Back to Archived Journals » Advances in Genomics and Genetics » Volume 4
Role of PTPN22 and VDR gene polymorphisms in susceptibility to rheumatoid arthritis: a study from central India
Authors Shukla S, Tripathi AK, Tripathi JK, Indurkar M, Chauhan UK
Received 29 January 2014
Accepted for publication 1 April 2014
Published 19 June 2014 Volume 2014:4 Pages 79—85
DOI https://doi.org/10.2147/AGG.S61526
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Smriti Shukla,1 Arvind Kumar Tripathi,1 Jitendra Kumar Tripathi,1 Manoj Indurkar,2 Ugam Kumari Chauhan1
1Centre for Biotechnology Studies, Awadhesh Pratap Singh University, Rewa, Madhya Pradesh, India; 2Department of Medicine, Shyam Shah Medical College, Rewa, Madhya Pradesh, India
Abstract: Rheumatoid arthritis (RA) is an autoimmune disorder which can be characterized by chronic inflammation of the joints. All joints of the body are prime targets for RA. Several genes which are associated with immunomodulatory functions could be associated with RA as well. In the present investigation we selected the genes which are involved in immunoregulation such as PTPN22 (protein tyrosine phosphatase, non-receptor type 22 (lymphoid)), and VDR (vitamin D receptor), and investigated the impacts of their polymorphism in RA susceptibility. One hundred and twelve patients were enrolled and 125 controls from similar ethnicity were selected for the present investigation. Genetic polymorphism was detected through the PCR-RFLP (polymerase chain reaction restriction fragment length polymorphism) method. The pattern of genotype, allele distribution, and carriage rate in the disease and control groups suggested a significant association of PTPN22-1858 T (rs2476601) carriage (carriage of “TT” and “CT”) in RA susceptibility. The pattern of genotype and allele distribution in the disease and control groups suggested a lack of association of VDR FokI (rs10735810) in RA susceptibility. Our findings revealed that the PTPN22 polymorphism is associated with RA, while, the VDR polymorphism is not associated with RA susceptibility.
Keywords: rheumatoid arthritis, autoimmune disorder, genetic polymorphism, VDR, PTPN22
Introduction
Rheumatoid arthritis (RA) is an autoimmune disorder which can be characterized by chronic inflammation in joints. All joints of the body are prime targets of RA. In general, the immune system of the human body consists of a variety of cells and proteins to combat foreign antigens and infection. When something goes wrong with the immune system, it loses its ability to distinguish our body cells from foreign particles and antigens. At this point the condition of autoimmunity arises and the immune system starts to destroy cells, and chronic inflammation results. RA is a debilitating and common disease, affecting approximately 1% of the world population,1 including India,2 with women affected three times as often as men. The etiology of RA is still unclear, and it is considered to be a multifactorial disease with interacting contributions from genetics, infections, and environmental and hormonal factors. Transmittance to progeny and increased susceptibility in monozygotic twins suggest that RA has a strong genetic base. As it is an autoimmune disorder, many genes which are associated with immunomodulatory functions could be associated with RA as well. In the present investigation, we selected the genes which are involved in immunoregulation such as PTPN22 (protein tyrosine phosphatase, non-receptor type 22 (lymphoid)) and VDR (vitamin D receptor), and investigated the impact of their polymorphism in RA susceptibility. PTPN22 is located in chromosome 1p13.3–p13.1, and encodes an intracellular enzyme that associates with the molecular adapter protein CBL, and may be involved in regulating CBL function in the T-cell receptor (TCR) signaling pathway that dephosphorylates Src family kinases Lck and Fyn, as well as components of the TCR/CD3 complex, and other key signaling molecules,3,4 so it works as an important negative regulator of T-cell responses. The 1858C-T (Arg620Trp) single nucleotide polymorphism (rs2476601) was found to be associated with autoimmune diseases, including RA, in many case control studies.2–5 The VDR gene is located on chromosome 12q13.11, and contains eleven exons, with three exons (1a, 1b and 1c) found in the 5′ noncoding sequence and eight exons2–8 encoding the structural portion of the protein.5 The gene contains a restriction fragment length polymorphism defined as FokI. The FokI polymorphism (an ACG to ATG transition) introduces an initiation codon leading to the addition of three amino acids to the VDR protein.6 There are many association studies from west that confirm the functional significance of these polymorphisms and their potential effects on disease susceptibility.4,9–13 No such studies are available from India. Therefore there is a need for further data to determine whether these polymorphisms can act as possible genetic markers of RA susceptibility, in our central Indian population.
Materials and methods
Patient recruitment
Rheumatoid arthritis patients were recruited from Sanjay Gandhi hospital, Rewa, Madhya Pradesh, Government. Ayurveda Medical College Rewa, Birla Hospital, Satna during the year 2008–2011. One hundred and twelve patients were enrolled in the study and 125 respective controls having similar ethnicity and socioeconomic status were recruited, to achieve more than 80% power in additive and 91% in multiplicative model (CaTs Model) for present investigation. This study was ethically approved by the institutional ethical committee of Awadhesh Pratap Singh University, in 2008.
The diagnosis of RA was based on the case history of patients and clinical tests (ie, rheumatoid factor, erythrocyte sedimentation rate, hematocrit, synovial fluid analysis, citrulline antibody, antinuclear antibodies, C-reactive protein, anti-cyclic citrullinated peptides antibodies) and radiological criteria (diagnosed strictly based on American Rheumatism Association 1987 Criteria). All patients participating in this study provided informed consent. Approximately 5 mL of blood sample was collected in 0.5 M EDTA (ethylenediaminetetraacetic acid) tubes from each of the RA patients as well as from the healthy controls (HC).
DNA isolation and quantification
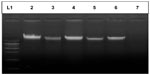
Genomic DNA was extracted from whole blood by the modification of salting out procedure described by Miller et al.6 The isolated genomic DNA samples were then tested for purity by measuring their absorbance values at 230 nm, 260 nm, 280 nm, and 300 nm using an ultraviolet visible spectrophotometer (Systronic (India) Ltd, Bhopal, India). Gel electrophoresis of the genomic DNA was carried out for qualitative estimation of samples prepared. Horizontal agarose slab gel electrophoresis apparatus (Bangalore Genei Pvt, Ltd, Bangalore, India) was used.
Detection of PTPN22-1858 single nucleotide polymorphism
The nucleotide position -1858 (at codon 620) gene has a single nucleotide polymorphism (SNP) that results in change of nucleotide from cytosine (C) to thymine (T). The oligonucleotide sequences (primers) were designed to create a recognition site for the restriction enzyme XcmI in the T allele.
Primer sequences
The oligonucleotide sequences (primers) used were those described by Kyogoku et al.7 Sequences are as follows:
Sense oligonucleotide 5′-TCACCAGCTTCCTCAACCACA-3′.
Antisense oligonucleotide 5′-GATAATGTTGCTTCAA-CGGAATTTA-3′.
Polymerase chain reaction mix
The polymerase chain reaction (PCR) was carried out in a total volume of 25 μL containing 100 ng of genomic DNA, 10 pM of each primer, 2 mM MgCl2, 0.2 mM dNTPs, 1× buffer and 2 U of Taq polymerase. PCR amplifications were performed on each sample in a 25 μL reaction volume consisting of 10 μL PCR buffer (Perkin Elmer Inc., Boston, MA, USA), 0.5 mM of each primer, 0.1 mM dNTPs (Bangalore Genei, Pvt, Ltd), 1.25 U of Taq Gold DNA polymerase (Perkin Elmer), 3 mM MgCl2 and 50 ng of genomic DNA, diluted to the final volume with H2O.
Thermal profile
The PCR cycles included initial denaturation at 94°C for 5 minutes, followed by 35 cycles of denaturation at 94°C for 60 seconds, annealing 59°C for 30 seconds, and amplification at 72°C for 1 minute, and one final cycle of extension at 72°C for 7 minutes.8 The PCR products were separated by 2% agarose gel stained with ethidium bromide, using a 100 bp molecular weight marker to confirm the PCR product size.
Restriction digestion by XcmI and genotyping
The PCR product was digested using 0.4 U XcmI restriction enzyme (New England BioLabs Inc., Ipswich, MA, USA) for 4 hours at 37°C. Digestion of the amplified 215 bp PCR product gave two fragments of 174 bp and 41 bp, respectively, if the product was digested by XcmI. Depending on the digestion pattern, individuals were scored as genotype CC when homozygous for presence of the XcmI site, genotype TT when homozygous for absence of the XcmI site and genotype CT in the case of heterozygosity.
VDR polymorphism screening
The VDR gene is ≈75 kb long, and is made up of eleven exons together with intervening introns. The gene encoding the VDR is on chromosome 12q, and has several known allelic variants including a FokI restriction fragment length polymorphism in intron 2.
Primers
The oligonucleotide sequences (primers) used were those described by Bid and Mittal.8 Sequences are as follows:
Forward primer – 5′ AGCTGGCCCTGGCACTGACTCTGCTCT 3′.
Reverse primer – 5′ ATGGAAACACCTTGCTTCTTCTCCCTC 3′.
PCR mix
The PCR reaction was carried out in a total volume of 25 μL containing 100 ng of genomic DNA, 10 pM of each primer, 2 mM MgCl2, 0.2 mM dNTPs, 1× buffer and 2 U of Taq polymerase. PCR amplifications were performed on each sample in a 25 μL reaction volume consisting of 10 μL PCR buffer (Perkin Elmer, Boston, MA, USA), 0.5 mM of each primer, 0.1 mM of each dNTPs (Bangalore Genei Pvt, Ltd), 1.25 U of Taq Gold DNA polymerase (Perkin Elmer Inc.), 3 mM MgCl2 and 50 ng of genomic DNA, diluted to the final volume with H2O.
Thermal profile
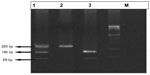
The PCR cycles included initial denaturation at 94°C for 5 minutes, followed by 35 cycles of denaturation at 94°C for 30 seconds, annealing at 61°C for 30 seconds, and amplification at 72°C for 1 minute, and one final cycle of extension at 72°C for 7 minutes.8 The PCR products were separated by 9% Page stained with ethidium bromide using a 100 bp molecular weight marker to confirm the PCR product size (Figure 1).
  | Figure 1 Gel 1 representative image of isolated DNA of case and control. |
Restriction digestion by FokI and polymorphism study
The PCR product was digested with 1.0 U of FokI restriction enzyme, and the reaction buffer, incubated at 37°C for 4 hours. Digestion of the amplified 265 bp PCR product gave two fragments of 169 bp and 96 bp respectively if the product was digested by FokI. Depending on the digestion pattern, the FF genotype (homozygote of common allele) lacked a FokI site and showed only one band of 265 bp. The ff genotype (homozygote of infrequent allele) generated two fragments of 169 and 96 bp. The heterozygote had three fragments of 265, 169, and 96 bp, designated as Ff.
Statistical analysis
Statistical analysis was done by comparing the distribution of genotype frequencies, allele frequencies and carriage rates of all the polymorphisms in the disease and control groups. Disease group included RA patients whereas control group included all HC enrolled in the study. Both the chi-square test and Fisher’s exact test were performed using Prism version 5.0 (GraphPad Software, Inc., La Jolla, CA, USA).
Results
The genotype frequencies of each gene in each study group were tested to be in accordance with Hardy Weinberg equilibrium using the chi-square (χ2) test for independence. The calculated value of χ2 was less than tabulated value (3.84) of χ2 at degree of freedom 1 (df=1) and level of significance (P=0.05).
Detection of PTPN22-C1855T polymorphism
The genotype distribution of PTPN22 (rs2476601) was significantly different in the case and control groups (χ2=6.897, P=0.0318). The HC group showed a significant increase in CC genotype as compared to the RA group (93.6% versus 83.0%). The heterozygous genotype CT was significantly lower in the HC group as compared to the RA group (6.4% in case versus 16.1% in control). Genotype TT was present in significantly less frequency in the case group and not present in the control group. The odds ratio (OR) of heterozygousity was 2.80, and thus associated with higher risk. Overall allele “C” was found to be significantly less in frequency in the disease group as compared to the HC group (91.1% versus 96.8%), whereas allele “T” was present in significantly high frequency in the disease group (χ2=6.975, P=0.0083). The T allele shows an odds ratio of 2.96, which indicates its possible association with RA susceptibility. The carriage rate of allele C was slightly higher in HC, whereas the carriage rate of allele T was significantly higher in the disease group compared to the control group (16.9% in case versus 6.4% in control), and the difference was statistically significant (χ2=4.543, P=0.0331). The pattern of genotype, allele distribution and carriage rate in the disease and control groups suggested a significant association of PTPN22-1858 T (rs2476601) carriage (carriage of TT and CT) in RA susceptibility. CT genotype and T allele carriage have each shown a strong susceptibility to RA (P=0.0216, OR=2.801, 95% confidence interval [CI] 1.166–6.726; and P=0.0104, OR 2.966, 95% CI 1.279–6.877, respectively), meanwhile CC genotype and C allele carriage were found to be significantly protective (P=0.0134, OR 0.3347, 95% CI 0.1402–0.7988; and P=0.0104, OR 0.3372, 95% CI 0.1454–0.7819, respectively), as shown in Table 1 and Figure 2.
Detection of VDR FokI gene polymorphism
Overall genotype pattern of VDR gene was not significantly different between the case and control groups (χ2 =2.266, df =2, P-value =0.3221). The RA group showed an increase in the “FF” genotype as compared to the control group (51.7% versus 43.2%), but was not significantly different. The Genotype “ff” was not significantly higher in the HC group as compared to the RA group (6.4% vs 3.6%). The overall allele distribution was also not found to be significant, but less common “f” allele was found in higher frequency in the controls as compared to the RA patients (31.6% versus 25.9%), and the F allele was found at a higher frequency in the cases as compared to the control group (74.1% versus 68.4%), but the difference was not statistically significant (χ2=1.873, P=0.1712). Carriage rate of the F allele was equivalent to the HC group and the RA group. Whereas carriage rate of allele f was higher in the control group (56.8% versus 48.2%), but not significantly different between cases and controls (χ2=0.7447, P=0.3882). The pattern of genotype and allele distribution in the disease and control groups suggested a lack of association of VDR FokI (rs10735810) in RA susceptibility, as shown in Table 2 and Figure 3.
Discussion
RA is one of the most common autoimmune diseases. It is a chronic, progressive, systemic inflammatory disorder affecting the synovial joints and typically producing symmetrical arthritis.14,15 RA is a complex disease in which environmental agents are thought to interact with genetic factors that influence susceptibility. This interaction triggers immunologic events that eventually result in the clinical signs of arthritis. Knowledge of the chain of etiological events that lead to the development of RA is incomplete.9 The exact etiology of RA remains unknown, but is likely to be multifactorial. The pathogenesis of the disease is largely determined by environmental and immunological factors in a predisposed animal system.9 The present study is the first case control association study from India in patients with RA and unaffected controls assessing the putative genetic association with PTPN22 and VDR gene polymorphisms. This study was conducted in a central part of India, so it can not extrapolate its results on other parts of the country, as the population in India differs on the basis of ethnicity, age, diet, lifestyle and other environmental factors. Moreover, the population within central India has mixed ethnicity, but there is a common sociocultural background and dietary habits.
The PTPN22 gene maps to chromosome 1p13.3–p13.1 and encodes a lymphoid-specific phosphatase (Lyp). Lyp is an intracellular protein tyrosine phosphatase and is physically bound through a proline-rich motif to the SH3 domain of CBL, which is an important suppressor of kinases that mediate T-cell activation. PTPN22 downregulates TCR signaling by dephosphorylating Src family kinases, such as Lck or Fyn. Evidence indicates that the PTPN22 risk allele affects the enzymatic activity of the encoded phosphatase.16 The ability of Csk and Lyp to inhibit TCR signaling requires their physical association.17 Bottini et al,3 described a SNP in the PTPN22 gene, 1858C>T, changing the arginine to a tryptophan residue in position 620 of the mature protein. The 1858T (620W) variant was found to be associated with insulin-dependent diabetes mellitus. Subsequently, association with 1858T was reproducibly described for other autoimmune diseases, including RA.4 In the present investigation we have found that PTPN22 1858C-T polymorphism is associated with the risk of RA. Our population contains a significantly lower frequency of the T allele as compared to Caucasian and other populations. In our present investigation, the TT genotype was present at much less frequency in patients with arthritis, and it was totally absent in the control group. The carriage of the C allele is higher in both the cases and controls, and it is much higher than any other population of the world. In a study carried out in Poland,10 CT + TT genotypes showed an OR of 1.96, which clearly indicates that the T allele is a susceptibility factor, although the frequency of the C genotype is higher in Indians as compared to any other population. Another study with participants from the southern Indian population is consistent with our study, but the relative risk of T allele (5.87) is higher than our study.11 A meta-analysis performed on the basis of 13 different RA case control studies suggested an overall OR of the T allele at 1.34.12 Steer et al18 established the association of this polymorphism with RA in a UK population, and found OR of 2.05, while Begovich et al19 showed a significant association of the PTPN22 polymorphism with RA in a USA Caucasian population.
The VDR gene is located on chromosome 12q13.11. VDR is a potent regulator of bone and calcium homeostasis, as well as in immunomodulation, cellular differentiation and replication in different target tissues. VDR gene polymorphisms have been associated with multiple traits and disease phenotypes like primary hyperparathyroidism, Grave’s disease, type 1 diabetes mellitus, osteoporosis, and RA. The FokI polymorphism (an ACG to ATG transition) introduces an initiation codon leading to the addition of three amino acids to the VDR protein. The pattern of genotype and allele distribution in the disease and control groups suggested a lack of association of VDR FokI in RA susceptibility. In our study, the RA group has shown an increase in the FF genotype as compared to the control group (51.8% versus 41.6%). In the case of genotype ff, it was not found to be significantly higher in the HC group as compared to the RA group (7.2% versus 3.6%). The heterozygous genotype Ff was nonsignificantly distributed in the HC group as compared to the RA group (51.2% versus 44.6%). An odds ratio of 0.708 for Ff showed weak or no association with RA susceptibility. Our results are consistent with a case control study in the German population, which showed no evidence of RA association with VDR.20 A previous study performed in a French population showed significance of the f allele with protection from RA,13 and VDR polymorphisms may contribute to the high prevalence of RA in North American native populations.21 The reason behind the inconsistencies may be due to the different origins and populations. The BsmI and/or FokI VDR polymorphisms have also been recognized as risk factors for some other autoimmune diseases, including RA, Behçet’s, Grave’s disease, Addison’s disease, psoriasis, multiple sclerosis, type 1 diabetes, and others.22–28 In our study the allele frequency of the f allele was higher in cases as compared to controls, but not enough to reach the 5% level of significance. According to our study, we found a clear association of the PTPN22 polymorphism with RA susceptibility. Meanwhile, our study did not suggest any association of the VDR Fok1 polymorphism with RA susceptibility.
Disclosure
The authors have no conflicts of interest to disclose in this work.
References
Lawrence JS. Prevalence of rheumatoid arthritis. Ann Rheum Dis. 1961; 20(1):11–17. | |
Malaviya AN, Kapoor SK, Singh RR, Kumar A, Pande I. Prevalence of rheumatoid arthritis in the adult Indian population. Rheumatol Int. 1993;13(4):131–134. | |
Bottini N, Vang T, Cucca F, Mustelin T. Role of PTPN22 in type 1 diabetes and other autoimmune diseases. Semin Immunol. 2006;18(4):207–213. | |
Gregersen PK, Lee HS, Batliwalla F, Begovich AB. PTPN22: setting thresholds for autoimmunity. Semin Immunol. 2006;18(4):214–223. | |
Miyamoto K, Kesterson RA, Yamamoto H, et al. Structural organization of the human vitamin D receptor chromosomal gene and its promoter. Mol Endocrinol. 1997;11(8):1165–1179. | |
Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Research. 1988;16(3):1215. | |
Kyogoku C, Langefeld CD, Ortmann WA, et al. Genetic association of the R620W polymorphism of protein tyrosine phosphatase PTPN22 with human SLE. Am J Hum Genet. 2004;75(3):504–507. | |
Bid HK, Mittal RD. Study of vitamin-D receptor (VDR) gene start codon polymorphism (Fok I) in healthy individuals from North India. Indian Journal of Human Genetics. 2003;9(2):51–54. | |
Klareskog L, Padyukov L, Lorentzen J, Alfredsson L. Mechanisms of disease: genetic susceptibility and environmental triggers in the development of rheumatoid arthritis. Nat Clin Pract Rheumatol. 2006;2(8):425–433. | |
Majorczyk E, Jasek M, Ploski R, et al. Association of PTPN22 single nucleotide polymorphism with rheumatoid arthritis but not with allergic asthma. Eur J Hum Genet. 2007;15(10):1043–1048. | |
Mastana S, GilmourA, Ghelani A, Smith H, Samanta A. Association of PTPN22 with rheumatoid arthritis among South Asians in the UK. J Rheumatol. 2007;34(10):1984–1986. | |
Lee YH, Kim HJ, Rho YH, Choi SJ, Ji JD, Song GG. Interleukin-1 receptor antagonist gene polymorphism and rheumatoid arthritis. Rheumatol Int. 2004;24(3):133–136. | |
Maalej A, Petit-Teixeira E, Michou L, Rebai A, Cornelis F, Ayadi H. Association study of VDR gene with rheumatoid arthritis in the French population. Genes and Immunity. 2005;6(8):707–711. | |
Feldmann M, Brennan FM, Maini RN. Rheumatoid arthritis. Cell. 1996;85(3):307–310. | |
Buch M, Emery P. The etiology and pathogenesis of rheumatoid arthritis. Hospital Pharmacist. 2002;9. | |
Viatte S, Plant D, Raychaudhuri S. Genetics and epigenetics of rheumatoid arthritis. Nature Reviews Rheumatology. 2013;9(3):141–153. | |
Cloutier JF, Veillette A. Co-operative inhibition of T-cell antigen receptor signaling by a complex between a kinase and a phosphatase. J Exp Med. 1999;189(1):111–121. | |
Steer S, Lad B, Grumley JA, Kingsley GH, Fisher SA. Association of R602W in a protein tyrosine phosphatase gene with a high risk of rheumatoid arthritis in a British population: evidence for an early onset/disease severity effect. Arthritis Rheum. 2005;52:358–360. | |
Begovich AB, Victoria E, Carlton H, et al. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am J Hum Genet. 2004;75(2):330–337. | |
Garcia-Lozano JR, Gonzalez-Escribano MF, Valenzuela A, Garcia A, Nunez-Roldan A. Association of vitamin D receptor genotypes with early onset rheumatoid arthritis. Eur J Immunogenet. 2001;28(1):89–93. | |
Hitchon CA, Sun Y, Robinson DB, et al. Vitamin D receptor polymorphism rs2228570 (Fok1) is associated with rheumatoid arthritis in North American natives. J Rheumatol. 2012;39(9):1792–1797. | |
Penna G, Adorini L. 1 Alpha, 25-dihydroxyvitamin D3 inhibits differentiation, maturation, activation, and survival of dendritic cells leading to impaired alloreactive T cell activation. J Immunol. 2000;164(5):2405–2411. | |
Karray EF, Ben Dhifallah I, Ben Abdelghani K, et al. Associations of vitamin D receptor gene polymorphisms FokI and BsmI with susceptibility to rheumatoid arthritis and Behçet’s disease in Tunisians. Joint Bone Spine. 2012;79:144–148. | |
Panierakis C, Goulielmos G, Mamoulakis D, Petraki E, Papavasiliou E, Galanakis E. Vitamin D receptor gene polymorphisms and susceptibility to type 1 diabetes in Crete, Greece. Clin Immunol. 2009;133:276–281. | |
Ramos-Lopez E, Kurylowicz A, Bednarczuk T, Paunkovic J, Seidl C, Badenhoop K. Vitamin D receptor polymorphisms are associated with Graves’ disease in German and Polish but not in Serbian patients. Thyroid. 2005;15:1125–1130. | |
Pani MA, Seissler J, Usadel KH, Badenhoop K. Vitamin D receptor genotype is associated with Addison’s disease. Eur J Endocrinol. 2002;147:635–640. | |
Lee YH, Choi SJ, Ji JD, Song GG. Vitamin D receptor ApaI, TaqI, BsmI, and FokI polymorphisms and psoriasis susceptibility: a Meta-analysis. Mol Biol Rep. 2012;39:6471–6478. | |
Tajouri L, Ovcaric M, Curtain R, et al. Variation in the vitamin D receptor gene is associated with multiple sclerosis in an Australian population. J Neurogenet. 2005;19:25–38. |
 © 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.