Back to Journals » OncoTargets and Therapy » Volume 9
Ruguo key genes and tumor driving factors identification of bladder cancer based on the RNA-seq profile
Authors Zhang M, Li H, Zou D, Gao J
Received 16 July 2015
Accepted for publication 11 November 2015
Published 5 May 2016 Volume 2016:9 Pages 2717—2723
DOI https://doi.org/10.2147/OTT.S92529
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Daniele Santini
Minglei Zhang,1 Hongyan Li,2 Di Zou,3 Ji Gao2
1Department of Orthopedics, Division of Tumor and Trauma Surgery, 2Department of Urology, China–Japan Union Hospital of Jilin University, 3Department of Nephrology, The First Affiliated Hospital of Changchun University of Chinese Medicine, Changchun, People’s Republic of China
Aim: This study aimed to select several signature genes associated with bladder cancer, thus to investigate the possible mechanism in bladder cancer.
Methods: The mRNA expression profile data of GSE31614, including ten bladder tissues and ten control samples, was downloaded from the Gene Expression Omnibus. The differentially expressed genes (DEGs) in bladder cancer samples compared with the control samples were screened using the Student’s t-test method. Functional analysis for the DEGs was analyzed using the Database for Annotation, Visualization, and Integrated Discovery from the Gene Ontology database, followed by the transcription function annotation of DEGs from Tumor-Associated Gene database. Motifs of genes that had transcription functions in promoter region were analyzed using the Seqpos.
Results: A total of 1,571 upregulated and 1,507 downregulated DEGs in the bladder cancer samples were screened. ELF3 and MYBL2 involved in cell cycle and DNA replication were tumor suppressors. MEG3, APEX1, and EZH2 were related with the cell epigenetic regulation in bladder cancer. Moreover, HOXB9 and EN1 that have their own motif were the transcription factors.
Conclusion: Our study has identified several key genes involved in bladder cancer. ELF3 and MYBL2 are tumor suppressers, HOXB9 and EN1 are the main regulators, while MEG3, APEX1, and EZH2 are driving factors for bladder cancer progression.
Keywords: bladder cancer, differentially expressed genes, tumor driving factor, function analysis
Introduction
Bladder cancer is one of the most common malignancies in epithelial and mesenchymal tissues of human’s urinary system and contributes to the increasing morbidity and mortality.1 Statistics data refer that approximately 74,000 new cases were diagnosed as bladder cancer in 2014.2 Treatment methods such as surgery, chemotherapy, and drug use play significant role in improving quality of life of patients with bladder cancer.3,4 However, prognosis of bladder cancer is unsatisfactory, with the poor 5-year survival rate due to difficulty in diagnosing and easy metastasis.4 Therefore, it will be of great significance to explore several signature genes and biomarkers for the target treatment and diagnosis of bladder cancer.
Recently, research on the signature genes and transcription factors (TFs), which are closely associated with bladder cancer, is becoming the hot spot. For instance, Dyrskjøt et al5 investigated that 12 gene signatures were associated with nonmuscle invasive bladder cancer prognosis, such as UBE2C, COL18A1, SKAP2, and NEK1. According to Riester et al,6 several tumor metastasis, tumor stage, and progression genes such as FN1, NNMT, POSTN, and SMAD6 are the prognostic signatures for high-risk bladder cancer. Dancik et al7 identified that genetic alterations of TP53, HRAS, KEM6A, and FGFR3 were the diagnostic signature genes that will drive the muscle invasive bladder cancer. Besides, increasing evidences suggest that motifs with the conserved short sequences in the upstream regions of gene promoters play crucial roles in gene transcription process in many cancers, such as bladder cancer,8 prostate cancer,9 and colorectal cancer.10 Rachakonda et al11 reported that mutation of the telomerase reverse transcriptase (TERT) promoter was a potential prognostic target for bladder cancer in clinical application. Despite many studies on the exploration of the prognostic biomarkers in bladder cancer, the mechanism for poor diagnosis and prognoses of bladder cancer still remain largely unknown.
In this study, bioinformatics approaches were used to screen the differentially expressed genes (DEGs) and driving factors that may be helpful for the bladder cancer treatment and prognosis based on the RNA sequencing profile data. The significant functions of the DEGs and the motifs of crucial genes in bladder cancer were analyzed to predict the key genes and driving factors for bladder cancer. This study aimed to explore several signature genes for the target treatment of bladder cancer and to investigate the mechanism of bladder cancer. Our study may provide theoretical basis for the diagnostic and therapeutic exploration for bladder cancer treatment.
Methods
Data resources and preprocessing
The mRNA expression profile data of GSE31614 was downloaded from the Gene Expression Profile in National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/geo/) based on the platform of GPL9115 Illumina Genome Analyzer II (Homo sapiens). The study contains the mRNA expression of cancer and matched adjacent tissues of seven testicular germ cell tumors and ten transitional cell carcinomas of bladder. In this study, the ten bladder cancer samples matched with ten matched normal samples were chosen for subsequent analysis.
Reads alignment and transcript expression analysis
The reads of genes sequences in the genome of patients with bladder cancer were aligned with the hg19 genome sequence deposited in the University of California Santa Cruz Genome Browser Database using the Bowtie software.12 Read with only one alignment result and with the base mismatch no more than two was selected. In addition, the expression values of genes in each transcript in each sample were calculated using the Cufflinks tool,13 based on the fragments per kilobase per million mapped fragments method.
Screening of DEGs
The DEGs in bladder cancer samples compared with matched normal samples were identified using the Student’s t-test method14 in R. P<0.05 and |log 2 fold change| ≥2 were chosen as the threshold.
Function annotation of DEGs
The gene ontology functions of the selected DEGs, including biological process (BP) and cellular component, were analyzed using the Database for Annotation, Visualization, and Integrated Discovery software.15 The false discovery rate <0.005 was chosen as the cutoff criterion. Besides, the genes that have the transcription regulatory functions were analyzed and selected based on the information of transcript database. Finally, genes that functions as oncogenes or tumor suppressors were selected based on the information of Tumor-Associated Gene database (http://sourceforge.jp/projects/sfnet_tdt/).
Prediction of the upstream regulatory elements of DEGs
The region in one gene from upstream 1 kb to the downstream 0.5 kb of the transcription start site was considered as the promoter region in this study. The motif of the selected DEGs in the promoter region was selected using the Seqpos.16 P<0.00001 and z score <−7 were chosen as the threshold.
Results
DEGs screening
With P<0.05 and |log 2 fold change| ≥2 as the threshold, a total of 1,571 upregulated and 1,507 downregulated DEGs in the bladder cancer samples were compared with matched normal samples using Student’s t-test method. Besides, 57 upregulated DEGs and 79 downregulated DEGs were analyzed as the TFs based on the transcript database analysis. Three upregulated genes such as E74-like factor 3 (ELF3), v-myb avian myeloblastosis viral oncogene homologue-like 2 (MYBL2), and MYCN were the known oncogenes, while three downregulated genes, including EHF, FNACB, and PML were the known tumor suppressors.
Functional analysis for the DEGs
The selected downregulated DEGs significantly participated in the BP terms such as cell adhesion, extracellular matrix organization, response to endogenous stimulus, and chemical homeostasis (Table 1A). Also, the upregulated genes were mainly involved in BP terms related to cell cycle, such as mitotic cell cycle, cell division, DNA replication, and cell cycle checkpoint (Table 1B).
  | Table 1 The significantly enriched GO terms by the differentially expressed genes in bladder cancer |
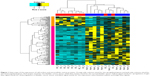
In addition, the heat maps of DEGs showed that there were 35 upregulated genes and 138 downregulated genes in tumor tissues that were associated with the cell surface and extracellular matrix (Figure 1). Genes, such as ITGA6, LAMB3, MMP1, TGFA, VEGFA, WNT10A, and WNT7A were all involved in the cancer-related pathways, suggesting these genes might be involved in the bladder cancer progression.
Finally, we selected 12 genes that were associated with the cell epigenetic regulation in bladder cancer tissues (Figure 2). Genes such as maternally expressed 3 (MEG3), PLD6, SMCHD1, and ZFP36 were downregulated, while APEX nuclease multifunctional DNA repair enzyme 1 (APEX1), APOBEC3C, APOBEC3F, ATF7IP, CHEK1, CNOT6, enhancer of zeste homologue 2 Drosophila (EZH2), and GATAD2A were upregulated in the cancer tissues.
  | Figure 2 Abnormal expression values of genes associated with the apparent regulation in bladder cancer tissues. |
Enrichment analysis of motif in TFs
A total of nine genes from the upregulated DEGs and ten genes from the downregulated DEGs were respectively selected, which have the motifs in the upstream sequence based on the motif enrichment analysis of the TFs (Figure 3). We selected the DNA motif for upregulated genes, such as CAATAAAA for homeobox B9 (HOXB9), TAATTA for engrailed homeobox 1 (EN1), and AGGAAGGGAAAGGA for PRDM1 (Figure 3A), while the motif for downregulated genes were TTTCT for NR3C1, TGTTCT for E2F6, and ACATGCGACATG for TP53 (Figure 3B), suggesting their potential tumor driving roles in bladder cancer (Figure 4).
  | Figure 4 Motif enrichment of transcription factors in bladder cancer. |
Discussion
Bladder cancer is one of the most common malignancies of the human urinary system, with increasing morbidity and mortality due to difficulty in diagnosing and easy metastasis. To explore several signature genes and biomarkers for the target treatment, diagnosis of bladder cancer will be of great significance. In this study, we analyzed the mRNA expression profile of bladder cancer to screen the signature genes and key driving factors for bladder cancer diagnosis and target treatment based on the RNA sequencing. The upregulated ELF3 and MYBL2 involved in cell cycle and DNA replication were the tumor suppressors. Downregulated MEG3 and upregulated APEX1 and EZH2 were involved in the cell epigenetic regulation in bladder cancer. Moreover, HOXB9 and EN1 that have their own motifs were the TFs in bladder cancer samples.
ELF3 is a member of the E26 transformation-specific (ETS) family that can bind and transactivate ETS sequences containing the consensus nucleotide core sequence GGA.17 Seth and Watson18 proved that ETS TFs could activate or repress the expression of genes that are involved in the cell proliferation, differentiation, and development and apoptosis of tumor cells in many cancers. Overexpression of ELF3 induces the endogenous transforming growth factor beta (TGF-β) type II receptor expression in human breast cancer cells,19 and the TGF-β type II receptor is necessary for mediating the effects of TGF-β on tumor cell growth inhibition.20 Thus, ELF3 may be associated with tumor cell growth. Also, ELF3 has been reported as the candidate transcription regulator involved in human urinary cytodifferentiation.21 In this study, the upregulated ELF3 was selected as the tumor suppressor gene, indicating the inhibitory roles in bladder cancer. Meanwhile, MYBL2 is a member of the MYB family, which is a nuclear protein involved in cell cycle progression.22 Sala23 proved that MYBL2 regulated the cell cycle of cancer cells to affect the tumorigenesis. Also, Wu et al24 reported that genetic variation in cell cycle control genes were related to the increased risk of bladder cancer. Our data showed that the upregulated MYBL2 was involved in the cell cycle function in bladder cancer samples, suggesting that MYBL2 might be the tumor suppressor gene for bladder cancer risk through cell cycle.
Our findings showed that the upregulated HOXB9 and EN1 that have their own motifs were the TFs in bladder cancer samples. HOXB9 is a Abd-B homeobox family protein with a homeobox DNA-binding domain on chromosome 17, while EN1 has a role in controlling development during the central nervous system.25,26 However, it has been reported that overexpression of HOXB9 promoted the cell proliferation and angiogenesis that was related to breast cancer.27 Also, high level of HOXB9 in breast cancer induces the expressions of several angiogenic factors such as interleukin-8, vascular endothelial growth factor, and ErbB to activate their respective pathways, leading to the lung cancer metastasis.28 Besides, Bell et al29 demonstrated that significant hypermethylation of EN1 at the transcriptional start site was observed, suggesting it may be a novel biomarker for poor prognosis of human salivary gland adenoid cystic carcinoma. Therefore, HOXB9 may contribute to tumorigenesis and EN1 may be related to cancer diagnosis. On the other hand, E2F1 is the candidate gene that binds to the promoter region of HOXB9 to induce its overexpression and then accelerated breast cancer progression.30 EN1 has the Pax5 binding site on the promoter region and Pax2 could regulate the enhancer of Pax5 at the midbrain–hindbrain boundary.31,32 Also, inhibited Pax2 results in cell death of prostate cancer.33 Based on our results, we speculate that HOXB9 with the conserved motif CAATAAAA may promote bladder cancer development, while EN1 with DNA sequence TAATTA might be related to bladder cancer diagnosis.
Meanwhile, our results displayed that the downregulated MEG3 and upregulated APEX1 and EZH2 were the genes involved in the cell epigenetic regulation in bladder cancer samples compared with the controls, suggesting their important roles in bladder cancer. Ying et al34 proved that downregulation of MEG3 activated autophagy and increased cell proliferation of bladder cancer by affecting the chromosome. Role of APEX1 and EZH2 in bladder cancer have not been fully reported in previous researches. However, Kim et al35 proved that APEX1 was the driving factor for colon cancer progression, while Varambally et al36 referred that EZH2 was involved in prostate cancer progression. Based on our study, we speculate that MEG3, APEX1, and EZH2 may be the tumor driving factors for bladder cancer progression.
Conclusion
Our findings indicate that ELF3, MYBL2, HOXB9, EN1, MEG3, APEX1, and EZH2 are identified as key genes involved in bladder cancer progression or development. ELF3 and MYBL2 are the key genes that play crucial roles in suppressing bladder cancer, while MEG3, APEX1, and EZH2 are the driving factors for bladder cancer progression. Also, HOXB9 and EN1 are the TFs that play key roles in bladder cancer development and progression. Our study may provide theoretical basis for the future bladder cancer investigation. However, there were still some limitations in the current study. First, the sample size enrolled in our study was small. Second, in vivo and in vitro experimental verification were not performed to validate the functions of crucial genes in bladder cancer samples. Further studies based on a larger sample size and experiments, such as expression validation or knockdown assay in bladder cancer cell lines, are still needed to confirm our results.
Disclosure
Hongyan Li and Di Zou are regarded as co-second authors. The authors report no conflicts of interest in this work.
References
Chromecki TF, Bensalah K, Remzi M, et al. Prognostic factors for upper urinary tract urothelial carcinoma. Nat Rev Urol. 2011;8(8):440–447. | ||
Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64(1):9–29. | ||
Chung SD, Huang CY, Yu HJ. Radiotherapy plus chemotherapy in muscle-invasive bladder cancer. N Engl J Med. 2012;367(4):379. | ||
James ND, Hussain SA, Hall E, et al. Radiotherapy with or without chemotherapy in muscle-invasive bladder cancer. N Engl J Med. 2012;366(16):1477–1488. | ||
Dyrskjøt L, Reinert T, Novoradovsky A, et al. Analysis of molecular intra-patient variation and delineation of a prognostic 12-gene signature in non-muscle invasive bladder cancer; technology transfer from microarrays to PCR. Br J Cancer. 2012;107(8):1392–1398. | ||
Riester M, Taylor JM, Feifer A, et al. Combination of a novel gene expression signature with a clinical nomogram improves the prediction of survival in high-risk bladder cancer. Clin Cancer Res. 2012;18(5):1323–1333. | ||
Dancik GM, Owens CR, Iczkowski KA, Theodorescu D. A cell of origin gene signature indicates human bladder cancer has distinct cellular progenitors. Stem Cells. 2014;32(4):974–982. | ||
Hiwasa K, Nagaya H, Terao S, et al. Improved gene transfer into bladder cancer cells using adenovirus vector containing RGD motif. Anticancer Res. 2012;32(8):3137–3140. | ||
Zhang J, Lu Y, Pienta KJ. Multiple roles of chemokine (CC motif) ligand 2 in promoting prostate cancer growth. J Natl Cancer Inst. 2010;102:522–528. | ||
Xu C, Yang M, Tian J, Wang X, Li Z. MALAT-1: a long non-coding RNA and its important 3′ end functional motif in colorectal cancer metastasis. Int J Oncol. 2011;39(1):169–175. | ||
Rachakonda PS, Hosen I, de Verdier PJ, et al. TERT mutations in bladder cancer affect patient survival and disease recurrence through modification by a common polymorphism. Proc Natl Acad Sci U S A. 2013;110(43):17426–17431. | ||
Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. | ||
Trapnell C, Roberts A, Goff L, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–578. | ||
Ruxton GD. The unequal variance t-test is an underused alternative to Student’s t-test and the Mann–Whitney U test. Behav Ecol. 2006;17(4):688–690. | ||
Huang DW, Sherman BT, Tan Q, et al. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. | ||
Vitorino R, Alves R, Barros A, et al. Finding new posttranslational modifications in salivary proline-rich proteins. Proteomics. 2010;10(20):3732–3742. | ||
Hollenhorst PC, McIntosh LP, Graves BJ. Genomic and biochemical insights into the specificity of ETS transcription factors. Annu Rev Biochem. 2011;80:437–471. | ||
Seth A, Watson DK. ETS transcription factors and their emerging roles in human cancer. Eur J Cancer. 2005;41(16):2462–2478. | ||
Chang J, Lee C, Hahm KB, Yi Y, Choi SG, Kim SJ. Over-expression of ERT (ESX/ESE-1/ELF3), an ETS-related transcription factor, induces endogenous TGF-beta type II receptor expression and restores the TGF-beta signaling pathway in Hs578t human breast cancer cells. Oncogene. 2000;19(1):151–154. | ||
Geiser AG, Burmester JK, Webbink R, Roberts AB, Sporn MB. Inhibition of growth by transforming growth factor-beta following fusion of two nonresponsive human carcinoma cell lines. Implication of the type II receptor in growth inhibitory responses. J Biol Chem. 1992;267(4):2588–2593. | ||
Böck M, Hinley J, Schmitt C, Wahlicht T, Kramer S, Southgate J. Identification of ELF3 as an early transcriptional regulator of human urothelium. Dev Biol. 2014;386(2):321–330. | ||
Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15(10):573–581. | ||
Sala A. B-MYB, a transcription factor implicated in regulating cell cycle, apoptosis and cancer. Eur J Can. 2005;41(16):2479–2484. | ||
Wu X, Gu J, Grossman HB, et al. Bladder cancer predisposition: a multigenic approach to DNA-repair and cell-cycle–control genes. Am J Hum Genet. 2006;78(3):464–479. | ||
Neil Sarkar I, Thornton JW, Planet PJ, Figurski DH, Schierwater B, DeSalle R. An automated phylogenetic key for classifying homeoboxes. Mol Phylogenet Evol. 2002;24(3):388–399. | ||
Hatten ME. Central nervous system neuronal migration. Annu Rev Neurosci. 1999;22(1):511–539. | ||
Seki H, Hayashida T, Jinno H, et al. HOXB9 expression promoting tumor cell proliferation and angiogenesis is associated with clinical outcomes in breast cancer patients. Ann Surg Oncol. 2012;19(6):1831–1840. | ||
Hayashida T, Takahashi F, Chiba N, et al. HOXB9, a gene overexpressed in breast cancer, promotes tumorigenicity and lung metastasis. Proc Natl Acad Sci. 2010;107(3):1100–1105. | ||
Bell D, Bell A, Roberts D, Weber RS, El-Naggar AK. Developmental transcription factor EN1 – a novel biomarker in human salivary gland adenoid cystic carcinoma. Cancer. 2012;118(5):1288–1292. | ||
Zhussupova A, Hayashida T, Takahashi M, Hiromitsu J, Kitagawa Y. Abstract P1-07-06: E2F-1 directly induces HOXB9 accelerating breast cancer progression. Can Res. 2013;73(24 Suppl):P1-07. | ||
Wurst W, Bally-Cuif L. Neural plate patterning: upstream and downstream of the isthmic organizer. Nat Rev Neurosci. 2001;2(2):99–108. | ||
Pfeffer PL, Bouchard M, Busslinger M. Pax2 and homeodomain proteins cooperatively regulate a 435 bp enhancer of the mouse Pax5 gene at the midbrain-hindbrain boundary. Development. 2000;127(5):1017–1028. | ||
Gibson W, Green A, Bullard RS, Eaddy AC, Donald CD. Inhibition of PAX2 expression results in alternate cell death pathways in prostate cancer cells differing in p53 status. Cancer Lett. 2007;248(2):251–261. | ||
Ying L, Huang Y, Chen H, et al. Downregulated MEG3 activates autophagy and increases cell proliferation in bladder cancer. Mol Biosyst. 2013;9(3):407–411. | ||
Kim MH, Kim HB, Yoon SP, et al. Colon cancer progression is driven by APEX1-mediated upregulation of Jagged. J Clin Invest. 2013;123(8): 3211–3230. | ||
Varambally S, Dhanasekaran SM, Zhou M, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419(6907):624–629. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.