/jcla.22402
42. da Silva AM, de Araújo JN, de Freitas RC, Silbiger VN. Circulating microRNAs as potential biomarkers of atrial fibrillation. Biomed Res Int. 2017;2017:7804763. doi:10.1155/2017/7804763
43. Buonaiuto G, Desideri F, Taliani V, Ballarino M. Muscle regeneration and RNA: new perspectives for ancient molecules. Cells. 2021;10(10):2512. doi:10.3390/cells10102512
44. Condorelli G, Latronico MV, Dorn GW
45. Lusardi TA, Murphy SJ, Phillips JI, et al. MicroRNA responses to focal cerebral ischemia in male and female mouse brain. Front Mol Neurosci. 2014;7:11. doi:10.3389/fnmol.2014.00011
46. Ha T, Liu L, Kelley J, Kao R, Williams D, Li C. Toll-like receptors: new players in myocardial ischemia/reperfusion injury. Antioxid Redox Signal. 2011;15(7):1875–1893. doi:10.1089/ars.2010.3723
47. Hally KE, La Flamme AC, Larsen PD, Harding SA. Platelet Toll-like receptor (TLR) expression and TLR-mediated platelet activation in acute myocardial infarction. Thromb Res. 2017;158:8–15. doi:10.1016/j.thromres.2017.07.031
48. Zhang S, Liu W, Liu X, Qi J, Deng C. Biomarkers identification for acute myocardial infarction detection via weighted gene co-expression network analysis. Medicine. 2017;96(47):e8375. doi:10.1097/MD.0000000000008375
49. Stillie R, Farooq SM, Gordon JR, Stadnyk AW. The functional significance behind expressing two IL-8 receptor types on PMN. J Leukoc Biol. 2009;86(3):529–543. doi:10.1189/jlb.0208125
50. Katayama T, Nakashima H, Takagi C, Honda Y, Suzuki S, Yano K. Predictors of mortality in patients with acute myocardial infarction and cardiogenic shock. Circ J. 2005;69(1):83–88. doi:10.1253/circj.69.83
51. Katayama T, Nakashima H, Honda Y, Suzuki S, Yano K. Relationship between adrenomedullin and left-ventricular systolic function and mortality in acute myocardial infarction. Angiology. 2005;56(1):35–42. doi:10.1177/000331970505600105
52. Khan SQ, O’Brien RJ, Struck J, et al. Prognostic value of midregional pro-adrenomedullin in patients with acute myocardial infarction: the LAMP (Leicester Acute Myocardial Infarction Peptide) study. J Am Coll Cardiol. 2007;49(14):1525–1532. doi:10.1016/j.jacc.2006.12.038
53. Nguyen NT, Lindsey ML, Jin YF. Systems analysis of gene ontology and biological pathways involved in post-myocardial infarction responses. BMC Genomics. 2015;16(Suppl 7):S18. doi:10.1186/1471-2164-16-S7-S18
54. Watson CN, Kerrigan SW, Cox D, Henderson IR, Watson SP, Arman M. Human platelet activation by Escherichia coli: roles for FcγRIIA and integrin αIIbβ3. Platelets. 2016;27(6):535–540. doi:10.3109/09537104.2016.1148129
55. Arbesu I, Bucsaiova M, Fischer MB, Mannhalter C. Platelet-borne complement proteins and their role in platelet-bacteria interactions. J Thromb Haemost. 2016;14(11):2241–2252. doi:10.1111/jth.13495
56. Dewald O, Zymek P, Winkelmann K, et al. CCL2/Monocyte Chemoattractant Protein-1 regulates inflammatory responses critical to healing myocardial infarcts. Circ Res. 2005;96(8):881–889. doi:10.1161/01.RES.0000163017.13772.3a
57. Dobaczewski M, Xia Y, Bujak M, Gonzalez-Quesada C, Frangogiannis NG. CCR5 signaling suppresses inflammation and reduces adverse remodeling of the infarcted heart, mediating recruitment of regulatory T cells. Am J Pathol. 2010;176(5):2177–2187. doi:10.2353/ajpath.2010.090759
58. Anzai A, Choi JL. The infarcted myocardium solicits GM-CSF for the detrimental oversupply of inflammatory leukocytes. J Exp Med. 2017;214(11):3293–3310. doi:10.1084/jem.20170689
59. Zhang Q, Zheng Y, Ning M, Li T. KLRD1, FOSL2 and LILRB3 as potential biomarkers for plaques progression in acute myocardial infarction and stable coronary artery disease. BMC Cardiovasc Disord. 2021;21(1):344. doi:10.1186/s12872-021-01997-5
60. Frangogiannis NG. The immune system and cardiac repair. Pharmacol Res. 2008;58(2):88–111. doi:10.1016/j.phrs.2008.06.007
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


