Back to Journals » Veterinary Medicine: Research and Reports » Volume 6
Schmallenberg virus infection of ruminants: challenges and opportunities for veterinarians
Authors Claine F, Coupeau D, Wiggers L, Muylkens B, Kirschvink N
Received 27 February 2015
Accepted for publication 27 April 2015
Published 29 June 2015 Volume 2015:6 Pages 261—272
DOI https://doi.org/10.2147/VMRR.S83594
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 6
Editor who approved publication: Professor Takashi Agui
François Claine, Damien Coupeau, Laetitia Wiggers, Benoît Muylkens, Nathalie Kirschvink
Veterinary Department, Faculty of Sciences, Namur Research Institute for Life Sciences (NARILIS), University of Namur (UNamur), Namur, Belgium
Abstract: In 2011, European ruminant flocks were infected by Schmallenberg virus (SBV) leading to transient disease in adult cattle but abortions and congenital deformities in calves, lambs, and goat kids. SBV belonging to the Simbu serogroup (family Bunyaviridae and genus Orthobunyavirus) was first discovered in the same region where bluetongue virus serotype 8 (BTV-8) emerged 5 years before. Both viruses are transmitted by biting midges (Culicoides spp.) and share several similarities. This paper describes the current knowledge of temporal and geographical spread, molecular virology, transmission and susceptible species, clinical signs, diagnosis, prevention and control, impact on ruminant health, and productivity of SBV infection in Europe, and compares SBV infection with BTV-8 infection in ruminants.
Keywords: Schmallenberg virus, Europe, ruminants, review
Introduction
In 2006, bluetongue virus serotype 8 (BTV-8) emerged in northern Europe causing economic losses to cattle and sheep farms.1 Five years later, a novel virus named Schmallenberg virus (SBV), in reference to the German city where the virus was first discovered, appeared in the same geographic region, but it remains uncertain whether these two viruses followed the same route of introduction.2 SBV from the family Bunyaviridae and the genus Orthobunyavirus affects domestic ruminants (mainly cattle and sheep) as well as wild ruminants (eg, buffalo, bison, red deer).3 A transient disease characterized by diarrhea, hyperthermia, and drop in milk production is described in adult animals, while important congenital musculoskeletal deformities such as arthrogryposis are observed in neonates if infection occurs during early pregnancy. SBV is transmitted by an insect vector: biting midges (Culicoides spp.) were identified as SBV vectors. However, transplacental and potential venereal transmissions were also described.4
In November 2011, the Friedrich-Loëffler Institute (FLI) in Germany was the first to detect RNA of SBV in blood samples collected among affected dairy cattle by metagenomic approach.5 Comparative genetic analysis indicated that this new virus was very similar to certain members of the Orthobunyavirus genus (eg, Akabane and Aino viruses). Even though the first SBV identification was realized in late autumn 2011, SBV emerged earlier in Europe: heads of Culicoides spp. caught in summer 2011 in Belgium were real-time polymerase chain reaction (RT-PCR) positive,6 and it was demonstrated that SBV has circulated in at least three Italian provinces since early September 2011.7 From 2011 to date, geographic distribution of SBV has steadily increased and now includes nearly all European countries.
This paper reviews current knowledge of temporal and geographical spread, molecular virology, transmission and susceptible species, clinical signs, diagnosis, prevention and control, impact on ruminant health, and productivity of SBV infection in Europe.
Temporal and geographical spread of SBV in Europe
In summer 2011, first reports of unidentified disease of dairy cattle were obtained from farms located in the Netherlands and Germany.5 In all the affected farms, the major clinical sign was diarrhea associated with drop in milk production and hyperthermia. In November 2011, FLI demonstrated that a novel pathogen was responsible for these clinical features: SBV in reference to the German city where the samples tested were collected. Shortly after, an epizootic outbreak of congenital malformations, featuring an arthrogryposis hydranencephaly syndrome, in newborn lambs, kids, and calves associated with SBV started in northwestern Europe.8 At the end of 2011, Belgium, as the Netherlands, reported cases of stillborn lambs or newborn lambs rapidly dying with arthrogryposis and brain abnormalities.9 The affected farms were mainly located in the north of Belgium close to the Dutch border. In early spring 2012, the total number of affected herds equaled 3,628 in eight countries (bovine 1,115, ovine 2,440, caprine 73).10
The high SBV within-flock seroprevalence (up to 98.03%) in geographic areas having reported SBV outbreaks in late 2011 and 2012 was expected to limit reemergence of the virus in 2012.11 However, a novel SBV episode occurred during summer 2012 in previously affected regions. A study conducted in a Belgian sheep farm demonstrated SBV reemergence in a sentinel flock of 50 ewe lambs bimonthly followed by serum neutralization test and RT-PCR between mid-July and mid-October.12 In Germany, new cases of SBV infection were observed in cattle, sheep, and goats from June 2012 proving the ability of the virus to recirculate after winter.13 In 2012 and 2013, SBV has spread across eastern Europe. In late September 2013, a total of 27 European countries were considered as SBV-infected.14
Figure 1 represents European countries having reported cases of SBV infection in cattle, sheep, or goat herds or having detected antibodies in serum or milk from summer 2011 until late 2014 presented as a cumulative list (Figure 1A) and colored on a map (Figure 1B).
A third SBV episode was evidenced in late 2014 in the Netherlands and Germany: SBV was detected in calves and lambs. In the Netherlands, increased antibody levels were observed in heifers.14 Even though three SBV episodes occurred in Europe since 2011, the case numbers of SBV-infected herds largely decreased from 2012 to 2013 and 2014. However, due to flock renewal and increased number of seronegative animals, malformations in newborn calves and lambs might be expected in the following months if heifers or ewes were infected during gestation.
The first outbreaks of both BTV-8 and SBV occurred in late summer. But SBV spread much more rapidly in 2011 than bluetongue in 2006 affecting more rapidly larger areas.15 Even though both viruses emerged in the same region, the question about their route of introduction remains. The cumulative proportion of infected farms over time from the start of each outbreak is similar, although BTV-8 case numbers were initially higher possibly due to the fact that BTV-8 was a notifiable disease.15 This was not the case for acute SBV infections. The highest point of interest is certainly that SBV spread more quickly than BTV-8 resulting in more difficulties to predict its extent and impact. Moreover, it was more difficult to give an accurate evaluation of the epidemiological situation after SBV infection in contrast to what could have been done after BTV infection. Indeed, except the observations of malformed calves and lambs, no clinical sign was detected in adult animals.
Molecular virology
First identification of SBV was performed by FLI in November 2011 from blood samples collected among cattle presenting clinical signs such as diarrhea associated with drop in milk production and hyperthermia. Metagenomic analysis and comparison of sequences indicated that the closest relatives were viruses from the Simbu serogroup (family Bunyaviridae and genus Orthobunyavirus).5 Viruses from the Simbu serogroup mainly affect ruminants, but none of these had been detected in Europe before. It is suggested by some authors that SBV could be a reassortant between Sathuperi and Shamonda viruses,16 while others think that SBV could be an ancestor of Shamonda virus.17
Structure and genome
SBV particles are enveloped and have a diameter of approximately 100 nm. SBV is a negative-sense single-stranded RNA virus whose genome is composed of three segments: a large (L), a medium (M), and a small (S) one. The L segment encodes the RNA-dependent RNA polymerase. The M segment encodes a precursor polyprotein co-translationally cleaved into the envelop proteins Gn and Gc and the nonstructural protein NSm. The S segment encodes the nucleoprotein N and the nonstructural protein NSs in an overlapping open-reading frame. A Belgian study conducted in 2013 showed that the SBV Gc protein contained an N-terminal domain that accumulates mutations in the natural course of SBV spread within flocks.18 This result suggests that the variability identified in this domain may support different biological processes. Among these processes, an immune evasion mechanism may occur in the actual context of infection and/or adaptation of the cell tropism by supporting a diversification of receptors usage and during in vitro cell culture adaptation in the absence of immune selective pressure.18,19 The impact of genetic variation is thus of first interest. Moreover, the introduction of another Simbu group virus in the future in Europe may give opportunity of reassortment with novel vector and virulence properties. Consequently, enhanced surveillance should be recommended.20
Viremia
In case of SBV infection, clinical signs in adult animals are observed during a very short period related to a short viremic period for the virus. These observations were confirmed after the first experimental infections in calves: the inoculated animals became infected and remained PCR positive from 2 to 5 days postinfection.5 After experimental infection in sheep, it was shown that the duration and the level of viremia seemed to be dose independent.21 However, a different pattern was observed in case of natural SBV infection: the duration of viremia in sheep appears to be longer under field conditions. Indeed, 20% of a group of sentinel ewe lambs naturally infected by SBV were found PCR positive 2 weeks apart.12 Interestingly, it appears that the highest SBV genome loads for tissue samples were found in mesenteric lymph nodes in most animals.22
Virus transmission and susceptible species
Vector transmission
Most of the bunyaviruses present in Asia or Africa are transmitted by an insect vector (biting midges or mosquitoes).23 When SBV emerged in 2011 in Europe, biting midges of the Obsoletus species complex of the ceratopogonid genus Culicoides were rapidly identified as SBV vectors. Indeed, viral genome was detected in different Culicoides spp. (Culicoides dewulfi, Culicoides chiopterus, Culicoides punctatus, etc) caught as early as summer and autumn 2011 in Belgium,6 Italy,7 the Netherlands,24 and Denmark.23 Under laboratory conditions, SBV replication and dissemination were detected in Culicoides sonorensis.25,26
Approximately 120 different species of Culicoides are described in Europe. The Obsoletus complex is the most abundant in European farms.27 Culicoides biting midges are implicated in transmission of several viruses of the Simbu serogroup. They were recognized as major vectors of BTV-8 in northern and central Europe during the 2006 outbreak of bluetongue disease.28 However, the vector competence for SBV may exceed rates recorded for BTV either in the number of species capable of transmitting the virus or in the proportion of individuals within a species.20 So, it is of first interest to better understand the ecology and the behavior of Culicoides spp. because these are elements to predict epidemiology of the pathogens they transmit.29 Many Culicoides life cycles are dependent on meteorological conditions. Appetitive flight is conditioned by temperature, humidity, light intensity, and wind speed.30 Moreover, the capability of Culicoides spp. to overwinter and serve as reservoir for new infections during the next year is of relevance to the transmission of BTV and SBV.2 Even though there is no evidence that other insect species (eg, mosquitoes) are implied in disease spreading,31 mechanic transmission by hematophagous insects could occur as for other viruses of the Simbu group.32
Transplacental and venereal transmission
If vector transmission is largely described for SBV in adult animals, fetuses can be infected transplacentally leading to observations of congenital deformities if viral infection occurs during the critical period of gestation. However, no evidence of horizontal (animal to animal) transmission has been detected.15 Besides vector-borne transmission being the most common route of SBV infection, researchers have recently demonstrated that SBV RNA-positive bovine semen could contain infectious SBV.4,33 If the risk of venereal transmission exists, the insemination of dams with SBV-containing semen would lead at worst to viremia of the dam facilitating vector transmission. No evidence of infection of the developing embryo has been reported so far.4 Many questions remain regarding the pathogenesis of SBV infection in pregnant animals, their transmission by embryo and/or gametes, and the dynamics of the virus toward and in the fetus.34
Susceptible species
Numerous species have been recognized as susceptible species to SBV infection. Table 1 summarizes the animal species in which direct and/or indirect SBV detection was performed and clinical expression in adults and/or their offspring was observed. It appears that clinical features of SBV infection have only been observed in domestic ruminants (cattle, sheep, and goats). The indirect detection of the pathogen (serological reaction to SBV) has only been performed in wild ruminants (eg, alpaca, buffalo, bison), zoo (eg, kudu, zebra, oryx), and some other mammalian species (eg, horse, wild boar), while in dogs, virological and serological evidences of SBV infection have been detected. Experimental SBV infection was performed in poultry, but the results indicate that the virus did not replicate.20 Due to their suggestive role in the epidemiology of Akabane virus, the role of pigs in the epidemiology of SBV was evaluated. Experimentally SBV-infected piglets showed only seroconversion underlining the inability of the virus to replicate.35
Clinical signs
SBV infection in adult ruminants is frequently associated with no specific clinical sign. It corresponds to a mild and transient disease, including reduced milk production (up to 50%), inappetence, hyperthermia, and diarrhea in cows.15 In sheep and goat, SBV infection remains subclinical. Only a small number of herds had reported acute cases in adults (6% for cattle, 1% for goats, and 3% for sheep).36 This nonspecific febrile syndrome was reported during summer and fall 2011 in adult dairy cows from farms in the Netherlands and Germany.37
In December 2011, the Netherlands reported the teratogenic effect of SBV infection in sheep with clinical manifestations comparable to those observed for Akabane and Aino viruses.38,39 Various congenital disorders were detected in aborted fetuses or stillborn ruminants (ovine, caprine, and bovine). Musculoskeletal deformities were commonly observed on the hind- and forelimbs, the vertebral column, and the neck. They consisted essentially of arthrogryposis, lordosis, scoliosis, torticollis, and brachygnathia inferior.40 Even if these atypical lesions could be associated with other factor, sacral spina bifida and cleft palate were observed in 2013 in two SBV-positive stillborn lambs.41 In case of twin gestation, one twin may present previously described malformations, while the other only showed neurological disorders or did not present any clinical sign.42
Macroscopically, common malformations of the central nervous system (CNS) in young ruminants observed during necropsy were hydranencephaly, porencephaly, lissencephaly, hydrocephalus, cerebellar and cerebral hypoplasia, and micromyelia. Histological lesions included lymphohistiocytic meningoencephalomyelitis in some cases, glial nodules mainly in the mesencephalon and hippocampus of lambs and goats, and neuronal degeneration and necrosis mainly in the brain stem of calves. Myofibrillar hypoplasia could be diagnosed in lambs and calves.40 It was suggested that lesions observed on the spinal cord were responsible for fetal immobility promoting arthrogryposis.37
The presence of musculoskeletal lesions due to malformations of the CNS in SBV-infected aborted fetuses or neonates led to the expression “AG-HE syndrome” for arthrogryposis and hydranencephaly which is often used to describe the SBV-induced deformities. Figure 2 presents the typical clinical lesions of stillborn lambs affected by SBV. Intrauterine BTV infection may lead to similar lesions (Figure 3).
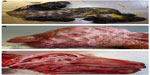  | Figure 2 Schmallenberg virus infection in stillborn lambs presenting arthrogryposis, scoliosis, and torticollis (A), brachygnathia inferior (B), and cerebellar/cerebral hypoplasia (C). |
  | Figure 3 Bluetongue virus serotype 8 infection in stillborn lambs presenting arthrogryposis (A), brachygnathia superior and anophthalmia (B), and cerebellar hypoplasia (C). |
The time of infection relative to the stage of gestation, and so the stage of fetal development (CNS tissues, immune system), seems to determine the severity of fetal malformations.43 Due to numerous similarities between clinical features of SBV and Akabane virus infections in neonates, it is estimated that high-risk period of gestation in ruminants could be very comparable. For Akabane virus, it is estimated between the 28th and 36th day of gestation in sheep, the 30th and 50th day in goats, and the 76th and 174th day in cattle.44 A recent study tried to define more precisely the critical phase for cattle in case of natural SBV infection of pregnant cows. Malformed calves were born after SBV transplacental infection between days 60 and 144 of gestation, which corresponds to the critical timeframe described for viruses closely related to SBV.45
SBV infection of wild ruminants or other mammal species (eg, dogs) was not related to clinical manifestations or malformations. To date, no evidence of viral infection in humans has been reported.46
Diagnosis
The diagnosis of SBV infection in living adult animals is difficult for veterinarians. As previously mentioned, SBV infection is mostly subclinical in adult ruminants or characterized, especially in cattle, by a febrile syndrome. These clinical signs observed could be noticed in case of other viral infections (eg, bovine-herpes virus type 1, bluetongue virus, foot-and-mouth disease virus).5 However, in case of transplacental infection, congenital CNS and musculoskeletal malformations such as AG-HE syndrome, aplasia or hypoplasia of the cerebrum or cerebellum, and hydranencephaly presented by aborted fetuses, stillborn, or newborn ruminants are more relevant. However, due to similarities between clinical features of SBV and other ruminant-specific viruses, only laboratory virological and/or serological diagnosis may confirm hypothesis of SBV episode.
The direct diagnosis of SBV infection can be realized by performing real-time quantitative polymerase chain reaction (RT-qPCR) developed by FLI in 2011 on L segment of the SBV genome. A protocol targeting the S segment was later developed and showed higher sensitivity.47 Various organs were tested by RT-PCR, but only some of them are suitable for SBV detection.48,49 It was established that cerebrum, spinal cord, external placental fluid, and umbilical cord are the most appropriate organs to detect SBV in malformed lambs or calves.49 Low Cq values were also obtained in adrenals and gonads, meaning that higher initial copy numbers of the targeted viral sequence were present in these organs (Coupeau, personal communication). The brain stem also seemed to be a suitable organ to perform direct diagnosis of SBV infection.9
Besides RT-qPCR, the detection of anti-SBV antibodies present in serum of infected animals can be performed by indirect method of SBV infection diagnosis. Virus neutralization test (VNT) and enzyme-linked immunosorbent assay (ELISA) have been developed as tools for serological diagnosis. If the first method appears to be time-consuming, ELISA is more rapid, less expensive, and allows testing a larger number of samples,42 but the possibility of cross-reactions with other Orthobunyaviruses from the Simbu serogroup exists.50 Moreover, ELISA tests are also able to detect anti-SBV antibodies in milk.51 However, many discrepancies were observed between results obtained by performing VNT and those obtained by ELISA tests. It seemed that ELISA tests presented lower specificity and sensitivity.52
Figure 4 summarizes the most commonly used diagnostic methods of SBV infection in ruminants (clinical signs observed in adult animals and in aborted fetuses, stillborns, or newborns, direct virological diagnosis by RT-PCR, and indirect serological diagnosis by VNT or ELISA), advantages and disadvantages of each of them, as well as the required material.
Besides the diagnostic methods mentioned earlier, it is necessary to specify that SBV infection diagnosis can also be performed by immunofluorescence and virus isolation,53 but these two methods are not routinely used for diagnosis. The detection of SBV proteins and RNA in paraffin sections can be performed by immunochemistry and in situ hybridization.54
In vitro, baby hamster kidney (BHK-21) cells are the most convenient hosts for SBV. The SBV genome segments were cloned as cDNA, and a three-plasmid rescue system was established to recover infectious virus.55
Prevention and control
Monitoring the evolution of vector population and having surveillance of the virus frequency in the vector seem to be the most appropriate ways to predict future viral episodes. Besides, the control of insect populations and the vaccination of cattle and sheep flocks are described as the two other prophylactic measures.
In both livestock- and human-associated species, wide-scale control of larvae or adults of Culicoides spp. through treatment, removal, or covering of development or resting sites is considered unfeasible due to the broad range and abundance of habitats utilized.56 Moreover, to control biting midges by the use of insecticides is certainly a poor tool to prevent and/or control SBV spread within flocks because no products are specifically licensed against Culicoides spp.
Vaccination is a preventive measure able to reduce the impact of SBV infection.53 Vaccination costs on one hand and reduced impact of SBV infection on herds and flocks on the other hand raise questions about the real utility of vaccination. Moreover, due to the fact that SBV has rapidly spread among almost all European countries since its emergence in 2011 and that evidences of second SBV episode have not been detected in all these countries, the surveillance of the virus appears to be more important than to vaccinate. However, the detection of novel SBV episode in Germany and in the Netherlands in late 2014 may reverse this situation because SBV could circulate in flocks composed of an important number of seronegative animals. Two vaccines have been developed against SBV: Bovilis SBV (MSD Animal Health) and SBVvax (Merial). Recently, mutant viruses were tested as modified live vaccines in cattle.57 Vaccination usually focuses on breeding animals that receive vaccination before service in order to prevent fetal infection. However, there is a lack of information about long-term immunity against SBV in domestic ruminants as well as duration of colostral immunity in neonates. This last question is of primary interest because passive immunity received by ingestion and absorption of colostrum could interfere with vaccine efficacy if vaccine is administered in young ruminant under colostral protection. Recent studies focus on these points in cattle and sheep. It appears that in cattle, long-term immunity persists at least 2 years after natural SBV infection. Based on the decay of maternal antibodies, calves could efficiently be vaccinated against SBV at an age of 6 months.58 In sheep, neutralizing antibodies against SBV were detected 2 years after the first viral infection, and the lambs seem to lose maternal antibodies at an age of 4 months.59
Impact on ruminant health and productivity
SBV outbreak in Europe is considered as sanitary event of veterinary public health interest,60 but its impact remains low or limited.42 However, it seems to be difficult to estimate real impact of SBV episode on European livestock due to number of undeclared SBV-affected flocks.60,61 The highest proportion of SBV-confirmed herds in comparison with the total number of herds per region is 6.6% for sheep and 4% for cattle.42
At a farm level, the impact of SBV infection was highly variable and is likely to depend on the calving and lambing program used.20 In sheep flocks, especially in those where matings are spread over several periods, the percentages of SBV-affected stillborns or newborns and the clinical features of SBV infection observed were extremely variable. These observations could be related to the fact that ewes underwent SBV infection when they were at different stages of production. Mating patterns during an at-risk period coinciding with a period of high vector activity are more likely to experience higher impact.20 In a Belgian sheep farm where SBV emerged and spread between mid-September and mid-October 2011, ewes lambing in January 2012 gave birth to 17% (28/163) of stillborn or newborn lambs presenting typical deformities, while ewes lambing in March 2012 gave birth to only 5% (8/150) of aborted fetuses, and in May 2012, no impact on lambings was observed.62 Data obtained in France on a larger scale during the first SBV episode are more or less equivalent. In the Netherlands, a case-control study was designed to describe the effect of SBV infection on reproductive performance and on mortality rates in sheep. It appears that the impact of viral episode was limited.8
In dairy cattle, a recent study tried to evaluate the impact of 2011 SBV epidemic on milk production, reproductive performance, and mortality in the Netherlands and Kleve district in Germany. No impact was evidenced on milk production and on mortality rates in calves, and only a small decrease in fertility rate was observed indicating the limited impact of SBV on productivity of dairy cattle.63 Even though it seems very clear that impact of SBV on ruminant health and productivity was low at a herd scale, the situation may be different while considering the overall meat and milk industry. Indeed, restrictions made by governments concerning imports of animal products from countries declared SBV affected could have a significant economic impact.
Comparison between BTV and SBV
Similarities and differences between BTV-8 and SBV both affecting ruminants and sharing numerous similarities are summarized in Table 2. Even though BTV-8 and SBV strictly belong to different virus classification, both are RNA viruses mainly affecting ruminant species. SBV appeared 5 years after BTV-8 in northern Europe (Germany, the Netherlands, and Belgium)64 but spread more rapidly to numerous European countries, while the geographical spread of BTV-8 was more limited. Several hypotheses could explain the differences between BTV-8 and SBV spread. At the time of BTV-8 outbreak, due to clinical evidences of viral infection in animals, restrictions on animal movements were established in Europe. This was not the case for SBV certainly due to the fact that SBV infection in adult ruminants was frequently subclinical. Moreover, the average distances between infectious farms and infected farms were shorter for BTV than for SBV suggesting differences in ability of the vector to spread the disease.15 At herd or flock level, SBV spreads more rapidly than BTV-8, as suggested by higher herd or flock seroprevalence.11 As duration of viremia within the ruminant host equaled 1–2 weeks for both viruses, it is possible that viral replication in the vector differs between BTV-8 and SBV.5,65
These two viruses are transmitted by biting midges (Culicoides spp.), and transplacental and venereal ways of transmission are described for both of them.65 Transmission by placentophagia is only reported for BTV-8.66 If clinical signs induced by SBV infection consisted in mild and transient disease essentially detected in cattle and sheep, BTV-8 infection in sheep led to numerous and intense clinical signs. Congestion of the head associated with excessive salivation and ulceration of mucosa throughout the body were often observed. If viral infection occurred during pregnancy, this led to abortion or observation of neurological defects in newborns. With BTV-8 infection, some cases of “dummy” lambs were described for aborted fetuses or stillborns. Abnormal development of CNS tissues was described at necropsy.67 During the bluetongue epidemic, a significant increase in bovine abortions and stillborns as well as the birth of weak or deformed calves was reported. “Dummy” syndrome and hydranencephaly were observed as for lambs.68 Such abnormalities were detected in stillborn or newborn ruminants in case of SBV infection, but they were always associated with musculoskeletal deformities (arthrogryposis, scoliosis, lordosis, and/or torticollis).
For viral infection diagnosis, the most commonly used laboratory diagnosis was genome detection by RT-PCR and/or evaluation of animal seropositivity by performing ELISA or VNT. As for SBV, numerous mammalian species were detected BTV-8 seropositive, but in the absence of PCR-positive results, it cannot be concluded that these species are real viral hosts.
Both for BTV-8 and for SBV, herd vaccination seemed to be the only realistic preventive measure to take to prevent novel viral episode. Massive compulsory vaccination campaigns against BTV-8 were conducted in northern Europe,65 while for SBV, the use of vaccines is not compulsory and therefore limited. For both BTV-8 and SBV, virus surveillance in vector populations could be helpful to predict hypothetical reemergence.
Comparatively, the impact of BTV-8 infection on ruminant health and productivity was clearly higher than that of SBV. Severe clinical signs and high levels of morbidity and mortality were observed in case of BTV-8 infection. Moreover, BTV-8 infection led to decreased semen quality in bulls and rams69,70 as to poor reproductive performance in females.67
Conclusion
In 5 years, European flocks underwent two major viral episodes: BTV-8 and SBV emerged in the same regions and rapidly spread among European countries. Even though they belong to different families, both arboviruses are transmitted by biting midges (Culicoides spp.) and caused economic losses to the European livestock industry. To date, too little is known about these insects. Further studies could focus on life cycle, habitats, and distribution of insects and the impact of meteorological conditions on their survival and spread. Indeed, at a time of relatively easy control of animal movements within and between countries, a better understanding of the biology and ecology of these insects recognized as vectors for several animal diseases seems to be of first interest.
For BTV and for SBV, it is clear that veterinarians played and play an essential role in the early detection of clinical signs related to viral infection and in the transmission of field data to health authorities. They are the first players of our health surveillance networks, and in front of the risk of the unpredictable emergence of novel animal pathogens, it is sure that veterinarians in Europe and all around the world will act as “warning signals”. But they are only part of a team effort: only strong collaborations between field workers, laboratories, and authorities could lead to efficient and valuable work.
Disclosure
None of the authors of this paper has a financial or personal relationship with other people or organizations that could inappropriately influence or bias the content of the paper. The authors report no conflicts of interest with this work.
References
Velthuis AG, Saatkamp HW, Mourits MC, de Koeijer AA, Elbers AR. Financial consequences of the Dutch bluetongue serotype 8 epidemics of 2006 and 2007. Prev Vet Med. 2010;93(4):294–304. | |
Koenraadt CJ, Balenghien T, Carpenter S, et al. Bluetongue, Schmallenberg – what is next? Culicoides-borne viral diseases in the 21st century. BMC Vet Res. 2014;10:77. | |
Larska M, Krzysiak MK, Kęsik-Maliszewska J, Rola J. Cross-sectional study of Schmallenberg virus seroprevalence in wild ruminants in Poland at the end of the vector season of 2013. BMC Vet Res. 2014; 10(1):307. | |
Schulz C, Wernike K, Beer M, Hoffmann B. Infectious Schmallenberg virus from bovine semen, Germany. Emerg Infect Dis. 2014;20(2):338–340. | |
Hoffmann B, Scheuch M, Höper D, et al. Novel orthobunyavirus in cattle, Europe, 2011. Emerg Infect Dis. 2012;18(3):469–472. | |
De Regge N, Deblauwe I, De Deken R, et al. Detection of Schmallenberg virus in different Culicoides spp. by real-time RT-PCR. Transbound Emerg Dis. 2012;59(6):471–475. | |
Goffredo M, Monaco F, Capelli G, et al. Schmallenberg virus in Italy: a retrospective survey in Culicoides stored during the bluetongue Italian surveillance program. Prev Vet Med. 2013;111:230–236. | |
Luttikholt S, Veldhuis A, van den Brom R, et al. Risk factors for malformations and impact on reproductive performance and mortality rates of Schmallenberg virus in sheep flocks in the Netherlands. PLoS One. 2014;9(6):e100135. | |
De Regge N, van den Berg T, Georges L, Cay B. Diagnosis of Schmallenberg virus infection in malformed lambs and calves and first indications for virus clearance in the fetus. Vet Microbiol. 2013;162(2–4):595–600. | |
ProMED-mail [homepage on the Internet]. Schmallenberg virus – Europe (34): decline, update; 2012. Available from: http://www.promedmail.org/direct.php?id=1115024. Accessed January 14, 2015. | |
Elbers AR, Loeffen WL, Quak S, et al. Seroprevalence of Schmallenberg virus antibodies among dairy cattle, the Netherlands, winter 2011–2012. Emerg Infect Dis. 2012;18(7):1065–1071. | |
Claine F, Coupeau D, Wiggers L, Muylkens B, Kirschvink N. Schmallenberg virus among female lambs, Belgium, 2012. Emerg Infect Dis. 2013;19(7):1115–1117. | |
Conraths FJ, Kamer D, Teske K, Hoffmann B, Mettenleiter TC, Beer M. Reemerging Schmallenberg virus infections, Germany, 2012. Emerg Infect Dis. 2013;19(3):513–514. | |
ProMED-mail [homepage on the Internet]. Schmallenberg virus – Europe (35): Netherlands, Germany, bovine; 2014. Available from: http://www.promedmail.org/direct.php?id=2978286. Accessed January 14, 2015. | |
Sedda L, Rogers DJ. The influence of the wind in the Schmallenberg virus outbreak in Europe. Sci Rep. 2013;3:3361. | |
Yanase T, Kato T, Aizawa M. Genetic reassortment between Sathuperi and Shamonda viruses of the genus Orthobunyavirus in nature: implications for their genetic relationship to Schmallenberg virus. Arch Virol. 2012;157(8):1611–1616. | |
Goller KV, Hoper D, Schirrmeier H, Mettenleiter TC, Beer M. Schmallenberg virus as possible ancestor of Shamonda virus. Emerg Infect Dis. 2012;18(10):1644–1646. | |
Coupeau D, Claine F, Wiggers L, Kirschvink N, Muylkens B. In vivo and in vitro identification of a hypervariable region in Schmallenberg virus. J Gen Virol. 2013;94(pt 6):1168–1174. | |
Fischer M, Hoffmann B, Goller KV, Hoper D, Wernike K, Beer M. A mutation ‘hot spot’ in the Schmallenberg virus M segment. J Gen Virol. 2013;94(pt 6):1161–1167. | |
European Food Safety Authority [homepage on the Internet]. Schmallenberg virus: state of art; 2014. Available from: http://www.efsa.europa.eu/fr/efsajournal/doc/3681.pdf. Accessed January 10, 2015. | |
Poskin A, Martinelle L, Mostin L, et al. Dose-dependent effect of experimental Schmallenberg virus infection in sheep. Vet J. 2014;201(3):419–422. | |
Hechinger S, Wernike K, Beer M. Evaluating the protective efficacy of a trivalent vaccine containing Akabane virus, Aino virus and Chuzan virus against Schmallenberg virus infection. Vet Res. 2013;44:114. | |
Rasmussen LD, Kristensen B, Kirkeby C, et al. Culicoids as vectors of Schmallenberg virus. Emerg Infect Dis. 2012;18(7):1204–1206. | |
Elbers AR, Meiswinkel R, van Weezep E. Sloet van Oldruitenborgh-Oosterbaan MM, Kooi EA. Schmallenberg virus in Culicoides spp. biting midges, the Netherlands, 2011. Emerg Infect Dis. 2013;19(1):106–109. | |
De Regge N, Madder M, Deblauwe I, et al. Schmallenberg virus circulation in Culicoides in Belgium in 2012: field validation of a real time RT-PCR approach to assess virus replication and dissemination in midges. PLoS One. 2014;9(1):e87005. | |
Veronesi E, Henstock M, Gubbins S, et al. Implicating Culicoides biting midges as vectors of Schmallenberg virus using semi-quantitative RT-PCR. PLoS One. 2013;8(3):e57747. | |
Mellor PS. Infection of the vectors and bluetongue epidemiology in Europe. Vet Ital. 2004;40:167–174. | |
Lehmann K, Werner D, Hoffmann B, Kampen H. PCR identification of culicoid biting midges (Diptera, Ceratopogonidae) of the Obsoletus complex including putative vectors of bluetongue and Schmallenberg viruses. Parasit Vectors. 2012;5:213. | |
Sanders CJ, Shortall CR, Gubbins S, et al. Influence of season and meteorological parameters on flight activity of Culicoides biting midges. J Appl Ecol. 2011;48(6):1355–1364. | |
Burgin LE, Gloster J, Sanders C, Mellor PS, Gubbins S, Carpenter S. Investigating incursions of bluetongue virus using a model of long-distance Culicoides biting midge dispersal. Transbound Emerg Dis. 2013;60(3):263–272. | |
Wernike K, Jost H, Becker N, Schmidt-Chanasit J, Beer M. Lack of evidence for the presence of Schmallenberg virus in mosquitoes in Germany, 2011. Parasit Vectors. 2014;7:402. | |
Scholte EJ, Mars MH, Braks M, et al. No evidence for the persistence of Schmallenberg virus in overwintering mosquitoes. Med Vet Entomol. 2014;28:110–115. | |
Ponsart C, Pozzi N, Bréard E, et al. Evidence of excretion of Schmallenberg virus in bull semen. Vet Res. 2014;45:37. | |
Zientara S, Ponsart C. Viral emergence and consequences for reproductive performance in ruminants: two recent examples (bluetongue and Schmallenberg viruses). Reprod Fertil Dev. 2014;27(1):63–71. | |
Poskin A, Van Campe W, Mostin L, Cay B, De Regge N. Experimental Schmallenberg virus infection of pigs. Vet Microbiol. 2014;170(3–4):398–402. | |
Afonso A, Abrahantes JC, Conraths F, et al. The Schmallenberg virus epidemic in Europe-2011–2013. Prev Vet Med. 2014;116(4):391–403. | |
Bayrou C, Garigliany MM, Sarlet M, Sartelet A, Cassart D, Desmecht D. Natural intrauterine infection with Schmallenberg virus in malformed newborn calves. Emerg Infect Dis. 2014;20(8):1327–1330. | |
Kurogi H, Inaba Y, Goto Y, Miura Y, Takahashi H. Serologic evidence for etiologic role of Akabane virus in epizootic abortion-arthrogryposis-hydranencephaly in cattle in Japan, 1972–1974. Arch Virol. 1975;47(1):71–83. | |
Tsuda T, Yoshida K, Ohashi S, et al. Arthrogryposis, hydranencephaly and cerebellar hypoplasia syndrome in neonatal calves resulting from intrauterine infection with Aino virus. Vet Res. 2004;35(5):531–538. | |
Herder V, Wohlsein P, Peters M, Hansmann F, Baumgartner W. Salient lesions in domestic ruminants infected with the emerging so-called Schmallenberg virus in Germany. Vet Pathol. 2012;49(4):588–591. | |
Claine F, Coupeau D, Wiggers L, Muylkens B, Kirschvink N. Evidences for re-emergence of Schmallenberg virus during summer and autumn 2012 among naïve female lambs and immunized ewes. In: Proceedings of the 7th EPIZONE Annual Meeting; October 1–4, 2013; Brussels, Belgium. | |
Doceul V, Lara E, Sailleau C, et al. Epidemiology, molecular virology and diagnostics of Schmallenberg virus, an emerging orthobunyavirus in Europe. Vet Res. 2013;14:31. | |
Steukers L, Bertels G, Cay AB, Nauwynck H. Schmallenberg virus: emergence of an Orthobunyavirus among ruminants in Western Europe. Vlaams Diergeneeskd Tijdschr. 2012;81(3):119–127. | |
Kirkland PD, Barry RD, Harper PA, Zelski RZ. The development of Akabane virus-induced congenital abnormalities in cattle. Vet Rec. 1988;122(24):582–586. | |
Wernike K, Holsteg M, Schirrmeier H, Hoffmann B, Beer M. Natural infection of pregnant cows with Schmallenberg virus – a follow-up study. PLoS One. 2014;9(5):e98223. | |
Reusken C, van den Wijngaard C, van Beek P, et al. Lack of evidence for zoonotic transmission of Schmallenberg virus. Emerg Infect Dis. 2012;18(11):1746–1754. | |
Fischer M, Schirrmeier H, Wernike K, Wegelt A, Beer M, Hoffmann B. Development of a pan-Simbu real-time reverse transcriptase PCR for the detection of Simbu serogroup viruses and comparison with SBV diagnostic PCR systems. Virol J. 2013;10:327. | |
van der Poel WH. Diagnostics for Schmallenberg virus. Vet Rec. 2012;171(12):294–295. | |
Bilk S, Schulze C, Fischer M, Beer M, Hlinak A, Hoffmann B. Organ distribution of Schmallenberg virus RNA in malformed newborns. Vet Microbiol. 2012;159(1–2):236–238. | |
Bréard E, Lara E, Comtet L, et al. Validation of a commercially available indirect elisa using a nucleocapside recombinant protein for detection of Schmallenberg virus antibodies. PLoS One. 2013;8(1):e53446. | |
Humphries D, Burr P. Schmallenberg virus milk antibody ELISA. Vet Rec. 2012;171(20):511–512. | |
van der Poel WH, Cay B, Zientara S, et al. Limited interlaboratory comparison of Schmallenberg virus antibody detection in serum samples. Vet Rec. 2014;174(15):380. | |
Tarlinton R, Daly J, Dunham S, Kydd J. The challenge of Schmallenberg virus emergence in Europe. Vet J. 2012;194(1):10–18. | |
Gerhauser I, Weigand M, Hahn K, et al. Lack of schmallenberg virus in ruminant brain tissues archived from 1961 to 2010 in Germany. J Comp Pathol. 2014;150(2–3):151–154. | |
Elliott RM, Blakqori G, van Knippenberg IC, et al. Establishment of a reverse genetics system for Schmallenberg virus, a newly emerged orthobunyavirus in Europe. J Gen Virol. 2013;94(pt 4):851–859. | |
Carpenter S, Mellor PS, Torr SJ. Control techniques for Culicoides biting midges and their application in the UK and northwestern Palaearctic. Med Vet Entomol. 2008;22(3):175–187. | |
Kraatz F, Wernike K, Hechinger S, et al. Deletion mutants of Schmallenberg virus are avirulent and protect from virus challenge. J Virol. 2015;89(3):1825–1837. | |
Elbers AR, Stockhofe-Zurwieden N, van der Poel WH. Schmallenberg virus antibody persistence in adult cattle after natural infection and decay of maternal antibodies in calves. BMC Vet Res. 2014;10:103. | |
Claine F, Coupeau D, Wiggers L, Muylkens B, Kirschvink N. Passive immunity against Schmallenberg virus in lambs born from naturally infected ewes. In: Proceedings of the 8th EPIZONE Annual Meeting; September 23–25, 2014; Copenhagen, Denmark. | |
Martinelle L, Dal Pozzo F, Gauthier B, Kirschvink N, Saegerman C. Field veterinary survey on clinical and economic impact of Schmallenberg virus in Belgium. Transbound Emerg Dis. 2014;61(3):285–288. | |
Saegerman C, Martinelle L, Dal Pozzo F, Kirschvink N. Preliminary survey on the impact of Schmallenberg virus on sheep flocks in South of Belgium. Transbound Emerg Dis. 2014;61(5):469–472. | |
Claine F, Coupeau D, Wiggers L, Muylkens B, Kirschvink N. Emergence and time course of spreading of Schmallenberg virus in late summer 2011 in a Belgian naïve sheep flock. In: Proceedings of the 7th EPIZONE Annual Meeting; October 1–4, 2013; Brussels, Belgium. | |
Veldhuis AM, Santman-Berends IM, Gethmann JM, et al. Schmallenberg virus epidemic: impact on milk production, reproductive performance and mortality in dairy cattle in the Netherlands and Kleve district, Germany. Prev Vet Med. 2014;116(4):412–422. | |
Saegerman C, Mellor P, Uyttenhoef A, et al. The most likely time and place of introduction of BTV8 into Belgian ruminants. PLoS One. 2010;5(2):e9405. | |
Leemans J, Hamers C, Chery R, et al. Interference of colostral antibodies with response to a Bluetongue serotype 8 inactivated vaccine in lambs born from hyperimmune ewes. Vaccine. 2013;31(15):1975–1980. | |
Menzies FD, McCullough SJ, McKeown IM, et al. Evidence for transplacental and contact transmission of bluetongue virus in cattle. Vet Rec. 2008;163:203–209. | |
Saegerman C, Bolkaerts B, Baricalla C, et al. The impact of naturally-occurring, trans-placental bluetongue virus serotype-8 infection on reproductive performance in sheep. Vet J. 2011;187(1):72–80. | |
Vercauteren G, Miry C, Vandenbussche F, et al. Bluetongue virus serotype 8-associated congenital hydranencephaly in calves. Transbound Emerg Dis. 2008;55:293–298. | |
Kirschvink N, Raes M, Saegerman C. Impact of a natural bluetongue serotype 8 infection on semen quality of Belgian rams in 2007. Vet J. 2009;182(2):244–251. | |
Muller U, Kemmerling K, Straet D, Janowitz U, Sauerwein H. Effects of bluetongue virus infection on sperm quality in bulls: a preliminary report. Vet J. 2010;186(3):402–403. | |
Alexander KA, MacLachlan NJ, Kat PW, et al. Evidence of natural bluetongue virus among African carnivores. Am J Trop Med Hyg. 1994;51:568–576. | |
Jauniaux TP, De Clercq KE, Cassart D, et al. Bluetongue in Eurasian Lynx. Emerg Infect Dis. 2008;14(9):1496–1498. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


 colored on a map of Europe (
colored on a map of Europe (