Back to Journals » OncoTargets and Therapy » Volume 14
RPN2 Predicts Poor Prognosis and Promotes Bladder Cancer Growth and Metastasis via the PI3K-Akt Pathway
Authors Han C, Chen S, Ma H, Wen X, Wang Z , Xu Y, Jin X, Yu X , Wang M
Received 8 January 2021
Accepted for publication 20 February 2021
Published 3 March 2021 Volume 2021:14 Pages 1643—1657
DOI https://doi.org/10.2147/OTT.S300480
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Gaetano Romano
Chenglin Han,1 Shuxiao Chen,2 Haiyang Ma,3 Xiangchuan Wen,4 Zilong Wang,1 Yingkun Xu,1 Xunbo Jin,1,5 Xiao Yu,5 Muwen Wang1,5
1Department of Urology, Shandong Provincial Hospital, Cheeloo College of Medicine, Shandong University, Jinan, Shandong, People’s Republic of China; 2Department of Vascular Surgery, Shandong Provincial Hospital, Cheeloo College of Medicine, Shandong University, Jinan, Shandong, People’s Republic of China; 3Department of Thoracic Surgery, Shandong Provincial Hospital, Cheeloo College of Medicine, Shandong University, Jinan, Shandong, People’s Republic of China; 4Department of Neurology, Shandong Provincial Hospital, Cheeloo College of Medicine, Shandong University, Jinan, Shandong, People’s Republic of China; 5Department of Urology, Shandong Provincial Hospital Affiliated to Shandong First Medical University, Jinan, Shandong, People’s Republic of China
Correspondence: Xiao Yu
Department of Urology, Shandong Provincial Hospital Affiliated to Shandong First Medical University, Jingwu Road, Huaiyin District, Jinan City, Shandong Province, People’s Republic of China
Tel +86-15168889682
Email [email protected]
Muwen Wang
Department of Urology, Shandong Provincial Hospital, Cheeloo College of Medicine, Shandong University, 9677 Jingshidong Road, Jinan City, Shandong Province, People’s Republic of China
Tel +86-15168886899
Email [email protected]
Background: Ribophorin II (RPN2) is a highly conserved glycoprotein involved in the N-linked glycosylation of multiple proteins. RPN2 was reported to be associated with malignant phenotype in several tumors. However, the function of RPN2 in bladder cancer (BCa) remains unclear.
Methods: Expression of RPN2 in BCa and adjacent tissues was compared by bioinformatics analysis, immunohistochemistry, and Western blotting. qRT-PCR was performed to explore the correlation between RPN2 expression and various clinical features in 38 patients. We assessed the effects of RPN2 on the biological activity of BCa both in vitro and in vivo, and explored its potential mechanisms based on gene set enrichment analysis (GSEA).
Results: We found that RPN2 was highly expressed in human BCa compared with normal adjacent tissues. There was a significant positive correlation between higher RPN2 mRNA levels and tumor T stage, lymph node (LN) metastasis and the degree of pathological differentiation in 38 patients with BCa. We further demonstrated that RPN2 silencing inhibited the growth and metastasis of BCa both in vitro and in vivo. Western blotting revealed that RPN2 knockdown suppressed epithelial-mesenchymal transition (EMT) and inhibited the PI3K-Akt pathway.
Conclusion: These data suggest that RPN2 functions as an oncogene to promote tumor development and is a promising prognostic factor and therapeutic target in BCa.
Keywords: bladder cancer, RPN2, poor prognosis, EMT, PI3K-Akt pathway
Introduction
Bladder cancer (BCa) is the most common urinary system malignancy, with increasing morbidity and mortality.1 There were an estimated 550, 000 new BCa cases in 2018 and 200, 000 deaths associated with BCa worldwide.2 More than 70% of the newly diagnosed tumors are non-muscular invasive bladder cancer (NMIBC) that were primitively treated by transurethral resection and intravesical instillation of BCG. However, 20%-25% of NMIBCs can eventually progress to muscle-invasive bladder cancer (MIBC) with lymph node (LN) and distant metastasis, which results in poor prognosis.3 The overall five-year survival rate of MIBC is less than 50%.4 Additionally, due to its high recurrence rate, the therapeutic cost is prohibitive for most BCa patients.5 Therefore, it is essential to elucidate the molecular mechanisms underlying BCa development. Discovering new biomarkers is critical for clinically accurate diagnosis and therapy of BCa.6–8
The highly conserved glycoprotein ribophorin II (RPN2), located in rough endoplasmic reticulum (RER) membranes, mediates the translocation of secretory proteins and maintains the endoplasmic reticulum specificity.9,10 Early studies indicated that RPN2 is an essential component of the oligosaccharyltransferase complex responsible for the N‐glycosylation of numerous proteins.11,12 Further evidence has demonstrated that RPN2 is aberrantly upregulated in breast,13 gastric,14 and colorectal cancer,15 which could facilitate the malignant progression of tumors. RPN2 confers docetaxel resistance in breast cancer cells by mediating CD63 glycosylation and subsequent membrane localization of multidrug resistance protein 1 (MDR1).16 However, the expression and biological role of RPN2 in bladder cancer is currently unknown.
This study aimed to investigate the clinical relevance and regulatory mechanisms of RPN2 in BCa. We found that the expression of RPN2 in BCa tissues was higher than that in adjacent tissues and was positively associated with poor prognosis of patients with BCa. Furthermore, knockdown of RPN2 can dramatically inhibit the growth and metastasis of bladder cancer cells both in vitro and in vivo. Mechanistically, the cell cycle protein cyclinD1 and pro-apoptotic protein Bax were regulated by RPN2, as well as epithelial-mesenchymal transition (EMT)-related genes. Given that RPN2 knockdown by siRNA led to a decrease in phosphorylation levels of PI3K and Akt, we speculate that RPN2 may be implicated in bladder cancer progression by regulating PI3K-Akt signaling. Collectively, these findings reveal the biological functions of RPN2 in BCa for the first time, and suggest that RPN2 may represent a novel prognostic biomarker and target for RNAi-based therapeutics against BCa.
Materials and Methods
Cell Culture
Human normal bladder cancer line (SV-HUC-1) and bladder cancer cell lines (BIU-87, 5637 and T24) were derived from the Chinese Academy of Science (Shanghai, China). Cells were cultured in Ham’s F-12 and RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 μg/mL streptomycin at 37 °C in a humidified chamber with 5% CO2.
Clinical Samples
38 pairs of BCa and para-cancer tissues were obtained from patients diagnosed with BCa who required surgical resection at Shandong Provincial Hospital of Shandong University. Histopathological diagnosis of the clinical specimens was confirmed by two pathologists. All samples were frozen in liquid nitrogen for long-term storage. Some tissues were used to analyze gene expression, while others were fixed with formalin for immunohistochemistry (IHC). This research was approved by the institute’s ethics committee of Shandong Provincial Hospital (Jinan, China), and written informed consent was obtained from each study participant, in accordance with the Declaration of Helsinki.
Immunohistochemistry (IHC)
Paraffin-embedded sections (4 μm) were dewaxed with xylene, and then rehydrated with graduated ethanol solution. These samples were immersed in the antigen retrieval solutions with 10 nM citrate buffer (PH 6.0) and boiled for 3 minutes, followed by cooling at room temperature. The slides were incubated with 3% hydrogen peroxide for 30 minutes to block endogenous peroxidase activity. Subsequently, samples were incubated with the primary antibody against RPN2 (1:100, Proteintech) overnight at 4 °C. The next day, the sections were incubated with secondary biotinylated goat anti-rabbit IgG for 30 minutes at room temperature. Finally, the slides were stained with 3-amino-9-ethylcarbazole (AEC), counterstained with hematoxylin, and then sealed with waterborne resin. The positive staining was visualized under light microscopy, and the immunostaining results were evaluated by two pathologists in a blinded manner.
RNA Extraction and Quantitative Real-Time PCR (qRT-PCR)
Following the manufacturer’s instructions, total RNA was extracted from bladder tumor tissues and adjacent non-cancerous tissues, or cultured cells using Trizol reagent (Invitrogen). The cDNA was synthesized using the quantified RNA as a temple with reverse transcription kits (Takara, Japan). Subsequently, we performed the qRT-PCR with SYBR-Green reagent (Takata, Japan) to determine the relative expression of RPN2 mRNA. The comparative expression of RPN2 was calculated using the 2−ΔΔCt method, and GAPDH was used as the internal control. Primers used in the study were as follows: RPN2 forward 5ʹ-TGCCGAGCCAGACAACAAGAA-3ʹ, and reverse 5ʹ-AGTAGAGAGTGTAGGTGCCAGAGG-3ʹ; GAPDH forward 5ʹ-GCACCGTCAAGGCTGAGAAC-3ʹ, and reverse 5ʹ-TGGTGAAGACGCCAGTGGA-3ʹ. The above experiments were performed in parallel three times.
Cell Transfection
RPN2-specific (siRPN2) and negative control (siNC) siRNAs were purchased from Genomeditech (Shanghai, China). Cells were transfected with siRNA using LipofectamineTM 3000 reagent (Thermo Fisher Scientific) in accordance with the manufacturer’s protocol. After transfection for 48 h, qRT-PCR and Western blotting were performed to evaluate the transfection efficiency, and cells were harvested for subsequent functional assays.
Cell Viability
Briefly, transfected cells were seeded into 96-well plates at a density of 3000 cells/well and maintained for 24 h at 37°C in 5% CO2. At the specific time points (0, 24, 48, and 72 h), the supernatant was removed, and 110 μL serum-free medium containing 10 μL Cell Counting Kit-8 (CCK-8, Dojindo) was added to each well for 2 h incubation. Finally, the optical density (OD) value of each well was measured at a wavelength of 450 nm using a microplate reader. Each experiment was performed in triplicates.
Colony Formation Assay
The indicated cells were placed into triplicate wells of 6-well plates at a density of 700 cells/well, and cultured for 10–14 days until colonies were visible. Subsequently, cells were fixed with 4% paraformaldehyde and then stained with 0.1% crystal violet (Sigma-Aldrich) for 20 minutes. Photographs were taken, and colonies with more than 50 cells were counted using Image J.
5-Ethynyl-2ʹ-Deoxyuridine (EdU) Assay
The si-NC and si-RPN2 transfected cells were seeded on coverslips of 24-well plates for 36 h and then incubated with 50 μM EdU for 2 h at room temperature. After fixed with 4% paraformaldehyde for 20 minutes, cells were permeabilized with 0.5% Triton X-100 for 10 minutes, followed by incubation with 1✖Apollo ® (RiboBio) for 30 minutes in the dark. Subsequently, the cell nuclei were stained with Hoechst 33,342. Images were obtained by fluorescence microscopy (Olympus, Japan), and the ratio of EdU+ was calculated in 5 random fields. All experiments were repeated at least 3 times independently.
Wound Healing Assay
Cells were grown to 70% confluence in 6-well plates. After transfection, three separate, parallel wounds were scratched using a sterile 1 mL pipette tip and cells were washed with PBS to remove floating cells. The supernatant was then replaced with low serum medium (1%) to avoid the effect of proliferation. Representative images of wounds were captured at 0, 24, and 48 h under phase-contrast microscopy, and the percentage of wound closure was evaluated using ImageJ software.
Transwell Assay
After transfection, cells were resuspended in serum-free media and transferred into the bottom chamber of 24-well transwell chambers with 8 μm-pore (Corning, NY, USA) with or without Matrigel. Medium (600 μL) containing 20% FBS, as a chemoattractant, was added to the lower chamber. After 48 h incubation, cells in the upper membrane surface were fixed with 4% paraformaldehyde, stained with 0.1% crystal violet, and scraped off with a cotton tip. Photographs were captured under an inverted light microscope, and the cells were counted in 5 random fields. Each experiment was performed in triplicate.
Western Blotting
Tissues and cells were lysed with RIPA lysis buffer (Beyotime) supplemented with protease inhibitors and phosphatases inhibitors. Total protein concentration was detected using Enhanced BCA Protein Assay Reagent (Beyotime). Denatured proteins (25 μg) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transfected to PVDF membranes (Keygen Biotech, KGP113K). After blocking with 5% nonfat milk for 1 h, the membranes were incubated overnight at 4°C with the indicated primary antibodies, followed by incubation with a goat anti-rabbit/mouse antibody for 1 h at room temperature. Finally, the protein bands were visualized using the enhanced chemiluminescent kit (Yeasen), and the levels of proteins were normalized to the GAPDH expression.
Gene Set Enrichment Analysis (GSEA)
To explore the roles of RPN2 in the biological behavior of bladder cancer, we conducted GO function and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses. The KEGG is a collection of databases for correlating related genes with their metabolic pathways. A p-value < 0.05 was set as the cutoff standard.
Xenograft Tumor Model
The protocol for animal experiment was approved by the Institutional Animal Care and Use Committee of Shandong University, and manipulations were performed in accordance with the ethical regulations laid down by the Committee of Shandong Provincial Hospital. The 5637 human bladder cancer cell line was used to establish the xenograft tumor model. A suspension containing 107 cells transfected with either siNC or siRPN2 was injected subcutaneously into the right axilla of 4-week-old male BALB/c nude mice (n=4 for each group). Beginning 10 days after injection, the tumor volume was recorded every 4 days, and solid tumors were removed, weighed, and photographed at the end of the experiment.
Statistics
Data were presented as mean ± SD. We used GraphPad Prism and ggplot2 Package to draw diagrams. Paired Student’s t-tests were performed for continuous variables between two groups. Statistical significance was determined using a two-tailed Student’s t-test or one-way ANOVA. Chi-square analyses were used for categorical comparisons. The Kaplan–Meier method was used for survival analyses and the Log rank test to evaluate survival differences. P-values < 0.05 is identified as statistical significance.
Results
Identification and Screening of Differentially Expressed Genes (DEGs) in BCa
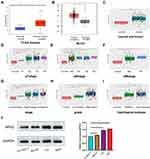
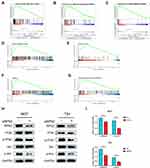
To identify the correlation between DEGs and BCa development, we analyzed the expression profile datasets GSE3167 and GSE7476 of BCa and normal bladder tissues, respectively. Of the consistently expressed 323 genes from the two datasets, 213 genes were downregulated and 110 genes were upregulated in BCa tissues compared to normal bladder tissues (Figure 1A). We downloaded the gene expression profile of bladder urothelial carcinoma from the USUS Xena website (https://xenabrowser.net/), and screened these 323 genes using univariate and multivariate Cox regression analyses (Supplemental Table 1.2). Finally, we identified 8 hub genes that were regarded as the dependent prognostic factors (Figure 1B). Subsequently, survival analyses of these 8 genes were performed using R packages “Survival” and “Surminer”. Interestingly, only RPN2 had significant differences in both overall survival (OS) and disease-free survival (DFS). The Kaplan–Meier curves suggested that BCa patients with high RPN2 expression had shorter OS and RFS than those with low RPN2 expression, which was consistent with results from the GEPIA website (http://gepia.cancer-pku.cn/) (Figure 1C–D). Therefore, RPN2 was chosen for further investigation. The STRING online database (https://string-db.org/) was used to establish a protein-protein interaction (PPI) network for RPN2. As shown in Figure 1E, RPN2 can regulate multiple genes such as RPN1, STT3A/B, and MAGT1, which are usually associated with tumor progression in various cancers.17–20
RPN2 Expression Was Upregulated in BCa and Negatively Correlated with the Prognosis
Both the UALCAN website (http://ualcan.path.uab.edu/) and GEPIA websites revealed that the expression of RPN2 was abnormally upregulated in BCa tissues (Figure 2A–B). We next conducted an R language analysis based on the expression profiles of BCa in The Cancer Genome Atlas (TCGA) datasets. Overexpression of RPN2 in BCa tissues was also validated (Figure 2C). RPN2 overexpression was closely associated with pT stage, lymphatic metastasis, distant metastasis, pTNM stage, tumor grade, and histological subtype in BCa (Figure 2D–I). Furthermore, we measured the RPN2 protein levels in a normal bladder cancer line (SV-HUC-1) and three bladder cancer cell lines (BIU-87, T24 and 5637) by Western blotting. RPN2 expression was relatively higher in 5637 and T24 cells compared to the BIU-87 and SV-HUC-1 cells (Figure 2J). Therefore, 5637 and T24 cells were selected for the subsequent experiments. The above data revealed that RPN2 might play a critical role in the malignant progression of BCa.
Clinical Significance of RPN2 in BCa
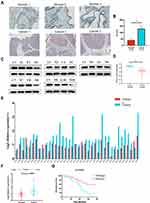
To further verify the difference in RPN2 expression in BCa, we measured RPN2 levels from tissue samples using immunohistochemistry. Our results suggested that RPN2 protein was mainly expressed in the cytoplasm, and its expression in human bladder cancer tissues was significantly higher than that in adjacent tissues, as quantified by H scores (Figure 3A–B). Consistent with the immunohistochemistry results, Western blotting showed that RPN2 protein levels were elevated in bladder cancer tissues (Figure 3C–D). qRT-PCR was performed to detect RPN2 mRNA levels in 38 pairs of BCa and adjacent tissues. We found that RPN2 expression was upregulated in 86.8% (33 of 38) of BCa tissues compared to adjacent tissues (Figure 3E–F). Subsequently, we analyzed the expression of RPN2 based on 38 patients’ gender and age as well as tumor T stage, N stage, pathology, number and invasion (Supplemental Figure 1A-G). Compared to the age (< 65 years), T1 stage, N0 stage and low-grade groups, RPN2 was highly expressed in the age (≥ 65 years) (p=0.0042), T2-T4 (p=0.0221), N1 stage (p=0.0002), and high-grade groups (p=0.0029). However, the levels of RPN2 were not significantly correlated with gender (p=0.6484), tumor number (p=0.2306), or invasion (p=0.3339).
To determine the clinical significance of RPN2 expression in BCa, we explored the correlation between RPN2 expression and clinicopathological features. According to the median value of relative RPN2 expression, 38 BCa patients with integrated clinical data were divided into RPN2 -high and -low groups. As shown in Table 1, increased RPN2 levels were positively correlated with age (p<0.01), pathological grade (p<0.001), T stage (p<0.05), and LN metastasis status (p<0.01) in BCa. Meanwhile, patients with high RPN2 expression exhibited a lower OS probability than those with low RPN2 expression (p=0.0244) (Figure 3G). Taken together, these data suggest that RPN2 may be an effective biomarker for poor prognosis in BCa.
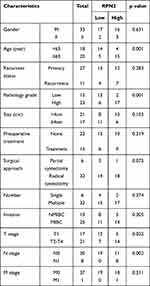 |
Table 1 The Correlation Between the Levels of RPN2 Expression and Clinicopathological Features of BC Patients |
RPN2 Knockdown Suppresses Bladder Cancer Cell Growth in vitro
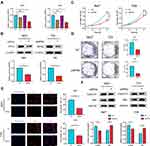
To illustrate the potential function of RPN2 in BCa progression, we transfected 5637 and T24 cells with siRNAs against RPN2, then performed qRT-PCR to evaluate the knockdown efficiency. As shown in Figure 4A, siRPN2-3 reduced RPN2 mRNA levels approximately 50% in both cell lines. Additionally, Western blotting confirmed that the expression of RPN2 protein was remarkably inhibited at 48 h post-siRPN2-3 transfection (Figure 4B). Therefore, we selected the siRPN2-3 sequence for subsequent experiments.
CCK-8 assay results suggested that RPN2 downregulation significantly inhibited cell viability (Figure 4C). Furthermore, a colony formation assay indicated that colony formation was substantially impaired by siRPN2 compared to NC (Figure 4D). EdU labeling showed that the percentage of EdU-positive cells transfected with siRPN2 was significantly decreased compared to control (Figure 4E). Immunoblotting results revealed that RPN2 silencing increased levels of the pro-apoptotic protein Bax and reduced the levels of cell cycle-related protein cyclin-D1 (Figure 4F). In summary, these data demonstrated that RPN2 plays a growth-promoting role in BCa.
RPN2 Facilitates BCa Metastasis by Driving EMT
As mentioned above, RPN2 was correlated with LN metastasis; therefore, we performed wound-healing and Transwell assays to examine the effects of RPN2 on the migration and invasion abilities of BCa cells. Intriguingly, downregulation of RPN2 inhibited wound healing rates (Figure 5A-B), indicating that RPN2 silencing attenuated the migration capability of bladder cancer cells. Additionally, RPN2 knockdown resulted in a significant decrease in the number of migrated or invasive cells in the Transwell assay (Figure 5C-D). Subsequently, we detected the expression of EMT-related markers by Western blotting. E-cadherin was upregulated, while N-cadherin and MMP-2 were downregulated after RPN2 silencing (Figure 5E-F). These findings demonstrate that RPN2 promotes cell migration and invasion by mediating EMT in bladder cancer cells.
RPN2 May Affect BCa Progression by Modulating the PI3K-Akt Pathway
To identify the potential signaling pathways mediated by RPN2, we first conducted a KEGG pathway enrichment analysis using the TCGA database. Cell cycle, p53 signaling, and mismatch repair pathways were significantly enriched in the RPN2-high group (Figure 6A–C). In addition, RPN2 expression was correlated with the cellular metabolic process, metabolic pathway, signaling, and cellular response to chemical stimulus, as revealed by GO function analysis using the R package ClusterProfiler (Figure 6D–G). These processes are usually regulated by the PI3K-Akt pathway. We next explored whether RPN2 influenced the PI3K-Akt pathway. Western blotting was performed to examine the expression of PI3K, p-PI3K, Akt, and p-Akt proteins in 5637 and T24 cells after 48 h siRPN2 transfection. As shown in Figure 6H–I, RPN2 knockdown clearly reduced the levels of p-PI3K and p-Akt, but had no significant effects on overall PI3K and Akt expression. Based on the above results, we speculated that RPN2 may accelerate BCa progression by activating the PI3K/Akt pathway.
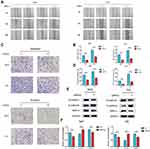
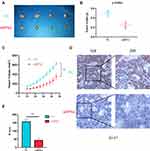
RPN2 Regulates the Tumorigenicity of BCa Cells in vivo
Finally, mouse xenograft models were constructed to further investigate the tumorigenic ability of RPN2 in BCa in vivo. As shown in Figure 7A, downregulation of RPN2 expression dramatically inhibited tumor growth on the 46 day. Moreover, mice injected with 5637-siRPN2 cells showed smaller weights and implanted tumor volume compared to those injected with 5637-siNC cells (Figure 7B–C). IHC indicated that the positive intensity of Ki-67 (a vital proliferative marker) was also suppressed in the siRPN2 group (Figure 7D–E). Collectively, these results demonstrated that RPN2 knockdown significantly impaired the tumorigenicity of BCa.
Discussion
Bladder cancer remains a primary clinical challenge because of frequent recurrence and tumor invasion. Therefore, it is becoming increasingly important to identify precise tumor-specific biomarkers for early BCa diagnosis and to apply them in clinical treatment.
High-throughput sequencing technology has been widely applied over the past few decades.21 It enables extensive amounts of sequencing data to be stored in public datasets, thus providing a more systematic and objective perspective for researchers.22,23 Currently, a growing number of molecules involved in bladder cancer progression have been selected by bioinformatic analysis. In our study, we first identified RPN2 as a potential oncogene in BCa by employing bioinformatics tools.24,25 Prior studies have suggested that RPN2 participates in tumor development. RPN2 mediates the glycosylation and membrane localization of epidermal growth factor receptor (EGFR), thus activating EGFR/ERK signaling to promote colorectal cancer cell proliferation.13 A fraction of breast cancer stem cells (CSCs) exhibited higher RPN2 expression, which could maintain the biological properties of CSCs via stabilization of mutant p53 and functional suppression of glycogen synthase kinase 3β (GSK3β).13,26 Recently, it was reported that RPN2 was implicated in drug resistance of tumors. RPN2 silencing inhibited glycosylation of the P-glycoprotein, thereby sensitizing breast cancer cells to docetaxel.27 RPN2 also conferred docetaxel and cisplatin resistance in gastric cancer.28 However, its biological function and prognostic significance of RPN2 in BCa remain unclear.
In the present study, we found that RPN2 expression was upregulated in BCa clinical tissues and samples listed in the TCGA datasets. Analysis of 38 clinical samples suggested that RPN2 overexpression is closely associated with age (≥65 years), pathology grade, T stage and lymphatic metastasis in BCa. Moreover, patients with high RPN2 had shorter survival, indicating that RPN2 may be involved in BCa progression. Next, we aimed to explore the detailed roles of RPN2 in BCa. Our results demonstrated that RPN2 promoted the proliferation and metastasis of BCa cells in vitro, whereas downregulation of RPN2 attenuated tumor growth in vivo. More importantly, we found that RPN2 could drive activation of EMT and the PI3K/Akt pathway, which may play a crucial role in the pro-oncogenic functions of RPN2. Given that RPN2 seems to be associated with cellular response to chemical stimulus, as revealed by GSEA, we intend to investigate the impact of RPN2 on the outcome of combined gemcitabine and cisplatin chemotherapy for advanced bladder cancer in further research. Taken together, these findings indicate that RPN2 has the prospect to serve as a potential biomarker and therapeutic target in BCa.
EMT is a crucial biological process involved in cellular transformation, while endowing metastatic and invasive properties to tumor cells in various malignancies, including BCa.29 In EMT, epithelial cells lose their adhesive features and acquire mesenchymal characteristics such as enhanced migratory capacity and increased production of extracellular matrix components.30 Typical EMT features at the molecular level include the upregulation of EMT-activating transcription factors Snail and Twist, acquisition of N-cadherin and vimentin, and the loss of E-cadherin. Furthermore, EMT has been reported to endow tumor cells with stem cell characteristics, thereby leading to drug resistance and poor prognosis in advanced bladder cancer stages. We found that RPN2 silencing induced elevated E-cadherin levels along with downregulation of N-cadherin and MMP-2 expression. These findings provide evidence that RPN2 functions as a vital regulator of EMT to promote the metastasis of BCa cells.
Bladder cancer exhibits profound metabolic abnormalities.31 Energy metabolism programming, a vital hallmark of many cancers, is indispensable for highly proliferating and invasive tumor cells.32 GSEA results indicated that cellular protein metabolic processes, metabolic pathways, and signaling were enriched in RPN2-high BCa. PI3K/Akt is regarded as a critical regulatory pathway for a wide range of cellular biological processes, including signaling transduction, metabolism, and survival.33 Previous studies have shown that the PI3K/Akt pathway is commonly activated in various human cancer types and participates in tumor progression.34 Hyper-activated PI3K/AKT in BCa functions as a central regulator of aerobic glycolysis to enable tumor cells to depend on glucose consumption for growth and aggression.35 Several proteins, including LDH, GLUT1, and PKM2, are implicated in PI3K/Akt-mediated aerobic glycolytic metabolism.31 Here, we explored the correlation between RPN2 and PI3K/Akt signaling. Western blotting revealed that RPN2 silencing induced the downregulation of p-PI3K and p-AKT expression. Furthermore, downstream targets of PI3K/Akt, including the cell cycle protein cyclin-D1 and the pro-apoptotic protein Bax, were modulated by RPN2. Therefore, we hypothesize that RPN2 activates the PI3K/Akt pathway to promote BCa progression, which deserves further research.
Conclusion
Our studies is the first to demonstrate the clinical significance of RPN2 and its effects on the biological functions of BCa. High RPN2 expression was strongly associated with a poor prognosis in patients with BCa. In addition, we found that downregulation of RPN2 could impair the proliferation and metastasis of BCa cells both in vitro and in vivo, which is possibly associated with inactivation of the PI3K-Akt pathway. Hence, RPN2 could represent a potential diagnostic and prognostic indicator in BCa and have value as a therapeutic target. Further studies are needed to clarify the broader mechanisms underlying the tumorigenicity of RPN2.
Data Sharing Statement
The data used to support the findings of this study are available from Xiao Yu and Muwen Wang upon request.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No.81572534), Natural Science Foundation of Shandong (Grant No. ZR2016HM32) and Traditional Chinese Medicine Science Foundation of Shandong Province (Grant NO. 2019-0300).
Disclosure
All authors declare that they have no conflicts of interest related to this paper.
References
1. Dong W, Bi J, Liu H, et al. Circular RNA ACVR2A suppresses bladder cancer cells proliferation and metastasis through miR-626/EYA4 axis. Mol Cancer. 2019;18(1):95. doi:10.1186/s12943-019-1025-z
2. Chen Z, Du Y, Liu X, et al. EZH2 inhibition suppresses bladder cancer cell growth and metastasis via the JAK2/STAT3 signaling pathway. Oncol Lett. 2019;18(1):907–915.
3. Chen M, Gao X, Huang D, et al. Knockdown of SLC35F2 inhibits the proliferation and metastasis of bladder cancer cells. Onco Targets Ther. 2019;12:10771–10786.
4. Wu Q, Zhou X, Li P, et al. High NRBP1 expression promotes proliferation and correlates with poor prognosis in bladder cancer. J Cancer. 2019;10(18):4270–4277.
5. Xia Y, Kang TW, Jung YD, Zhang C, Lian S. Sulforaphane inhibits nonmuscle invasive bladder cancer cells proliferation through suppression of HIF-1alpha-mediated glycolysis in hypoxia. J Agric Food Chem. 2019;67(28):7844–7854.
6. Kiyoshima K, Akitake M, Shiota M, et al. Prognostic significance of preoperative urine cytology in low-grade non-muscle-invasive bladder cancer. Anticancer Res. 2016;36(2):799–802.
7. Chen Z, Chen X, Xie R, et al. DANCR promotes metastasis and proliferation in bladder cancer cells by enhancing IL-11-STAT3 signaling and CCND1 expression. Mol Ther. 2019;27(2):326–341.
8. Chen X, Zhang J, Ruan W, et al. Urine DNA methylation assay enables early detection and recurrence monitoring for bladder cancer. J Clin Invest. 2020;130(12):6278–6289.
9. Fang Q, Liu H, Zhou A, Zhou H, Zhang Z. Circ_0046599 promotes the development of hepatocellular carcinoma by regulating the miR-1258/RPN2 network. Cancer Manag Res. 2020;12:6849–6860.
10. Huang L, Jian Z, Gao Y, et al. RPN2 promotes metastasis of hepatocellular carcinoma cell and inhibits autophagy via STAT3 and NF-κB pathways. Aging (Albany NY). 2019;11(17):6674–6690. doi:10.18632/aging.102167
11. Ono M, Tsuda H, Kobayashi T, et al. The expression and clinical significance of ribophorin II (RPN2) in human breast cancer. Pathol Int. 2015;65(6):301–308. doi:10.1111/pin.12297
12. Sun J, Chen Z, Xiong J, et al. MicroRNA422a functions as a tumor suppressor in glioma by regulating the Wnt/β‑catenin signaling pathway via RPN2. Oncol Rep. 2020;44(5):2108–2120.
13. Takahashi RU, Takeshita F, Honma K, Ono M, Kato K, Ochiya T. Ribophorin II regulates breast tumor initiation and metastasis through the functional suppression of GSK3beta. Sci Rep. 2013;3:2474.
14. Fujimoto D, Goi T, Hirono Y. Expression of ribophorine II is a promising prognostic factor in human gastric adenocarcinoma. Int J Oncol. 2017;50(2):448–456.
15. Zhou T, Wu L, Wang Q, et al. MicroRNA-128 targeting RPN2 inhibits cell proliferation and migration through the Akt-p53-cyclin pathway in colorectal cancer cells. Oncol Lett. 2018;16(6):6940–6949.
16. Tominaga N, Hagiwara K, Kosaka N, Honma K, Nakagama H, Ochiya T. RPN2-mediated glycosylation of tetraspanin CD63 regulates breast cancer cell malignancy. Mol Cancer. 2014;13:134.
17. Krasnov GS, Oparina N, Dmitriev AA, et al. [Novel reference gene RPN1 for normalization of quantitative data in lung and kidney cancer]. Mol Biol (Mosk). 2011;45(2):238–248.
18. Ruan Z, Liang M, Lai M, Shang L, Deng X, Su X. KYA1797K down-regulates PD-L1 in colon cancer stem cells to block immune evasion by suppressing the beta-catenin/STT3 signaling pathway. Int Immunopharmacol. 2020;78:106003.
19. Liu J, Ma T, Gao M, et al. Proteomic characterization of proliferation inhibition of well-differentiated laryngeal squamous cell carcinoma cells under below-background radiation in a deep underground environment. Front Public Health. 2020;8:584964.
20. Zheng K, Yang Q, Xie L, et al. Overexpression of MAGT1 is associated with aggressiveness and poor prognosis of colorectal cancer. Oncol Lett. 2019;18(4):3857–3862.
21. Kulasingam V, Diamandis EP. Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nat Clin Pract Oncol. 2008;5(10):588–599.
22. Gan Y, Li Y, Li T, Shu G, Yin G. CCNA2 acts as a novel biomarker in regulating the growth and apoptosis of colorectal cancer. Cancer Manag Res. 2018;10:5113–5124.
23. Zhang M, Di CY, Guo P, et al. Screening and identification of key biomarkers in pancreatic cancer: evidence from bioinformatic analysis. J Comput Biol. 2020;27(7):1079–1091.
24. Xie R, Chen X, Cheng L, et al. NONO inhibits lymphatic metastasis of bladder cancer via alternative splicing of SETMAR. Mol Ther. 2021;29(1):291–307.
25. Chen X, Xie R, Gu P, et al. Long noncoding RNA LBCS inhibits self-renewal and chemoresistance of bladder cancer stem cells through epigenetic silencing of SOX2. Clin Cancer Res. 2019;25(4):1389–1403.
26. Fujita Y, Yagishita S, Takeshita F, Yamamoto Y, Kuwano K, Ochiya T. Prognostic and therapeutic impact of RPN2-mediated tumor malignancy in non-small-cell lung cancer. Oncotarget. 2015;6(5):3335–3345.
27. Honma K, Iwao-Koizumi K, Takeshita F, et al. RPN2 gene confers docetaxel resistance in breast cancer. Nat Med. 2008;14(9):939–948.
28. Fujimoto D, Goi T, Koneri K, Hirono Y. RPN2 is effective biomarker to predict the outcome of combined chemotherapy docetaxel and cisplatin for advanced gastric cancer. Oncotarget. 2018;9(20):15208–15218.
29. Huang J, Lo UG, Wu S, et al. The roles and mechanism of IFIT5 in bladder cancer epithelial-mesenchymal transition and progression. Cell Death Dis. 2019;10(6):437.
30. Yun SJ, Kim WJ. Role of the epithelial-mesenchymal transition in bladder cancer: from prognosis to therapeutic target. Korean J Urol. 2013;54(10):645–650.
31. Massari F, Ciccarese C, Santoni M, et al. Metabolic phenotype of bladder cancer. Cancer Treat Rev. 2016;45:46–57.
32. Li R, Weng L, Liu B, et al. TRIM59 predicts poor prognosis and promotes pancreatic cancer progression via the PI3K/AKT/mTOR-glycolysis signaling axis. J Cell Biochem. 2020;121(2):1986–1997.
33. Weichhart T, Saemann MD. The PI3K/Akt/mTOR pathway in innate immune cells: emerging therapeutic applications. Ann Rheum Dis. 2008;67(Suppl 3):iii70–74.
34. Hu R, Wang MQ, Niu WB, et al. SKA3 promotes cell proliferation and migration in cervical cancer by activating the PI3K/Akt signaling pathway. Cancer Cell Int. 2018;18:183.
35. Elstrom RL, Bauer DE, Buzzai M, et al. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 2004;64(11):3892–3899.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.