Back to Journals » OncoTargets and Therapy » Volume 11
Role of miR-613 as a tumor suppressor in glioma cells by targeting SOX9
Received 9 November 2017
Accepted for publication 24 January 2018
Published 30 April 2018 Volume 2018:11 Pages 2429—2438
DOI https://doi.org/10.2147/OTT.S156608
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Yao Dai
Qiuling Sang,1 Xuejuan Liu,2 Daju Sun3
1Department of Neurology, The China–Japan Union Hospital of Jilin University, Changchun, People’s Republic of China; 2Department of Pathology, The First Hospital of Jilin University, Changchun, People’s Republic of China; 3Department of Pathology, The China–Japan Union Hospital of Jilin University, Changchun, People’s Republic of China
Background and objectives: MicroRNA-613 (miR-613), a novel cancer-related microRNA, has been shown to be responsible for the inhibition of tumor development and progression in various cancers. We aimed to investigate the biological function and regulatory mechanisms of miR-613 in gliomas.
Materials and methods: miR-613 expression were detected by qRT-PCR assays in glioma tissues and cell lines. Cell Counting Kit-8 (CCK-8) assay, colony formation analysis, wound healing and transwell invasion assays were performed to evaluate cell proliferation, colony formation, migration and invasion abilities. Luciferase reporter assays, qRT-PCR and Western blot were performed to explore the potential targets of miR-613. Xenograft mice model was established to evaluate the effect of miR-613 in vivo.
Result: The expression levels of miR-613 were significantly downregulated in the glioma tissues and cell lines, and the decreased level was significantly negatively associated with the overall disease-free survival of the patients. Functionally, ectopic expression of miR-613 in glioma cells suppressed the proliferation, colony formation, and migration and invasion of the cells. The sex-determining region Y-box 9 (SOX9) was identified as a direct functional target of miR-613, and its expression was inversely correlated with miR-613 expression in glioma tissues. Moreover, rescue of SOX9 could partially reverse the inhibitory effects of miR-613 on glioma cell proliferation, colony formation, migration, and invasion. Importantly, miR-613 also suppressed tumor growth in vivo by targeting SOX9.
Conclusion: Taken together, these findings demonstrate that miR-613 functions as a tumor suppressor in glioma cells by directly targeting SOX9.
Keywords: miR-613, glioma, SOX9, proliferation, invasion
Introduction
Malignant gliomas are the most common and aggressive type of primary tumors in the central nervous systems of adults worldwide, accounting for ~30% of central nervous system tumors and 80% of all malignant tumors in the brain.1 Despite aggressive therapies, including surgical resection, radiotherapy, and chemotherapy, the prognosis for patients with glioma remains poor mainly because of significant malignant proliferation and invasion.2 Accumulating evidence suggests that molecular heterogeneity is a key hurdle for improving the clinical outcomes of gliomas.3 Therefore, elucidation of the molecular mechanisms involved in glioma proliferation and invasion is urgently needed for identifying effective therapeutic targets for this disease.
MicroRNAs (miRNAs) are short (18–25 nucleotides long) single-stranded RNA molecules that regulate gene expression by binding to the 3′-untranslated region (3′-UTR) of target mRNAs.4,5 Because of their mediation of target gene expression, miRNAs have been implicated in the regulation of various biological processes, such as cell proliferation, invasion, apoptosis, and differentiation.6,7 miRNAs have been shown to be involved in various physiological and pathological processes of gliomas,8,9 suggesting that they could serve as diagnostic markers or therapeutic agents for this type of cancer.
Several research studies have shown that miRNA-613 (miR-613) was obviously downregulated in colorectal cancer,10 bladder cancer,11 osteosarcoma,12 thyroid cancer,13 breast cancer,14 hepatocellular carcinoma,15 nonsmall-cell lung cancer,16 and ovarian cancer.17 Although miR-613 has been shown to inhibit cell proliferation and colony formation, induce cell cycle arrest in the G0/G1 phase, and suppress the invasive ability of glioma cells, the function and underlying mechanism of miR-613 in gliomas remain largely unclear.18
Sex-determining region Y-box 9 (SOX9), a high-mobility group box-containing transcription factor, is a member of the sex-determining region Y family, which plays critical roles during embryogenesis, cell differentiation, tumor initiation and invasion, and stem cell self-renewal.19,20 The dysregulation of SOX9 has been shown to function like an oncogene, promoting cell proliferation, inhibiting senescence, and facilitating cellular transformation.21 In addition, elevated SOX9 expression has been observed in glioma cell lines and clinical specimens, and its increased level was significantly correlated with higher World Health Organization (WHO)-grade gliomas.22–24 Collectively, these studies imply that miR-613 and SOX9 are involved in glioma progression, but their roles in regulating glioma development remain to be explained.
In this study, the expression status and clinical significance of miR-613, as well as its role and underlying mechanism, were investigated using glioma tissues from patients and various glioma cell lines. It was found that the miR-613 levels were significantly downregulated in all the glioma tissues and cell lines tested and that the decreased level was significantly negatively associated with the overall disease-free survival of the patients. Functional assays showed that miR-613 inhibited glioma growth in vitro and in vivo. Mechanistic analysis revealed that miR-613 exerted its suppressive role in gliomas by targeting SOX9. These studies suggest that miR-613 might be a potential therapeutic target against gliomas.
Patients and methods
Clinical samples
Glioma tissues were collected from 30 patients (17 men and 13 women, mean age: 58.2 years) with gliomas, who were histologically diagnosed at the Department of Neurology, China–Japan Union Hospital of Jilin University (Changchun, People’s Republic of China). Normal brain specimens were collected from 5 patients (3 men and 2 women, mean age: 45.6 years) who had suffered traumatic brain injuries. The glioma specimens were categorized as either low-grade gliomas (WHO I–II, 18 cases) or high-grade gliomas (WHO III–IV, 12 cases) according to the diagnosis done by two experienced clinical pathologists from our hospital. Informed consent was signed by all the patients before surgery. Written informed consent was obtained from all the patients whose biological samples were used in this study. This study was approved by the research ethics committee of Jilin University.
Cell culture and transfection
Human glioma cell lines U251, T98, and U87MG and human umbilical vein endothelial cells were obtained from the Chinese Academy of Sciences Cell Bank (Shanghai, People’s Republic of China). All cells were grown in high-glucose Dulbecco’s Modified Eagle’s Medium (DMEM; Thermo Fisher Scientific, Waltham, MA, USA) containing 10% fetal bovine serum (Thermo Fisher Scientific) and 100 units/mL penicillin or 100 μg/mL streptomycin at 37°C in a humidified air atmosphere containing 5% carbon dioxide.
A negative control mimic (miR-NC) and the miR-613 mimic were purchased from GenePharma (Shanghai, People’s Republic of China). The SOX9 overexpression vector pCDNA3.1-SOX9, which lacks the 3′-UTR, was a kind gift from Dr Ju Peng (Jilin University).
The U87MG cells were transiently transfected with the aforementioned oligonucleotides (final concentration: 100 nM each) or the expression vector (100 g) using Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s instructions.
RNA preparation and real-time PCR
Total RNA was isolated from the frozen tissue samples and the cultured cells using TRIzol reagent (Thermo Fisher Scientific) according to the manufacturer’s protocol. Then, cDNA was synthesized from 100 ng of the RNA, using a BcaBEST RNA PCR kit (Takara, Dalian, People’s Republic of China) according to the manufacturer’s instruction. The cDNAs were subjected to the quantitative reverse transcription polymerase chain reaction (qRT-PCR) using SYBR Premix Ex Taq (Takara) to detect miR-613 and SOX9 mRNA with the ABI 7900 Fast system (Thermo Fisher Scientific). The primers used in this study have been described previously.15,22 The U6 gene was used as an endogenous control for miRNA expression,25 whereas the GAPDH gene was used as an endogenous control for mRNA expression. The relative expression levels of miR-613 and SOX9 mRNA were calculated using the comparative delta CT (2−ΔΔCT) method.26
Cell proliferation and colony formation assays
Cell proliferation was determined using Cell Counting Kit-8 (CCK-8; Dojindo Laboratories, Kumamoto, Japan) according to the manufacturer’s instructions. In brief, transfected cells were collected and seeded into 96-well plates (Corning Incorporated, Corning, NY, USA) at a density of 2,000 cells per well and cultured for 24–72 h. At indicated times (24, 48, and 72 h), 10 μL of CCK-8 solution was added to each well. Cell viability was determined by measuring the absorbance at 450 nm using a microplate reader (Bio-Tek Company, Winooski, VT, USA).
For the colony formation assay, 1,000 transfected cells per well were seeded into six-well plates (Corning Incorporated) and cultured for 14 days. The cell colonies were fixed with 4% paraformaldehyde for 30 min and then stained with 0.1% crystal violet for 10 min. The stained colonies were imaged and counted under a light microscope (Olympus Corporation, Tokyo, Japan).
Cell migration and invasion assays
To measure cell migration, 8 mm pore size culture inserts (Transwell; Corning Incorporated) for separating upper and lower chambers were placed into the wells of 24-well culture plates. In the lower chamber, 600 μL of DMEM containing 10% fetal bovine serum was added as the chemoattractant, whereas the upper chamber inserts were seeded with 2 × 105 transfected cells in serum-free DMEM. After incubation at 37°C with 5% CO2 for 24 h, the cells that had migrated through the pores were fixed with 4% paraformaldehyde for 30 min and then stained with 0.1% crystal violet for 10 min. For the cell invasion assay, the upper chamber was precoated with 30 μL of Matrigel solution (BD Biosciences, San Jose, CA, USA) and then seeded with 1 × 105 cells. The remainder of the procedure was similar to that of the cell migration assay. The number of cells was quantified by counting five independent visual fields under a light microscope (Olympus Corporation; 200×).
Luciferase assay
Four established bioinformatic prediction tools (miRDB, miRanda, TargetScan, and RNA2222) were used to predict potential miR-613 targets. Human SOX9 3′-UTRs, containing either the putative miR-613 binding site or a mutant site, were synthesized by GenePharma and inserted into the psiCHECK-2 vector (Ambion, Austin, TX, USA) between the NotI and XhoI sites. The resulting expression vectors were named wild type (WT)-SOX9 and mutated (MUT)-SOX9. All constructed plasmids were confirmed by sequencing. For the luciferase reporter assay, U87MG cells were seeded into 96-well plates (1 × 104 cells/well) for 24 h and were then co-transfected with 100 ng of either the WT-SOX9 or the MUT-SOX9 3′-UTR reporter plasmids, and 100 nM miR-613 mimic or miR-NC. At 48 h after transfection, the cells were lysed, and luciferase assays were carried out using the Dual-Luciferase Reporter Assay System (Promega Corporation, Fitchburg, WI, USA). The firefly luciferase activity was normalized to that of Renilla luciferase.
Western blot analysis
The glioma cells or tissues were incubated with radio immunoprecipitation assay buffer (Shanghai Gefan Biotechnology Co., Ltd., Shanghai, China) on ice for 30 min and then centrifuged at 20,000 × g at 4°C for 15 min. The supernatants were collected, and their protein concentrations were determined with a bicinchoninic Acid kit (Shanghai Gefan Biotechnology Co., Ltd.). Equal amounts of protein (30 μg) were separated by 10%–15% sodium dodecyl sulfate polyacrylamide gel electrophoresis under a constant voltage of 110 V and then transferred to polyvinylidene difluoride membranes. The membranes were blocked with 5% skim milk for 1 h and then incubated overnight at 4°C with the following antibodies: rabbit anti-SOX9 (1:1,000; Santa Cruz Biotechnology Inc., Dallas, TX, USA) and rabbit anti-GAPDH (1:2,000; Santa Cruz Biotechnology Inc.). The membranes were then incubated with horseradish peroxidase-conjugated goat-anti-rabbit antibody (1:5,000; Santa Cruz Biotechnology Inc.). The protein bands were observed by enhanced chemiluminescence amplification according to the manufacturer’s protocol (GE Healthcare UK Ltd, Little Chalfont, UK). The density of each band was quantified with Image-Pro Plus 6.0 software.
In vivo assay
BALB/c nude mice were purchased from Jilin Laboratory Animal Limited Liability Company (Jilin, People’s Republic of China) and maintained in our animal facility under standard conditions. Xenograft transplantation of glioma cells into the mice was performed as described previously.27 In brief, after pretransplant preparation and 5% chloral hydrate anesthetization of the nude mice, U87MG cells (2 × 106) transfected with miR-613 mimic or miR-NC were, respectively, implanted into the left and right flanks (cells per flank) of the mice (n = 10). The tumor volumes were determined every 5 days by measuring the tumor length (L) and width (W) and were calculated according to the formula V = LW2/2. After 30 days, the mice were sacrificed, and the tumors were extracted and weighed. The expression of SOX9 protein in the xenografted tumor tissues was examined by Western blot assay. All the experiments were performed according to the institutional guidelines and approved by the ethical committee of Jilin University.
Statistical analysis
The SPSS 17.0 for Windows (SPSS Inc., Chicago, IL, USA) was used for all statistical analyses. The data shown are presented as mean ± SD from at least three independent experiments. Differences between experimental groups were analyzed using Student’s t-test or one-way analysis of variance. Kaplan–Meier survival analysis was performed to analyze the survival distributions. The correlation between expression levels of miR-613 and SOX9 was assessed using Spearman’s correlation analysis. All differences were considered statistically significant at P < 0.05.
Results
miR-613 is downregulated in glioma cells and correlated with poor survival
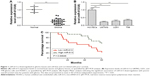
To determine the miR-613 status in gliomas, the expression level of miR-613 in human glioma tissues and normal brain tissues was first measured by qRT-PCR. It was found that miR-613 expression was significantly (P < 0.05) downregulated in the 30 glioma tissues relative to the levels in the five normal brain tissues (Figure 1A). In agreement with this result, miR-613 was also downregulated in the three glioma cell lines U251, T98, and U87MG compared with that in normal human umbilical vein endothelial cells (P < 0.05, Figure 1B). Because the U87MG cell line had the lowest expression of miR-613 among the glioma cell lines (Figure 1B), it was selected for the functional assays. On the basis of a median miR-613 expression level of 0.25, the patients with glioma were divided into two groups: high miR-613 expression group (>0.25, n = 15) and low miR-613 expression group (≤0.25, n = 15). Kaplan–Meier survival analysis revealed that patients in the low miR-613 expression group had shorter overall survival than those in the high miR-613 expression group (Figure 1C). These results suggested that the decreased expression of miR-613 might play an important role in glioma progression and development.
miR-613 inhibits glioma cell proliferation, colony formation, migration, and invasion
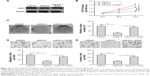
To further investigate the biological role of miR-613 in gliomas, U87MG cells were transfected with either miR-613 mimic or miR-NC. The cells transfected with miR-613 mimic had significantly higher expression levels of miR-613 than the miR-NC-transfected cells (Figure 2A). The CCK-8 assay results indicated that miR-613 had significantly inhibited U87MG cell proliferation (Figure 2B). In addition, the colony formation assay was used to investigate the role of miR-613 in the clonogenic survival of U87MG cells. Relative to that observed in the miR-NC group, the overexpression of miR-613 had decreased the clonogenic survival of U87MG cells by 60% (Figure 2C). Next, the potential effect of miR-613 on cell motility and invasiveness was investigated by the transwell chamber assay. It was found that miR-613 overexpression had significantly inhibited the migratory and invasive capabilities of the U87MG cells (Figure 2D and E). These data suggested that miR-613 might be a tumor suppressor in gliomas.
SOX9 is a direct target of miR-613 in glioma cells
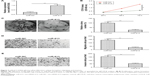
To predict the potential targets of miR-613, four established bioinformatic prediction tools (miRDB, miRanda, TargetScan, and RNA22) were used. SOX9 was subsequently selected as a target gene of miR-613 on the basis of its biological function in gliomas.22–24 SOX9 has previously been found to be upregulated in glioma tissues and cell lines and to be closely involved in glioma cell proliferation, migration, and invasion.22–24 As shown in Figure 3A, the SOX9 mRNA had one possible binding site for miR-613, at the 1,103–1,110 bp position. The mRNA expression of SOX9 was upregulated in the glioma tissues (Figure 3B), and its expression was inversely correlated with miR-613 expression (r = −0.638, P < 0.001, Figure 3C). To confirm if SOX9 interacts directly with miR-613, a Dual-Luciferase® Reporter System was used. The WT or MUT 3′-UTRs of SOX9 mRNA were inserted downstream of the luciferase reporter gene and then co-transfected with miR-613 mimic or miR-NC into U87MG cells. It was found that miR-613 significantly inhibited the luciferase activity of WT-SOX9 3′-UTR but not that of MUT-SOX9 3′-UTR (Figure 3D). Moreover, the SOX9 mRNA or protein levels were markedly lower in the U87MG cells transfected with miR-613 mimic than in the cells transfected with miR-NC (Figure 3E and F). These results indicate that miR-613 can directly target the 3′-UTR of SOX9 mRNA.
Restoration of SOX9 expression rescues the inhibitory effects caused by miR-613 in glioma cells
To further confirm whether miR-613 exerts its function by targeting SOX9 in the glioma cells, U87MG cells were transfected with either miR-613 mimic or miR-NC alone or co-transfected with miR-613 mimic plus pCDNA3.1-SOX9 (lacking the 3′-UTR) and then subjected to a series of functional assays. Western blot analysis revealed that co-transfection with the SOX9 overexpression vector had successfully mitigated the miR-613 overexpression-induced restriction of SOX9 protein expression (Figure 4A). CCK-8 assay showed that the reduced proliferation induced by miR-613 overexpression in U87MG cells was partly abolished by the introduction of SOX9 overexpression plasmid (Figure 4B). Consistent with these results, colony formation analysis revealed that inhibition role of miR-613 in colony formation was reversed by SOX9 overexpression treatment (Figure 4C). Meanwhile, transwell migration/invasion assay revealed that the inhibitory role of miR-613 in cell migration and invasion was also reversed by the SOX9 overexpression plasmid (Figure 4D and E). These data suggested that miR-613 exerts its antitumor effect in gliomas, at least in part, by repressing SOX9.
miR-613 suppresses tumor growth in vivo
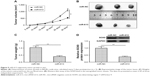
To investigate the effect of miR-613 on tumor growth in vivo, U87MG cells stably expressing miR-613 or miR-NC were implanted into nude mice by subcutaneous injection, and the growth of the tumors was measured every 5 days. Tumor growth was slower in the miR-613/U87MG group than in the miR-NC/U87MG group (Figure 5A). At 30 days postinjection, the mice were sacrificed, and the tumor tissues were extracted and weighed. The weight and size of the xenografted tumors of the miR-613/U87MG group were found to be smaller than those of the miR-NC/U87MG group (Figure 5B and C). Moreover, SOX9 protein expression was significantly lower in the miR-613/U87MG group than in the miR-NC/U87MG group (Figure 5D).
Discussion
miRNA deregulation has become a main characteristic of malignant gliomas, and some miRNAs are potentially novel biomarkers for glioma diagnosis and prognosis.8,9 In addition, some miRNAs have been shown to be involved in the inhibition of glioma progression and development and therefore proposed as potential therapeutic targets against gliomas.8,9 Therefore, it is important to find novel miRNAs in gliomas in order to gain more prognostic biomarkers and attractive therapeutic targets.
miR-613 has been reported to be downregulated in several types of cancers and to function as a tumor suppressor.10–17 For example, miR-613 suppressed colorectal cancer cell proliferation, migration, and invasion and induced cell cycle arrest at the G1 phase by targeting formin-like protein 2.10 Overexpression of miR-613 in osteosarcoma cells significantly suppressed tumor cell proliferation and colony formation by regulating cell cycle arrest at the G0/G1 phase, impaired the migratory and invasive abilities of the cells, and subsequently suppressed epithelial–mesenchymal transition through the repression of c-MET.12 miR-613 significantly suppressed thyroid cancer cell growth, migration, and invasion in vitro and inhibited the tumor growth in vivo by targeting sphingosine kinase 2.13 Although a study showed that overexpression of miR-613 significantly inhibits glioma cell growth,18 the role and underlying mechanism of miR-613 in gliomas remain largely unclear. In this study, miR-613 expression in glioma tissues and cell lines was investigated, and it was found that its expression was strongly downregulated in all the glioma cells tested, which was consistent with previous studies.18 It was also found that miR-613 expression was negatively associated with the overall disease-free survival of the patients with gliomas. With regard to the biological function of miR-613, it was found that its overexpression suppressed glioma cell growth, migration, and invasion in vitro and inhibited tumor growth in vivo. Collectively, these data strongly suggested that miR-613 might be a tumor suppressor miRNA in gliomas.
It has been demonstrated that miRNAs regulate various biological and pathological processes, including tumor growth, cell invasion, and tumor metastasis, by repressing target genes.28 To further investigate the biomolecular mechanism underlying the miR-613 effects in glioma cells, potential targets of miR-613 were screened for using four bioinformatic prediction tools. Subsequently, SOX9 was selected as a target gene of miR-613. Through luciferase reporter assay, qRT-PCR, and Western blotting, it was further confirmed that SOX9 is a direct target of miR-613 in gliomas. It has been shown that SOX9 overexpression significantly increased glioma cell proliferation, migration, and invasion by regulating several pathways, such as the Wnt29 and AKT signaling pathways.30,31 Importantly, SOX9 has been reported to be regulated by miR-101, miR-145, and miR-105 in gliomas.32–34 These results implied that SOX9 functions as an oncogene in gliomas. In this study, it was shown that SOX9 mRNA expression was upregulated and inversely correlated with miR-613 expression in glioma tissues. It was also demonstrated that the overexpression of SOX9 effectively rescued the miR-613 overexpression-induced inhibitory effects on glioma cell proliferation, colony formation, migration, and invasion. Moreover, in vivo assay showed that miR-613 suppressed tumor growth in nude mice by repressing SOX9. These results suggest that miR-613 plays a tumor suppressor role in gliomas, at least in part, by targeting SOX9.
Conclusion
Evidence for the role of miR-613 as a tumor-suppressive miRNA has been provided, where it was strongly downregulated and negatively correlated with the overall disease-free survival of patients with glioma. It has been shown that miR-613 overexpression suppressed glioma cell proliferation, colony formation, migration, and invasion in vitro, as well as tumor growth in vivo, by directly targeting SOX9. Taken together, these findings suggest that miR-613 might be a novel therapeutic target against gliomas.
Acknowledgment
This study was funded by The First Affiliated Hospital of Jilin University Foundation (No 2016B05) and the National Natural Science Foundation of China (No 81402062).
Disclosure
The authors report no conflicts of interest in this work.
References
Buckner JC, Brown PD, O’Neill BP, Meyer FB, Wetmore CJ, Uhm JH. Central nervous system tumors. Mayo Clin Proc. 2007;82(10):1271–1286. | ||
Wang Y, Jiang T. Understanding high grade glioma: molecular mechanism, therapy and comprehensive management. Cancer Lett. 2013;331(2):139–146. | ||
Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. | ||
Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835–840. | ||
McManus MT. MicroRNAs and cancer. Semin Cancer Biol. 2003;13(4):253–258. | ||
Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113(1):25–36. | ||
Sun K, Lai EC. Adult-specific functions of animal microRNAs. Nat Rev Genet. 2013;14(8):535–548. | ||
Tivnan A, McDonald KL. Current progress for the use of miRNAs in glioblastoma treatment. Mol Neurobiol. 2013;48(3):757–768. | ||
Barciszewska AM. MicroRNAs as efficient biomarkers in high-grade gliomas. Folia Neuropathol. 2016;54(4):369–374. | ||
Li B, Xie Z, Li Z, Chen S, Li B. MicroRNA-613 targets FMNL2 and suppresses progression of colorectal cancer. Am J Transl Res. 2016;8(12):5475–5484. | ||
Yu H, Duan P, Zhu H, Rao D. miR-613 inhibits bladder cancer proliferation and migration through targeting SphK1. Am J Transl Res. 2017;9(3):1213–1221. | ||
Li X, Sun X, Wu J, Li Z. MicroRNA-613 suppresses proliferation, migration and invasion of osteosarcoma by targeting c-MET. Am J Cancer Res. 2016;6(12):2869–2879. | ||
Qiu W, Yang Z, Fan Y, Zheng Q. MicroRNA-613 inhibits cell growth, migration and invasion of papillary thyroid carcinoma by regulating SphK2. Oncotarget. 2016;7(26):39907–39915. | ||
Wu J, Yuan P, Mao Q, et al. miR-613 inhibits proliferation and invasion of breast cancer cell via VEGFA. Biochem Biophys Res Commun. 2016;478(1):274–278. | ||
Wang W, Zhang H, Wang L, Zhang S, Tang M. miR-613 inhibits the growth and invasiveness of human hepatocellular carcinoma via targeting DCLK1. Biochem Biophys Res Commun. 2016;473(4):987–992. | ||
Li D, Li DQ, Liu D, Tang XJ. MiR-613 induces cell cycle arrest by targeting CDK4 in non-small cell lung cancer. Cell Oncol (Dordr). 2016;39(2):139–147. | ||
Zhang X, Zhang H. Diminished miR-613 expression as a novel prognostic biomarker for human ovarian cancer. Eur Rev Med Pharmacol Sci. 2016;20(5):837–841. | ||
Li Q, Zhou L, Wang M, et al. MicroRNA-613 impedes the proliferation and invasion of glioma cells by targeting cyclin-dependent kinase 14. Biomed Pharmacother. 2017;98:636–642. | ||
Larsimont JC, Youssef KK, Sanchez-Danes A, et al. Sox9 controls self-renewal of oncogene targeted cells and links tumor initiation and invasion. Cell Stem Cell. 2015;17(1):60–73. | ||
Sekido R, Lovell-Badge R. Sex determination and SRY: down to a wink and a nudge? Trends Genet. 2009;25(1):19–29. | ||
Matheu A, Collado M, Wise C, et al. Oncogenicity of the developmental transcription factor Sox9. Cancer Res. 2012;72(5):1301–1315. | ||
Gao J, Zhang JY, Li YH, Ren F. Decreased expression of SOX9 indicates a better prognosis and inhibits the growth of glioma cells by inducing cell cycle arrest. Int J Clin Exp Pathol. 2015;8(9):10130–10138. | ||
Hiraoka K, Hayashi T, Kaneko R, et al. SOX9-mediated upregulation of LGR5 is important for glioblastoma tumorigenicity. Biochem Biophys Res Commun. 2015;460(2):216–221. | ||
Wang L, He S, Yuan J, et al. Oncogenic role of SOX9 expression in human malignant glioma. Med Oncol. 2012;29(5):3484–3490. | ||
Ludwig N, Leidinger P, Becker K, et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016;44(8):3865–3877. | ||
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. | ||
Cheng L, Wu Q, Guryanova OA, et al. Elevated invasive potential of glioblastoma stem cells. Biochem Biophys Res Commun. 2011;406(4):643–648. | ||
Kushlinskii NE, Fridman MV, Braga EA. Molecular mechanisms and microRNAs in osteosarcoma pathogenesis. Biochemistry (Mosc). 2016;81(4):315–328. | ||
Liu H, Liu Z, Jiang B, Peng R, Ma Z, Lu J. SOX9 overexpression promotes glioma metastasis via Wnt/beta-Catenin signaling. Cell Biochem Biophys. 2015;73(1):205–212. | ||
Swartling FJ, Ferletta M, Kastemar M, Weiss WA, Westermark B. Cyclic GMP-dependent protein kinase II inhibits cell proliferation, Sox9 expression and Akt phosphorylation in human glioma cell lines. Oncogene. 2009;28(35):3121–3131. | ||
Garros-Regulez L, Aldaz P, Arrizabalaga O, et al. mTOR inhibition decreases SOX2-SOX9 mediated glioma stem cell activity and temozolomide resistance. Expert Opin Ther Targets. 2016;20(4):393–405. | ||
Liu N, Zhang L, Wang Z, et al. MicroRNA-101 inhibits proliferation, migration and invasion of human glioblastoma by targeting SOX9. Oncotarget. 2017;8(12):19244–19254. | ||
Rani SB, Rathod SS, Karthik S, Kaur N, Muzumdar D, Shiras AS. MiR-145 functions as a tumor-suppressive RNA by targeting Sox9 and adducin 3 in human glioma cells. Neuro Oncol. 2013;15(10):1302–1316. | ||
Liu X, Wang H, Zhu Z, Ye Y, Mao H, Zhang S. MicroRNA-105 targets SOX9 and inhibits human glioma cell progression. FEBS Lett. 2016;590(23):4329–4342. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.