Back to Journals » OncoTargets and Therapy » Volume 12
RRM2 Regulated By LINC00667/miR-143-3p Signal Is Responsible For Non-Small Cell Lung Cancer Cell Progression
Authors Yang Y, Li S, Cao J, Li Y, Hu H, Wu Z
Received 29 June 2019
Accepted for publication 4 October 2019
Published 20 November 2019 Volume 2019:12 Pages 9927—9939
DOI https://doi.org/10.2147/OTT.S221339
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Leo Jen-Liang Su
Yanbing Yang,1,* Sensen Li,2,* Juan Cao,1 Yaojun Li,1 Haiying Hu,1 Zhuyu Wu1
1Department of Respiratory Medicine, Luohe Central Hospital, The First Affiliated Hospital of Luohe Medical College, Luohe, Henan 462000, People’s Republic of China; 2Department of Pharmacy, Luohe Central Hospital, The First Affiliated Hospital of Luohe Medical College, Luohe, Henan 462000, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Haiying Hu; Zhuyu Wu Email [email protected]; [email protected]
Background: Non-small cell lung cancer (NSCLC) is a common and fatal cancer worldwide with a very low 5-year overall survival rate. Ribonucleotide reductase M2 subunit (RRM2), a small subunit of the ribonucleotide reductase complex, has been found to be an oncogenic role in a variety of tumors including NSCLC. However, the regulatory mechanism of RRM2 in NSCLC is not clear. Increasing evidence suggests that non-coding RNAs (ncRNAs) including miRNAs and lincRNAs may promote or inhibit tumor initiation and development through regulating the expression of oncogenic genes. It is interesting to find ncRNAs which play important role in regulating RRM2 expression.
Materials and methods: The expression levels of RRM2, LINC0066 and miR-143-3p in NSCLC tumor tissues and cell lines were detected using qRT-PCR. The regulatory relationships among RRM2, LINC0066 and miR-143-3p were predicted using database analysis and verified by luciferase reporter assay and RIP analysis. The proliferation ability of NSCLC cells was assessed using CCK8 and colony formation assays. The expression of related proteins was determined by Western blot. In vivo effect of RRM2, LINC0066 and miR-143-3p to NSCLC were detected through xenograft experiments.
Results: In this study, we found RRM2 was upregulated in NSCLC tumor and cell lines, and the aberrant upregulation predicted a poor prognosis. Then, we predicted and confirmed that RRM2 was negatively regulated by miR-143-3p. Further study implied that LINC00667 acted as a ceRNA by sponging miR-143-3p and regulated RRM2 expression indirectly. Moreover, we found that the growth of NSCLC was regulated by LINC00667/miR-143-3p/RRM2 signal pathway both in vitro and in vivo. LINC00667 and RRM2 promoted the tumor growth while miR-143-3p inhibited it.
Conclusion: Our study revealed a LINC00667/miR-143-3p/RRM2 signal pathway that played an important role in the progress of NSCLC, which might be potential therapeutic targets for NSCLC.
Keywords: NSCLC, RRM2, miR-143-3p, LINC00667
Introduction
Lung cancer which could be divided into small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC) is the most common cancer worldwide.1,2 NSCLC initiating from non-small cells of the lung accounts for approximately 85% of all lung cancer cases.3 Although plenty of therapeutic strategies such as surgical therapy, chemotherapy, radiotherapy have been used, the 5-year overall survival rate is still lower than 20%.4 Therefore, the molecular mechanism in the occurrence and progression of NSCLC should be further explored for developing novel therapeutic strategy.
Ribonucleotide reductase M2 subunit (RRM2), a small subunit of the ribonucleotide reductase complex, is a rate-limiting enzyme responsible for dNTP producing.5 The expression of RRM2 is cell-cycle dependent and reaches its highest level during S-phase.6 RRM2 acts as a oncogenic role under pathological conditions and overexpressed RRM2 has been found in various cancers, including gastric carcinoma, clear-cell renal cell carcinoma and melanoma.7–9 In NSCLC, RRM2 upregulation was associated with poor prognosis and low survival.10 Increasing evidences demonstrate that RRM2 could be a novel target for cancer treatment. However, the regulatory mechanism of RRM2 remains unclear.
Non-coding RNAs (ncRNAs) represent RNA molecules which are transcribed from genome but do not encode proteins, including microRNAs (miRNAs), long non-coding RNAs (lincRNAs), circle RNA (circRNA) and so on.11 Although ncRNAs do not encode proteins, they can directly or indirectly regulate gene expression at epigenetic, transcriptional and post-transcriptional levels. MiRNAs, a small endogenous ncRNA containing about 21–24 nucleotides, can negatively regulate gene expression through targeting the 3ʹ-untranslated region (UTR) of mRNAs.12 Several miRNAs such as miRNA-92a, miR-204, and miRNA-455 have been reported to regulate NSCLC carcinogenesis.13–15 LincRNAs, another kind of ncRNAs containing about more than 200 nucleotides, participate in various epigenetic regulatory processes.16 Recent studies have raised a new regulatory theory that lincRNAs can act as competing endogenous RNA (ceRNA) to regulate the expression of miRNA targeted genes through sponging miRNAs.17 Several lincRNA/miRNA/target gene signal pathways have been reported in NSCLC. For example, Lu et al revealed linc00673/miR-150-5p/ZEB1 regulated NSCLC proliferation, migration, invasion and epithelial–mesenchymal transition.18 Jia et al reported that lincRNA MAFG-AS1/miR-339/MMP15 signal pathway regulated NSCLC cells growth and metastasis.19
In this study, we found that RRM2 expression was upregulated in NSCLC tumor tissue and cell lines which predicted poor prognosis. RRM2 knockdown could significantly inhibit the growth of NSCLC cell lines. The expression of RRM2 was negatively regulated by miR-143-3p. Further study demonstrated that LINC00667 could act as a ceRNA to modulate RRM2 expression via competitively binding to miR-143-3p. Moreover, we confirmed that LINC00667/miR-143-3p could affect the growth of NSCLC cell lines through regulating the expression of RRM2. Our study elaborated a new LINC00667/miR-143-3p/RRM2 regulatory pathway in the progress of NSCLC, providing a potential therapeutic target for NSCLC.
Materials And Methods
Tissue Specimens
NSCLC tumors were collected from 30 patients by surgical resection in Luohe Central Hospital from April 2017 to October 2018. The tumor mass was isolated from the center and the surrounding lung tissue was isolated from adjacent tissues. The fresh specimens were immediately frozen in liquid nitrogen and stored at −80℃ prior to RNA extraction. This study was approved by the Ethics Committee of Luohe Central Hospital. Informed consents were signed by all patients prior to participating in this study.
Histology
Partial tissue samples were fixed with 4% paraformaldehyde for 2 days, followed dehydrated, embedded in paraffin, and sliced. The slices were stained with hematoxylin and eosin (H&E) and were viewed under the light microscope.
RT-qPCR Analysis
Total RNA was isolated from tissues or cells using TRNzol (TianGen, Beijing, People’s Republic of China). For mRNAs and lincRNA quantification, the reverse transcription was performed using HiScript® II Reverse Transcriptase (Vazyme, Nanjing, People’s Republic of China) in accordance with manufacturer’s instructions. Quantitative real-time PCR was performed using specific primers with GAPDH as an internal control. For miRNA quantification, reverse transcription was performed using miRNA First-Stand cDNA Synthesis Kit (GeneCopoeia, Guangzhou, People’s Republic of China) with specific primers for miRNA-143-3p and U6. Primer sequences were provided in Supplementary Table S1. Real-time PCR reactions were performed on Applied Biosystems 7500 Real-time PCR Systems (Thermo Fisher Scientific, USA). The primer sequences were listed in Supplementary Table S1.
Cell Culture
Human bronchiolar epithelial cell line (BEAS-2B), four human non-small-cell lung cancer cell lines (Calu-3, SPCA-1, H1975, A549) were purchased from Chinese Academy of Sciences. All cell lines were cultured in RPMI 1640 medium (Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum at 37°C and 1% penicillin/streptomycin in a humidified air with 5% CO2.
siRNA, miRNA And Plasmid DNA Transfection
Negative control siRNA, siRNAs targeting RRM2 and LINC00667, miRNA mimics and inhibitors were designed and synthesized by Sangon Biotech (Shanghai, People’s Republic of China). The siRNA sequences were listed in Supplementary Table S1. sh-RNAs targeting RRM2 and LINC00667, sh-NCs, sequences of LINC00667 cDNA were constructed in pcDNA3.1 by GenePharma (Shanghai, People’s Republic of China). The 3ʹUTR of RRM2 mRNA and LINC00667 containing the wild-type sequence (WT) or containing mutations in the putative miR-143-3p binding site (Mut) were amplified and cloned into pmirGLO luciferase vector (Promega, Madison, WI, USA), respectively. The generated vectors were named as WT-RRM2-3′UTR or MUT-RRM2-3′UTR, WT-LINC00667-pGLO or MUT-LINC00667-pGLO. SPCA-1 and A549 cells were plated in 24-well culture plates at a concentration of 1×105 cells/well for 24 hrs, then siRNAs, miRNA mimics or plasmid DNAs were transfected with lipofectamine 2000 (Invitrogen, USA) according to the manufacturer′s instruction. The sequences of miRNA are listed in Supplementary Table S2.
Western Blot
Cells were collected and lysed with RIPA Lysis Buffer (Beyotime Biotechnology, Beijing, People’s Republic of China). Protein samples were separated on 8% SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis) and transferred to polyvinylidenedifluoride membrane (Millipore, Billerica, USA) for 3 hrs with transfer buffer (25 mM Tris, 192 mM glycine and 20% [v/v] methanol). The membranes were blocked in 5% non-fat dry milk in TBS with 0.1% Tween 20 for 1 hr at room temperature and incubated with primary antibodies at 4°C overnight, including RRM2 (1:1000, ab57653, Abcam), Cyclin A1 (1:1000, ab53699, Abcam), Cyclin B1 (1:1000, ab72, Abcam), CDK2 (1:1000, ab32174, Abcam), p-AKT (1:2000, 4060, CST), total-AKT (1:2000, 4691, CST), GAPDH (1:2000, 2118S, CST), followed by incubation with an HRP-conjugated anti-mouse or anti-rabbit secondary antibody separately. Finally, the bands were imaged by Tanon 5200 using the BeyoECL Moon (Beyotime Biotechnology, Beijing, People’s Republic of China).
Cell Cytoplasm/Nucleus Fraction Isolation
Isolation of nuclear and cytosolic RNA in SPCA-1 and A549 cells was performed with the PARIS Kit (Thermo Fisher Scientific, USA). U6 and GAPDH served as the nuclear control and cytosolic control separately.
Cell Proliferation And Colony Formation Assays
SPCA-1 and A549 cells were seeded in 24-well culture plates at a concentration of 5×103 cells/well and transfected with indicated plasmids and miRNA mimic for the indicated time points. Cell proliferation analysis was performed using the Cell Counting Kit‑8 (CCK‑8; Dojindo Laboratories, Kumamoto, Japan) according to the manufacturer’s instructions. For colony-forming assays, cells were transfected with indicated plasmids and miRNA mimic and were seeded into 12-well plates (600 cells/wells). Fourteen days later, colonies were fixed by 4% paraformaldehyde and stained by 0.4% crystal violet solution. Pictures were obtained by a camera and number of colonies (diameter >0.5 mm) was counted.
Dual‑luciferase Reporter Assay
SPCA-1 and A549 cells were seeded in 24-well culture plates for 24 hrs, followed by transfected with WT-RRM2-3′UTR or MUT-RRM2-3′UTR, WT-LINC00667-pGLO or MUT-LINC00667-pGLO reporter plasmids, together with miR-143-3p or anti-miR-143-3p mimics. Twenty-four hourslater, cells were lysed and the luciferase activity was measured by the Dual-Luciferase Reporter Assay System (Promega Corp, Madison, WI, USA).
RNA Immunoprecipitation (RIP)
RIP experiments were performed in SPCA-1 and A549 cells using Magna RIPTM RNA Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA, USA). Subsequently, RNA was isolation for the detection of LINC00667 and miR-143-3p.
Xenograft Experiments In Vivo
Sequences of sh-RNA targeting LINC00667 (sh-LINC00667), LINC00667 and miR-143-3p were subcloned into the pLenti6/V5-D-TOPO vector (Invitrogen). The nonsense sequences were also subcloned into the pLenti6/V5-D-TOPO vector, and formed the control sh-NC for sh- LINC00667, the control NC for LINC00667, the control Mock for miR-143-3p. Then, the constructed Lenti-sh-HOTAIR vectors were transfected into A549 cells. A total of 1×107 transfected A549 cells were subcutaneously injected into male BALB/c nude mice (6–7 weeks old, Beijing, People’s Republic of China). There were two mice in our each group. Tumor volumes were measured every 3 days. At 18 days after transplantation, mice were sacrificed. Tumors were harvested, weighed, photographed and frozen for RRM2, Cyclin A1, Cyclin B1, CDK2 and p-AKT expression analysis. The experimental procedures were performed following the Guidelines for Care and Use of Laboratory Animal with the approval of Ethics Committee of Luohe Central Hospital.
Statistical Analysis
Statistical analysis was performed using SPSS 20.0 software (SPSS, Chicago, IL, USA) with Student′s t-test and one-way ANOVA to estimate the significant group differences. P values <0.05 were considered significant. These data are expressed as the mean ± SEM (standard error of mean).
Results
RRM2 Expression Is Upregulated In NSCLC Tissues And Cell Lines
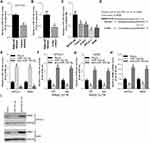
The expression of RRM2 in microarray database was analyzed by STARBASE. As shown in Figure 1A, RRM2 expression level in lung adenocarcinoma patients was remarkably higher than normal controls. The TCGA (Tumor Cancer Genome Atlas) database analysis demonstrated that RRM2 expression was negatively associated with overall survival of NSCLC patients (Figure 1B). We collected the tumor issues from NSCLC patients and the representative images of HE stain of cancer tissues and adjacent tissues are shown in Figure 1C. The mRNA level of RRM2 in cancer tissues and adjacent tissues from NSCLC patients was examined using RT-qPCR analysis. As shown in Figure 1D, the expression of RRM2 was significantly higher in the tumor than atacand tissues. The expression of RRM2 in 4 NSCLC cell lines was also higher than control bronchial epithelial cell line BEAS-2B.
RRM2 Expression Is Negatively Regulated By miR-143-3p
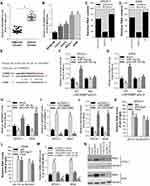
Using the miRNA targets prediction tool PITA, we predicted the 3′UTR of RRM2 could be targeted by miR-143-3p (Figure 2D). The expression of miR-143-3p in cancer tissues and adjacent tissues from NSCLC patients was evaluated by GEO2R analysis. The result demonstrated that miR-143-3p level was lower in tumor than adjacent tissues (Figure 2A). The expression of miR-143-3p in our collected cancer tissues and adjacent tissues was determined by qRT-PCR, and the tendency is consistent with the GEO2R analysis (Figure 2B). The expression of miR-143-3p was also relatively lower in NSCLC cell lines than control cell line (Figure 2C). To confirm that miR-143-3p could target the 3′UTR of RRM2, we constructed RRM2 3′UTR wild type or mutated luciferase plasmids (termed as WT-RRM2-3′UTR or MUT-RRM2-3′UTR) in the pmirGLO dual-luciferase reporter vector (the sequence information is shown in Figure 2D). We first evaluated the overexpression and inhibition effect of miR-143-3p and anti-miR-143-3p mimics in both SPCA-1 and A549 cells, and found that miR-143-3p transfection could significantly improve the level of miR-143-3p while anti-miR-143-3p significantly inhibited the expression of miR-143-3p (Figure 2E). And then luciferase activity was evaluated after co-transfection of miRNA mimics and luciferase plasmids. As Figure 2F and G shows, overexpression of miR-143-3p could reduce the luciferase activity significantly when co-transfection of miR-143-3p and WT-RRM2-3′UTR occurred while inhibition of miR-143-3p significantly increases the luciferase activity when co-transfection of anti-miR-143-3p and WT-RRM2-3′UTR, but the effects were disappeared when miRNAs were co-transfected with MUT-RRM2-3′UTR. We also determined the expression of RRM2 after miR-143-3p and anti-miR-143-3p mimics were transfected into cells. As Figure 2H and I shows, miR-143-3p remarkably inhibited the expression of RRM2 while anti-miR-143-3p had the opposite effect in the mRNA and protein level.
LINC00667 Acted As A ceRNA By Sponging miR-143-3p And Regulated RRM2 Expression Indirectly
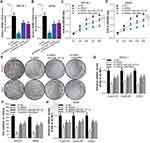
Recently raised ceRNA hypothesis revealed that lincRNA and miRNA could be regulated by each other.17 To find the regulatory lincRNAs of miR-143-3p, we used the STARBASE tool. The prediction results displayed the existence of recognition sites between miR-143-3p and LINC00667 (Figure 3E). We detected the expression of LINC00667 in cancer tissues and adjacent tissues from NSCLC patients. The results revealed that LINC00667 level was higher in cancer tissues compared with adjacent tissues (Figure 3A). The expression of LINC00667 in 4 NSCLC cell lines was also higher than control cell line BEAS-2B (Figure 3B). Additionally, LINC00667 mainly existed in the cytoplasm of SPCA-1 and A549 cells (Figure 3C and D). To validate the association between LINC00667 and miR-143-3p, we also constructed LINC00667 recognition sites wild type or mutated luciferase plasmids (termed as WT-LINC00667-pGLO or MUT-LINC00667-pGLO) in the pmirGLO dual-luciferase reporter vector (the sequence information is shown in Figure 3E), and luciferase activity was evaluated after co-transfection of miRNA mimics and luciferase plasmids. As Figure 3F and G shows, miR-143-3p mimic significantly reduces the luciferase activity when co-transfected with WT-LINC00667-pGLO while anti-miR-143-3p mimic had the opposite effect, but the effects were absent when co-transfected with MUT-LINC00667-pGLO plasmids. The overexpression of miR-143-3p significantly reduces the level of LINC00667 in SPCA-1 and A549 cells while inhibition of miR-143-3p had the opposite effect (Figure 3H). We transfected SPCA-1 and A549 cells with LINC00667 or shRNA plasmid to overexpress or inhibit the expression of LINC00667, as the Figure 3I shows, the expression of LINC00667 was indeedly overexpressed when transfected with LINC00667 plasmid and inhibited when transfected with shRNA plasmid. However, the expression of miR-143-3p was on the contrary (Figure 3J). RIP analysis was performed to evaluate the enrichment degree of LINC00667 and miR-143-3p by Ago2 antibody in SPCA-1 and A549 cells. The results further validated that association between LINC00667 and miR-143-3p (Figure 3K and L). We detected the mRNA and protein expression of RRM2 after LINC00667 and miR-143-3p was transfected alone or together. As Figure 3M and N shows, transfection with LINC00667 alone significantly promoted the expression of RRM2 while co-transfection with miR-143-3p inhibited it. These data reveal that RRM2 expression is regulated by LINC00667/miR-143-3p pathway.
RRM2 Promotes The Growth Of NSCLC Cells And Was Regulated By miR-143-3p And LINC00667
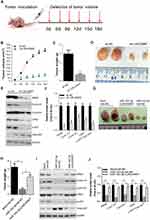
We next evaluate the role of LINC00667/miR-143-3p/RRM2 pathway in NSCLC cells′ growth. The expression of RRM2 was significantly inhibited after sh-RRM2 plasmid was transfected into SPCA-1 and A549 cells, but anti-miR-143-3p or LINC00667 co-transfection could rescue it (Figure 4A and B). The proliferation activity of SPCA-1 and A549 cells were assessed by CCK8 assay when RRM2 signal pathway was inhibited or rescued. As Figure 4C and D shows, RRM2 knockdown significantly inhibits the growth of SPCA-1 and A549 cells while anti-miR-143-3p or LINC00667 co-transfection partially rescues it. RRM2 knockdown also significantly reduces the colony formation ability, and anti-miR-143-3p or LINC00667 co-transfection partially recovered it (Figure 4E and F). Progression of cell cycle is mediated by a series of cyclin-dependent kinases (CDKs) and cyclins. Cyclin A1 and Cyclin B1 can bind to CDK2 and CDK1, respectively, and the formed complex plays an important role in the G2-M phase transition of the cell cycle.20 Overexpression of these genes has been found in various malignant tumors including NSCLC.21–23 AKT is an important signaling molecule involved in cellular survival pathways, and its overexpression is associated with the development of NSCLC.24 The expression of proliferation-related genes such as Cyclin A1, Cyclin B1 and CDK2 was detected by RT-qPCR. The results reveal that these genes were downregulated when RRM2 was suppressed; however, their expression was rescued when anti-miR-143-3p or LINC00667 was co-transfected. These results indicate that LINC00667/miR-143-3p can affect the growth of NSCLC cells through regulating the expression of RRM2.
Attenuation Of LINC00667 Expression Impaired Tumor Proliferation In Vivo
To explore the impact of LINC00667 on tumorigenesis of NSCLC in vivo, SPCA-1 cells infected with lenti-sh-HOTAIR or lenti-sh-NC were inoculated into the BALB/c nude mice to construct xenograft model. Tumor volume and weight was markedly decreased in LINC00667 knockdown group when compared to Lenti-sh-NC control group (Figure 5A and B). Representative images of resected tumor masses in LINC00667 knockdown and Lenti-sh-NC control groups are shown in Figure 5C. The expression of RRM2 and proliferation-related genes was detected by Western blot. As Figure 5D and E shows, LINC00667 knockdown significantly decreases the expression of RRM2, proliferation-related genes such as Cyclin A1, Cyclin B1, CDK2 and p-AKT were also significantly downregulated. In order to further explore the effect of miR-143-3p and the interaction between LINC00667 and miR-143-3p in vivo, we also constructed xenograft models using A549 cell lines with miR-143-3p solely overexpressed or both miR-143-3p and LINC00667 overexpressed. Representative images of resected tumor masses in miR-143-3p overexpression alone or together with LINC00667 and Mock+NC control groups are shown in Figure 5G. Tumor weight was markedly decreased in miR-143-3p overexpression group and the decline is reversed when LINC00667 was overexpressed together (Figure 5I). The expression levels of RRM2, Cyclin A1, Cyclin B1, CDK2 and p-AKT were detected by Western blot. These protumor genes were significantly inhibited when miR-143-3p was overexpressed; however, the inhibited expression is reversed when LINC00667 was overexpressed together. These results confirmed that LINC00667 knockdown or miR-143-3p overexpression impaired tumor proliferation in vivo.
Discussion
Since typically asymptomatic early stage, most of the lung cancers are diagnosed at advanced stage, and the 5-year survival rate for NSCLC remains poor.1 Although enormous research efforts have been put into the carcinogenesis and pathophysiology of NSCLC, effective treatment is still very scarce. Although several targeted and immunotherapy drugs such as angiogenesis inhibitors,25 PD-1 inhibitors were available,26 most of them are only useful in treating some certain types of NSCLC. It is urgent to develop novel therapeutic strategies. ncRNAs extensively participate in various biological processes including cancer. In the present study, we confirmed that LINC00667/miR-143-3p/RRM2 signal pathway played an important role in the tumorigenesis of NSCLC. Firstly, we found that the expression of RRM2 was upregulated in NSCLC tumor and cell lines, and the aberrant upregulation predicted a poor prognosis. Then, we predicted and confirmed that RRM2 was regulated by miR-143-3p, and miR-143-3p was regulated by LINC00667. Therefore, in this study we revealed three potential targets for developing new drugs including small molecule inhibitors or nucleotide drugs such as siRNAs.
Uncontrolled proliferation of cancers depends on sufficient dNTPs supply.5 As a key enzyme regulating dNTP pool, increased RRM2 expression has been found in several cancer types, including NSCLC, clear-cell renal cell carcinoma, gastric adenocarcinoma, breast cancer and melanoma.8–10,27,28 In the present study, we found the expression of RRM2 in lung cancer was upregulated and predicted a poor prognosis through analyzing microarray databases. We also detected the expression of RRM2 in cancer tissues of NSCLC patients and NSCLC cell lines and found its expression was significantly upregulated. These results further confirm the oncogenic role of RRM2 in NSCLC.
Then, we predicted and confirmed that RRM2 was regulated by miR-143-3p. Aberrant expression of miR-143-3p has been reported in several cancers. For example, Hou et al reported that miR-143-3p inhibits the tumorigenesis of human breast cancer cells by modulating the expression of MAPK7.29 Ding et al found that miR-143-3p can inhibit the development of colorectal cancer.30 Li et al performed an integrative analysis of 32 miRNAs between NSCLC tissue and non-cancerous lung tissue and found miR-143-3p were downregulated more than two folds in the NSCLC.31 The result of dual‑luciferase reporter assay further confirmed that miR-143-3p could target the 3′UTR of RRM2. What is more, the expression of RRM2 was suppressed when miR-143-3p was overexpressed while miR-143-3p knockdown significantly increased the expression of RRM2.
Emerging evidence reveals that lincRNAs can act as a ceRNA to sponge miRNAs and then regulate the expression of the targets of miRNA. In this study, LINC00667 was predicted and confirmed to be a ceRNA of miR-143-3p. LINC00667 has been reported to be associated with survival of small hepatocellular carcinoma patients,32 but its role in NSCLC remains unclear. Database analysis and RT-qPCR detection results demonstrated that LINC00667 was highly expressed in cancer tissues and NSCLC cell lines. The result of dual‑luciferase reporter assay further validated the association between LINC00667 and miR-143-3p. Moreover, we found that the expression of LINC00667 and miR-143-3p was antagonistic to each other in NSCLC cell lines. RIP analysis further confirmed the association between the two ncRNAs. The expression of RRM2 was regulated by both LINC00667 and miR-143-3p, and all three molecules synergistically control the growth of NSCLC cell lines. In vivo experiments also discovered that LINC00667 knockdown or miR-143-3p overexpression could decrease the expression of RRM2 and proliferation-related genes, therefore inhibiting the growth of tumor. Recently Zhou et al reported that LINC00667 was one of seven lincRNAs involved in the progression of NSCLS, which is consistent with our current findings.33
In conclusion, our study revealed the LINC00667/miR-143-3p/RRM2 signal pathway which played an important role in the progress of NSCLC. LINC00667 could act as a ceRNA regulating the expression of RRM2 through sponging miR-143-3p (Figure 6). Proper regulation of these molecules could inhibit the growth of NSCLC both in vitro and in vivo, indicating they might be potential therapeutic targets for NSCLC.
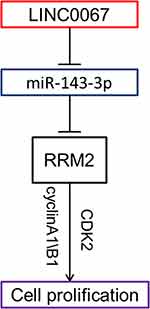 |
Figure 6 Schematic plot showed LINC00667/miR-143-3p/RRM2 signal pathway played an important role in the progress of NSCLC. |
Acknowledgments
This work is financially supported by research grants from Luohe Medical College 2019 Innovation and Entrepreneurship Development Ability Enhancement Engineering Research Project (2019-LYZKYYB009), Henan Medical Science and Technology Co-construction Project (2018021024), and Luohe Youth Top Talents Fund (2018QNBJRC01005).
Disclosure
The authors declare that they have no competing interests regarding this work.
References
1. Miller KD, Nogueira L, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin. 2019;69(5):363–385.
2. Laskin JJ, Sandler AB. State of the art in therapy for non-small cell lung cancer. Cancer Invest. 2005;23:427–442. doi:10.1081/CNV-67172
3. Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83:584–594. doi:10.1016/S0025-6196(11)60735-0
4. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–386. doi:10.1002/ijc.29210
5. Aye Y, Li M, Long MJ, Weiss RS. Ribonucleotide reductase and cancer: biological mechanisms and targeted therapies. Oncogene. 2015;34:2011–2021. doi:10.1038/onc.2014.155
6. Engstrom Y, Eriksson S, Jildevik I, Skog S, Thelander L, Tribukait B. Cell cycle-dependent expression of mammalian ribonucleotide reductase. Differential regulation of the two subunits. J Biol Chem. 1985;260:9114–9116.
7. Morikawa T, Hino R, Uozaki H, et al. Expression of ribonucleotide reductase M2 subunit in gastric cancer and effects of RRM2 inhibition in vitro. Hum Pathol. 2010;41:1742–1748. doi:10.1016/j.humpath.2010.06.001
8. Zou Y, Zhou J, Xu B, Li W, Wang Z. Ribonucleotide reductase subunit M2 as a novel target for clear-cell renal cell carcinoma. Onco Targets Ther. 2019;12:3267–3275. doi:10.2147/OTT.S196347
9. Zuckerman JE, Hsueh T, Koya RC. Davis ME and Ribas A. siRNA knockdown of ribonucleotide reductase inhibits melanoma cell line proliferation alone or synergistically with temozolomide. J Invest Dermatol. 2011;131:453–460. doi:10.1038/jid.2010.310
10. Grossi F, Dal Bello MG, Salvi S, et al. Expression of ribonucleotide reductase subunit-2 and thymidylate synthase correlates with poor prognosis in patients with resected stages I-III non-small cell lung cancer. Dis Markers. 2015;2015:302–649. doi:10.1155/2015/302649
11. ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57-74
12. Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi:10.1101/gr.082701.108
13. Wang P, Lv HY, Zhou DM, Zhang EN. miR-204 suppresses non-small-cell lung carcinoma (NSCLC) invasion and migration by targeting JAK2. Genet Mol Res. 2016;15:10–4238.
14. Li YJ, Ping C, Tang J, Zhang W. MicroRNA-455 suppresses non-small cell lung cancer through targeting ZEB1. Cell Biol Int. 2016;40:621–628. doi:10.1002/cbin.10584
15. Ren P, Gong F, Zhang Y, Jiang J, Zhang H. MicroRNA-92a promotes growth, metastasis, and chemoresistance in non-small cell lung cancer cells by targeting PTEN. Tumour Biol. 2016;37:3215–3225. doi:10.1007/s13277-015-4150-3
16. Novikova IV, Hennelly SP, Tung CS, Sanbonmatsu KY. Rise of the RNA machines: exploring the structure of long non-coding RNAs. J Mol Biol. 2013;425:3731–3746. doi:10.1016/j.jmb.2013.02.030
17. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi:10.1016/j.cell.2011.07.014
18. Lu W, Zhang H, Niu Y, et al. Long non-coding RNA linc00673 regulated non-small cell lung cancer proliferation, migration, invasion and epithelial mesenchymal transition by sponging miR-150-5p. Mol Cancer. 2017;16:118. doi:10.1186/s12943-017-0685-9
19. Jia YC, Wang JY, Liu YY, Li B, Guo H, Zang AM. LncRNA MAFG-AS1 facilitates the migration and invasion of NSCLC cell via sponging miR-339-5p from MMP15. Cell Biol Int. 2019;43:384–393. doi:10.1002/cbin.v43.4
20. Pagano M, Pepperkok R, Verde F, Ansorge W, Draetta G. Cyclin A is required at two points in the human cell cycle. Embo J. 1992;11:961–971. doi:10.1002/embj.1992.11.issue-3
21. Cho NH, Choi YP, Moon DS, et al. Induction of cell apoptosis in non-small cell lung cancer cells by cyclin A1 small interfering RNA. Cancer Sci. 2006;97:1082–1092. doi:10.1111/cas.2006.97.issue-10
22. Soria JC, Jang SJ, Khuri FR, et al. Overexpression of cyclin B1 in early-stage non-small cell lung cancer and its clinical implication. Cancer Res. 2000;60:4000–4004.
23. Cai F, Zhu Q, Miao Y, Shen S, Su X, Shi Y. Desmoglein-2 is overexpressed in non-small cell lung cancer tissues and its knockdown suppresses NSCLC growth by regulation of p27 and CDK2. J Cancer Res Clin Oncol. 2017;143:59–69. doi:10.1007/s00432-016-2250-0
24. Fumarola C, Bonelli MA, Petronini PG, Alfieri RR. Targeting PI3K/AKT/mTOR pathway in non small cell lung cancer. Biochem Pharmacol. 2014;90:197–207. doi:10.1016/j.bcp.2014.05.011
25. Malapelle U, Rossi A. Emerging angiogenesis inhibitors for non-small cell lung cancer. Expert Opin Emerg Drugs. 2019;24:71–81. doi:10.1080/14728214.2019.1619696
26. Liu T, Ding S, Dang J, Wang H, Chen J, Li G. First-line immune checkpoint inhibitors for advanced non-small cell lung cancer with wild-type epidermal growth factor receptor (EGFR) or anaplastic lymphoma kinase (ALK): a systematic review and network meta-analysis. J Thorac Dis. 2019;11:2899–2912. doi:10.21037/jtd
27. Kang W, Tong JH, Chan AW, et al. Targeting ribonucleotide reductase M2 subunit by small interfering RNA exerts anti-oncogenic effects in gastric adenocarcinoma. Oncol Rep. 2014;31:2579–2586. doi:10.3892/or.2014.3148
28. Chen WX, Yang LG, Xu LY, et al. Bioinformatics analysis revealing prognostic significance of RRM2 gene in breast cancer. Biosci Rep. 2019;39.
29. Hou Y, Feng H, Jiao J, et al. Mechanism of miR-143-3p inhibiting proliferation, migration and invasion of osteosarcoma cells by targeting MAPK7. Artif Cells Nanomed Biotechnol. 2019;47:2065–2071. doi:10.1080/21691401.2019.1620252
30. Ding X, Du J, Mao K, Wang X, Ding Y, Wang F. MicroRNA-143-3p suppresses tumorigenesis by targeting catenin-delta1 in colorectal cancer. Onco Targets Ther. 2019;12:3255–3265. doi:10.2147/OTT.S184118
31. Li C, Yin Y, Liu X, Xi X, Xue W, Qu Y. Non-small cell lung cancer associated microRNA expression signature: integrated bioinformatics analysis, validation and clinical significance. Oncotarget. 2017;8:24564–24578. doi:10.18632/oncotarget.15596
32. Gu J, Zhang X, Miao R, et al. A three-long non-coding RNA-expression-based risk score system can better predict both overall and recurrence-free survival in patients with small hepatocellular carcinoma. Aging. 2018;10:1627–1639. doi:10.18632/aging.v10i7
33. Zhou W, Liu T, Saren G, Liao L, Fang W, Zhao H. Comprehensive analysis of differentially expressed long non-coding RNAs in non-small cell lung cancer. Oncol Lett. 2019;18:1145–1156. doi:10.3892/ol.2019.10414
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.