Back to Journals » OncoTargets and Therapy » Volume 14
Role of Long Non-Coding RNAs in the Chemoresistance of Gastric Cancer: A Systematic Review
Authors Li Z , Lü M, Zhou Y, Xu L, Jiang Y, Liu Y, Li X, Song M
Received 27 November 2020
Accepted for publication 31 December 2020
Published 18 January 2021 Volume 2021:14 Pages 503—518
DOI https://doi.org/10.2147/OTT.S294378
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Tohru Yamada
Zonglin Li,1 Muhan Lü,2 Yejiang Zhou,1 Linxia Xu,1 Yifan Jiang,1 Yi Liu,1 Xin Li,1 Min Song3
1Department of Gastrointestinal Surgery, The Affiliated Hospital of Southwest Medical University, Luzhou 646000, People’s Republic of China; 2Department of Gastroenterology, The Affiliated Hospital of Southwest Medical University, Luzhou 646000, People’s Republic of China; 3Department of Laboratory Medicine, The Affiliated Hospital of Southwest Medical University, Luzhou 646000, People’s Republic of China
Correspondence: Min Song
Department of Laboratory Medicine, The Affiliated Hospital of Southwest Medical University, Luzhou 646000, Sichuan, People’s Republic of China
Tel +8613982424099
Email [email protected]
Purpose: Accumulating evidence demonstrates that long non-coding RNAs (lncRNAs) play a vital role in the chemoresistance of gastric cancer (GC). The present systematic review summarises the emerging role, potential targets or pathways and regulatory mechanisms of lncRNAs involved in chemoresistance and proposes a number of clinical implications of lncRNAs as novel therapeutic targets for GC.
Methods: Studies on lncRNAs involved in the chemoresistance of GC published until July 2020 in the PubMed and Web of Science databases were systematically reviewed and the expression form, role in chemoresistance, targets or pathways, corresponding drugs and potential mechanisms of relevant lncRNAs were summarised in detail.
Results: A total of 48 studies were included in this systematic review. Amongst these studies, 32 involved single drug resistance and 16 involved in multidrug resistance (MDR). The 48 studies collected described 38 lncRNAs in the drug-resistant cells of GC, including 33 upregulated and 5 downregulated lncRNAs. Cisplatin (DDP) was the most studied drug and lncRNA MALAT1 was the most studied lncRNA related to the chemoresistance of GC. The potential mechanisms of chemoresistance for lncRNAs in GC mainly included, amongst others, reduction of apoptosis, induction of autophagy, repair of DNA damage, promotion of epithelial–mesenchymal transition (EMT) and regulation of the related signalling pathways.
Conclusion: LncRNAs play a vital role in the chemoresistance of GC and are novel therapeutic targets for the disease. Detailed chemoresistance mechanisms, translational studies and clinical trials on lncRNAs in GC are urgently needed.
Keywords: gastric cancer, GC, long non-coding RNAs, lncRNAs, chemoresistance, multidrug resistance, MDR
Introduction
Gastric cancer (GC) is amongst the most common malignant tumours, with over 1,000,000 new cases reported annually, and the third leading cause of cancer-related deaths worldwide.1 Radical surgical resection is the best option for GC patients in the early stages of the disease; however, many GC patients are not diagnosed early on account of the insidious symptoms of the disease and a lack of suitable early-detection techniques. Besides surgical resection, advanced-stage GC is mainly treated with chemotherapy. Unfortunately, intrinsic or acquired chemoresistance may interfere with the clinical treatment of GC and lead to treatment failure.
Long non-coding RNAs (lncRNAs) are a class of RNA transcripts with lengths longer than 200 nucleotides and no or limited protein-coding potential; these RNAs perform vital regulatory roles in many aspects of genome function, including gene epigenetics and transcriptional and posttranscriptional levels, as well as a variety of biological processes related to the cell cycle, differentiation, migration, angiogenesis, apoptosis, autophagy and the immune response.2–4 Recent studies have revealed that lncRNAs play an important role in the occurrence and development of all types of cancers.5 For instance, lncRNA HOXB-AS3 could promote the proliferation and migration of lung cancer cells by activating the PI3K-AKT pathway.6 Another study revealed that lncRNA GAS5 affects epithelial–mesenchymal transition (EMT) and the invasion of breast cancer cells by regulating miR-216b.7 Lan et al8 revealed that lncRNA OCC-1 could promote the ubiquitination and degradation of HuR protein, thereby inhibiting the proliferation of colorectal cancer cells. Accumulated evidence indicates that lncRNAs modulate the chemoresistance of GC by means of various mechanisms, such as reduction of apoptosis, induction of autophagy, promotion of EMT, alteration of drug concentrations and regulation of the related signalling pathways.9
In this article, we performed a systematic literature review of lncRNAs involved in the chemoresistance of GC and their underlying molecular mechanisms. Our findings reveal that lncRNAs may be potential therapeutic targets of GC.
Materials and Methods
Data Source and Search Strategy
This review was conducted according to Preferred Reporting Items for Systematic reviews and Meta-Analyses (PRISMA) guidelines.10 A systematic search of the PubMed and Web of Science databases for lncRNAs associated with the chemoresistance of GC published in English up to July 2020 was conducted. The keywords used for the search were “gastric cancer”, “long non-coding RNA” and “chemoresistance”. So the following search string was used across both databases: “gastric cancer” AND “long non-coding RNA (lncRNA)” AND “chemoresistance” OR “drug resistance” OR “multidrug resistance (MDR)” OR “cisplatin (DDP)” OR “oxaliplatin (OXA)” OR “5-fluorouracil (5-FU)” OR “adriamycin (ADR)” OR “doxorubicin (DOX)” OR “paclitaxel (PTX)” OR “vincristine (VCR)”.
Study Selection
The inclusion criteria were as follows: (1) Full papers of original research on lncRNAs related to the chemoresistance of GC and (2) studies including basic information, such as lncRNAs’ expression form, role in chemoresistance, targets or pathways, corresponding drugs and potential mechanisms.
The exclusion criteria were as follows: (1) Studies published in abstract form only, reviews, comments and letters and (2) studies related to lncRNAs involved in targeted drugs or non-GC chemotherapeutic drugs.
Two authors (ZL and ML) independently selected the papers for inclusion on the basis of their titles, abstracts and full texts. A third author (MS) evaluated all papers with disagreement, and consensus was achieved through discussion.
Data Extraction
Two authors (ZL and ML) independently extracted the data of all eligible articles. The following information from each article was recorded: (1) paper title, first author, journal and publication year and (2) lncRNA name, expression form, role in chemoresistance, targets or pathways, corresponding drugs and potential mechanisms. The articles and their information were sorted by drug classification, and lncRNAs were sorted in ascending order by the first letter of their names for each type of drug. LncRNAs related to MDR were sorted separately. The results were checked by a third author (MS).
Results
A total of 48 studies were included in this systematic review (Figure 1); amongst these studies, 32 involved single drug resistance and 16 involved in MDR. A total of 32, 5, 11, 13, 4 and 4 studies were related to the chemoresistance of GC to DDP, OXA, 5-FU, ADR or DOX, PTX and VCR, respectively. The 48 studies collected described 38 lncRNAs in the drug-resistant cells of GC, including 33 upregulated and 5 downregulated lncRNAs. DDP was the most studied drug and lncRNA MALAT1 was the most studied lncRNA related to the chemoresistance of GC. The potential mechanisms of chemoresistance for lncRNAs in GC mainly involved, amongst others, reduction of apoptosis, induction of autophagy, repair of DNA damage, promotion of EMT and regulation of the related signalling pathways (Figure 2).
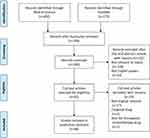 |
Figure 1 Flow diagram representing the systematic review process performed according to PRISMA guidelines. |
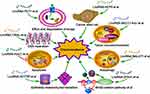 |
Figure 2 Mechanism diagram of long non-coding RNAs in the chemoresistance of gastric cancer. |
LncRNAs Involved in DDP or OXA Resistance in GC
DDP and OXA represent a class of cell-cycle non-specific platinum-based chemotherapeutic drugs that mainly promote the necrosis or apoptosis of cancer cells. Platinum drugs are widely used in all types of cancer therapy, and platinum-based chemotherapy is a standard treatment for GC. A large number of lncRNAs have been reported to be involved in DDP or OXA resistance in GC (Table 1).
 |
Table 1 LncRNAs Involved in DDP or OXA Resistance in GC |
LncRNA ADAMTS9-AS2 acts as a tumour suppressor and could enhance DDP sensitivity in GC cells by activating NLRP3-mediated pyroptotic cell death through sponging miR-223-3p.11 LncRNA ANRIL is highly upregulated in drug-resistant GC tissues and cells and could promote GC cell resistance to DDP by upregulating the expression of MDR1 and MRP1.12 Notch 1 could upregulate lncRNA AK022798 expression and promote DDP-resistant GC cell formation; thus, targeting this lncRNA may provide a new means to treat terminal-stage GC.13 LncRNA ARHGAP5-AS1 is upregulated in chemoresistant GC cells by impaired autophagy, and inhibiting this lncRNA could reverse the chemoresistance of GC cells to DDP.14 LncRNA ASB16‑AS1 is upregulated in DDP-resistant GC cells; this lncRNA could phosphorylate TRIM37 to activate the NF‑κB pathway and promote the proliferation, stemness and DDP resistance of GC.15 LncRNA BCAR4 could regulate the expression of β-catenin by modulating the Wnt signalling pathway and then promote GC cell resistance to DDP.16 Downregulation of lncRNA CASC2 contributes to DDP resistance in GC by sponging miR-19a.17 LncRNA CRAL could function as a competing endogenous RNA to reverse DDP resistance in GC via the miR-505/CYLD/AKT axis.18 LncRNA DANCR contributes to the development of DDP resistance by regulating P-gp and MRP1 expression.19 LncRNA FAM84B-AS is closely associated with the differentiation level, T stage and N stage of GC; this lncRNA offers a certain predictive function for GC prognosis and could promote DDP resistance by inhibiting FAM84B expression.20 LncRNA GHET1 promotes DDP resistance related to the expression of Bax, Bcl-2, MDR1 and MRP1 in GC.21 LncRNA HOTAIR promotes DDP resistance in GC by downregulating miR-34a and miR-126 and activating the PI3K/Akt and Wnt/β-catenin signalling pathways.22,23 The exosome-mediated transfer of lncRNA HOTTIP contributes to DDP resistance in GC cells by regulating the HMGA1/miR-218 pathway,24 while salinomycin reduces EMT-mediated DDP resistance by modifying the expression of lncRNA HOTTIP in GC cells.25 Patients with high expression of lncRNA HULC often have low survival rates; METase could suppress autophagy to reduce DDP resistance in GC cells by regulating the HULC/FoxM1 pathway.26,27 LncRNA MALAT1 contributes to the DDP resistance of GC cells by increasing the level of ATG12, regulating the PI3K/AKT, miR-30b/ATG5 and miR-30e/ATG5 pathways, increasing autophagy and reducing apoptosis.28–31 LncRNA PCAT-1 contributes to DDP resistance in GC by epigenetically silencing PTEN via recruitment of EZH2 and modulating the miR-128/ZEB1 axis. Knockdown of lncRNA PCAT-1 could improve DDP sensitivity.32,33 LncRNA PVT1 may promote the development of DDP resistance by regulating the mTOR/HIF-1a/P-gp and MRP1 signalling pathways; silencing of this lncRNA could reverse DPP resistance in GC via downregulation of TBL1XR1 by sponging miR-3619-5p.34,35 LncRNA ROR could regulate cell growth, metastasis and DDP resistance in GC via the miR-519d-3p/HMGA2 axis.36 Overexpression of lncRNA SNHG5 could upregulate Bax, MDR1 and MRP1, downregulate Bcl-2 and increase the Bcl-2/Bax ratio in GC cells, thereby resulting in DDP resistance in GC.37 LncRNA UCA1 promotes DDP resistance in GC by recruiting EZH2 and activating the PI3K/AKT pathway; the UCA1-miR-27b pathway has also been implicated in the regulation of chemoresistance of GC cells.38,39 LncRNA XIST contributes to DDP resistance by regulating MDR1 and MRP1; downregulation of this lncRNA may promote chemotherapeutic drug-induced apoptosis and partially reverse DDP resistance in GC cells.40 LncRNA XLOC_006753 could promote DDP resistance in GC by inducing cell proliferation and reducing apoptosis via the PI3K/AKT/mTOR pathway.41 LncRNA ZFAS1 is upregulated in GC tissues, and silencing of this lncRNA could inhibit DDP resistance by blocking the Wnt/β-catenin signalling pathway.42
LncRNA BLACAT1 epigenetically targets miR-361 to promote ABCB1 protein expression and is involved in OXA resistance in GC.43 LncRNA DDX11-AS1 facilitates cell progression and OXA resistance by regulating the miR-326/IRS1 axis in GC.44 LncRNA HCP5 drives fatty acid oxidation through the miR-3619-5p/AMPK/PGC1α/CEBPB axis to promote the stemness and OXA resistance of GC.45 Mesenchymal stem cells secrete TGF-β1 and induce lncRNA MACC1-AS1 expression in GC cells, thereby promoting the latter’s stemness and OXA resistance.46 LncRNA MALAT1 contributes to OXA resistance in GC cells by downregulating miR-22-3p and upregulating ZFP91.47
LncRNAs Involved in 5-FU Resistance in GC
Similar to platinum-based drugs, 5-FU is a basic cell cycle-specific drug commonly used to treat GC in the clinical setting. This drug impairs DNA replication by suppressing thymidylate synthase, thereby inhibiting the cell cycle and inducing the apoptosis of cancer cells. Multiple lncRNAs have been proven to be involved in 5-FU resistance in GC (Table 2).
 |
Table 2 LncRNAs Involved in 5-FU Resistance in GC |
LncRNA ANRIL is highly expressed in 5-FU resistant cells, and knockdown of this lncRNA in GC cells increases the percentage of apoptotic tumour cells and inhibits the development of 5-FU resistance.12 LncRNA ARHGAP5-AS1 contributes to chemoresistance in GC; downregulation of this lncRNA could reverse resistance to 5-FU, enhance drug-induced apoptosis and increase the intracellular drug concentration.14 LncRNA HCP5 could abrogate the anti-tumour effect of 5-FU in in vivo assays of nude mice, thus indicating that HCP5 may represent a new therapeutic target for GC.45 LncRNA HOTTIP is upregulated in 5-FU resistant cells, and downregulation of this lncRNA could markedly reduce the 50% inhibitory concentration of 5-FU in GC cells.25 GC patients with high expression of lncRNA HULC often show poor prognosis; silencing of this lncRNA could reverse chemoresistance by inducing apoptosis in GC cells.27 LncRNA LEIGC functions as a tumour suppressor gene by inhibiting EMT and could enhance chemosensitivity to 5-FU in the MGC-803 cell line of GC.48 Overexpression of lncRNA MACC1-AS1 in GC cells could facilitate clone formation and reduce growth inhibition during treatment with 5-FU; the expression of this lncRNA gradually increases with increasing 5-FU concentration during the establishment of chemotherapy-resistant cells.46 LncRNA MALAT1 is significantly upregulated in chemoresistant GC patients, and high expression of this lncRNA results in poor disease-free survival and overall survival in patients who had received 5-FU-based adjuvant therapy.28 LncRNA PVT1 could regulate Bcl2 expression and enhance 5-FU resistance in GC cells. Patients with high PVT1 expression do not experience survival-related benefits from 5-FU-based chemotherapy.49 LncRNA UCA1 is negatively correlated with miR-27b expression in GC tissues, and this lncRNA could increase 5-FU resistance in GC cells by downregulating miR-27b.39 LncRNA XLOC_006753 is upregulated in 5-FU-resistant cells, and overexpression of this lncRNA could promote MDR in GC by inducing cell proliferation metastasis and apoptosis reduction through the PI3K/AKT/mTOR pathway.41
LncRNAs Involved in ADR (or DOX) Resistance in GC
ADR (or DOX) is a cell-cycle nonspecific anticancer drug that can inhibit the synthesis of nucleic acids by embedding into DNA and exert strong cytotoxic effects on cancer cells. Several lncRNAs have been reported to be involved in ADR resistance in GC (Table 3).
 |
Table 3 LncRNAs Involved in ADR (or DOX) Resistance in GC |
LncRNA ARHGAP5-AS1 is a new chemoresistance-promoting antisense lncRNA that is upregulated in ADR-resistant GC cells due to impaired autophagy.14 High expression levels of lncRNA CASC9 are associated with aggressive pathological characteristics, including deep invasion, poor differentiation and increased lymph node metastases. Knockdown of CASC9 in GC cells could restore chemosensitivity to ADR.50 LncRNA D63785 is highly expressed in GC tissues and cells. Silencing of this lncRNA inhibits cell proliferation, migration and invasion in a number of GC cell lines, reduces tumour volumes in mice and increases the sensitivity of GC cells to DOX.51 LncRNA GACAT1 shows low expression in ADR-resistant GC cells. Overexpression of this lncRNA inhibits tumour growth and enhances ADR-mediated antitumour effects in xenograft tumour models of GC.52 The expression of lncRNA HOTAIR is upregulated in GC tissues. Ectopic expression of HOTAIR promotes DOX resistance and increases GC cell proliferation and migration by targeting miR-217.53 LncRNA HOTTIP is a crucial determinant of EMT-mediated ADR resistance in GC cells. Downregulation of this lncRNA could markedly attenuate EMT, thereby reversing ADR resistance.25 LncRNA HULC is involved in ADR resistance in GC, and silencing of this lncRNA could enhance chemotherapy-induced apoptosis in GC cells.27 LncRNA MRUL promotes ABCB1 expression in ADR-resistant GC cell lines, and knockdown of this lncRNA in GC cells increases apoptosis and reduces ADR release in the presence of ADR.54 LncRNA NEAT1 is highly expressed in GC cells and functions as an oncogene that could modulate malignant behaviours and chemoresistance to ADR in GC.55 LncRNA ROR promotes MRP1 expression and ADR resistance in GC cells and is correlated with the poor prognosis of GC patients.56 LncRNA UCA1 is highly expressed in GC tissues and an independent prognostic biomarker of disease-free survival for GC patients. Silencing of this lncRNA could significantly inhibit GC cell proliferation and depress chemoresistance to ADR by upregulating PARP expression and downregulating Bcl-2 expression.57 LncRNA UCA1 also increases ADR resistance in GC by downregulating miR-27b. Overexpression of this lncRNA or inhibition of miR-27b could increase ADR-induced cell apoptosis.39 LncRNA XIST contributes to the ADR resistance of GC cells, and silencing of this lncRNA could downregulate the related genes of MDR, such as MDR1 and MRP1.40
LncRNAs Involved in PTX Resistance in GC
PTX is a new type of antimicrotubule drug that promotes tubulin polymerisation, maintains tubulin stability, inhibits mitosis and arrests cancer cells in the G2 and M phases. Several oncogenic lncRNAs can promote PTX resistance in GC (Table 4).
 |
Table 4 LncRNAs Involved in PTX Resistance in GC |
LncRNA CASC9 is overexpressed in GC tissues and drug-resistant cells. Because it can promote cell growth and chemoresistance to PTX, CASC9 may be a potential therapeutic target for GC.50 LncRNA HOTAIR is upregulated in GC tissues, and higher expression of this lncRNA is associated with a later stage of GC. Overexpression of lncRNA HOTAIR enhances PTX resistance in GC cells by inhibiting miR-217 expression.53 LncRNA PVT1 exhibits high expression in both GC tissues and PTX-resistant GC cells; as such, this lncRNA may potentially be used as a new biomarker for GC to indicate lymph node invasion. Screening for PVT1 expression may be useful in improving early diagnosis and therapeutic effects, thereby enhancing the quality of life of patients with GC.58 LncRNA ZFAS1 is related to cell proliferation, invasion, migration, EMT and resistance to chemotherapeutic reagents, such as PTX and DDP; this finding suggests that the lncRNA may be considered a new diagnostic marker or novel therapeutic target of GC.42
LncRNAs Involved in VCR Resistance in GC
VCR is another type of antimicrotubule drug that mainly inhibits the polymerisation of tubulin and affects the formation of the spindle, ultimately leading to the mitosis arrest of malignant cells. Some lncRNAs cited in the included studies are involved in VCR resistance in GC (Table 5).
 |
Table 5 LncRNAs Involved in VCR Resistance in GC |
LncRNA GACAT1 suppresses tumorigenesis and alleviates VCR resistance by regulating the PTEN/AKT/mTOR/S6K1 pathway in GC. Thus, upregulation of this lncRNA could be considered a potential strategy to alleviate the chemoresistance of GC.52 LncRNA MALAT1 acts as a competing endogenous RNA for miR-23b-3p and attenuates the inhibitory effect of miR-23b-3p, thereby leading to chemo-induced autophagy and VCR resistance in GC cells.28 The expression level of lncRNA MRUL is negatively correlated with the chemosensitivity of GC cells; this lncRNA plays a positive role in the regulation of ABCB1 expression and is a potential target for reversing VCR resistance.54 The increased expression of lncRNA ROR in GC cells reflects the latter’s resistance to treatment with VCR. Inhibition of this lncRNA reduces MRP1 expression and increases apoptosis, which could reverse the VCR resistance of GC cells.56
LncRNAs Involved in MDR in GC
When cancer cells become resistant to a certain chemotherapeutic drug, they may develop cross-resistance to other chemotherapeutic drugs with different chemical properties and mechanisms. This cross-resistance is called MDR. The current studies included in this review revealed that a number of lncRNAs are involved in the MDR of GC (Table 6).
 |
Table 6 LncRNAs Involved in MDR in GC |
LncRNA ANRIL, HCP5, MACC1-AS1 and XLOC_006753 are involved in DDP (or OXA) and 5-FU resistance by upregulating MDR1 and MRP1, regulating the miR-3619-5p/AMPK/PGC1α/CEBPB axis, promoting fatty acid oxidation and antagonising miR-145-5p, and inducing cell proliferation as well as reducing apoptosis through the PI3K/AKT/mTOR pathway, respectively.12,41,45,46 LncRNA XIST promotes the chemoresistance of ADR and DDP by upregulating MDR1 and MRP1.40 LncRNA ZFAS1 could induce the chemoresistance of DDP and PTX by activating the Wnt/β-catenin signalling pathway.42 LncRNA CASC9 and HOTAIR induce ADR (or DOX) and PTX resistance in GC by increasing MDR1 expression and inhibiting miR-217 expression, respectively.50,53 LncRNA MRUL and ROR induce ADR and VCR resistance by promoting ABCB1 expression, increasing MRP1 expression and inhibiting apoptosis,54,56 while lncRNA GACAT1 could reverse the chemoresistance of DOX and VCR by regulating the PTEN/AKT/mTOR/S6K1 signalling pathway.52 LncRNA ARHGAP5-AS1, HOTTIP, HULC and UCA1 could promote the chemoresistance of ADR (or DOX), DDP and 5-FU by reducing apoptosis, downregulating miR-27b and increasing EMT.14,25,27,39 LncRNA MALAT1 induces the chemoresistance of DDP, 5-FU and VCR by promoting autophagy through sequestration of miR-23b-3p and then increasing ATG12 levels.28
Discussion
GC, a common malignant disease with high mortality, threatens human health worldwide and may place a massive financial burden on society.59 The diagnosis of GC is heavily dependent on gastroscopy plus biopsy, but the clinical applications of these techniques are limited because of their invasiveness and inconvenience. Additionally, because of a lack of obvious and specific symptoms at the early stages of the disease, most GC patients are diagnosed at advanced stages with poor prognosis; some patients even suffer from cancerometastasiswhen they are first diagnosed. Gastrectomy with D2 lymphadenectomy is an effective treatment for GC, but single surgical resection, even with enlarged lymphadenectomy, cannot effectively prolong the survival time of GC patients.60 Chemotherapy is as important as surgical resection for patients in advanced stages of GC. Indeed, chemotherapy is the first-line standard and the most important treatment method for patients who are not suitable for surgical treatment. Unfortunately, chemoresistance occurs frequently during the long-term chemotherapy of GC patients and results in poor prognosis, tumour recurrence and metastasis. Therefore, chemoresistance is a major clinical challenge for GC treatment.
The mechanisms of the chemoresistance of GC cells are complex and may include: (1) Acceleration of the drug efflux by ATP-binding cassette transporters and reduction of the effective drug concentration by inactivating drugs in cancer cells; (2) promotion of EMT in cancer cells; (3) repair of DNA damage; (4) induction of drug target mutations; (5) dysfunction of apoptosis and autophagy; (6) alteration of the tumour microenvironment, such as anoxia and cytokines; and (7) various mechanisms of cancer stem cells.61–63 The same GC cells may involve multiple or different mechanisms, and different GC cells may have the same mechanisms of drug resistance. Although the mechanisms of chemoresistance in GC have been broadly studied, details of the associated regulatory mechanisms are not completely understood.
LncRNAs are transcripts of over 200 nucleotides that interact with DNA, RNA and various proteins; these molecules have been determined to participate in various physiological and pathological processes.64 Advanced sequencing technology has revealed that numerous lncRNAs are aberrantly expressed in all types of cancers. LncRNAs have been reported to act as critical factors in the occurrence and development of cancers by regulating angiogenesis, cell proliferation and metastasis.65 Increasing evidence shows that many lncRNAs are related to the clinicopathological features of GC, such as tumour size, differentiation, TNM stage and metastasis.66,67 More importantly, lncRNAs are widely involved in the chemoresistance of GC by regulating various mechanisms. These findings provide novel insights into potential treatments for drug-resistant GC cells.
Treatments targeting abnormally expressed lncRNAs may potentially reverse drug resistance. The combination of lncRNA-based targeted therapy with traditional chemotherapy may be a promising option to address chemoresistance and prolong the overall survival and improve the life quality of GC patients.68 Nevertheless, considerable work must be done before lncRNA-based targeted therapy can be applied in clinical applications. Firstly, the mechanisms of chemoresistance are complicated, the role of lncRNAs remains poorly understood, and the selection of critical lncRNAs from numerous candidates is challenging. Secondly, the majority of the available research involves in vitro studies; animal experiments involving lncRNAs are relatively scarce, but effective functional studies rely on animal experiments prior to clinical trials. Thirdly, as the sequence conservation of lncRNAs is much poorer than that of protein-coding genes, even with a perfect lncRNA target, obtaining a desirable delivery mode is difficult because of off-target effects, low transfection efficacy and the short half-life of RNA.69 Anyhow, recent studies have provided sufficient data to sustain further research on the role of lncRNAs in the chemoresistance of GC. LncRNAs serve as either oncogenes or tumour suppressors and, thus, may provide promising therapeutic targets to help GC patients overcome the problem of chemoresistance and improve survival rates in the near future.
Conclusion
Overall, this systematic review provides compelling evidence for lncRNAs as a regulator of chemoresistance in GC. As shown in Figure 2, the underlying mechanisms of chemoresistance for lncRNAs in GC mainly involved degradation of drugs, reduction of apoptosis, induction of autophagy, repair of DNA damage, promotion of EMT, alteration of tumor microenvironment, stemness of cancer cell and regulation of the related signalling pathways. In conclusion, lncRNAs play a vital role in the chemoresistance of GC and are novel therapeutic targets for the disease. Detailed chemoresistance mechanisms, translational studies and clinical trials on lncRNAs in GC are urgently needed, which may eventually improve the prognosis of GC patients.
Abbreviations
GC, Gastric cancer; lncRNA, Long non-coding RNA; miR, microRNA; ceRNA, Competing endogenous RNA; HOXB-AS3, HOXB cluster antisense RNA 3; GAS5, Growth arrest specificity 5; EMT, Epithelial-mesenchymal transition; OCC-1, Overexpressed in colon carcinoma-1; MDR, Multidrug resistance; DDP, Cisplatin; OXA, Oxaliplatin; 5-FU, 5-fluorouracil; ADR, Adriamycin; DOX, Doxorubicin; PTX, Paclitaxel; VCR, Vincristine; ADAMTS9-AS2, ADAMTS9 antisense RNA 2; NLRP3, NLR family pyrin domain containing 3; ANRIL, Antisense non-coding RNA in the INK4 locus; MDR1, MDR gene 1; MRP1, MDR protein 1; ARHGAP5-AS1, ARHGAP5 antisense RNA 1; ASB16-AS1, ASB16 antisense RNA 1; TRIM37, Tripartite motif containing 37; BCAR4, Breast cancer anti-estrogen resistance 4; CASC2, Cancer susceptibility candidate 2; CRAL, Cisplatin Resistance-Associated LncRNA; DANCR, Differentiation antagonizing non-protein coding RNA; P-gp, P-glycoprotein; FAM84B-AS, Family with sequence similarity 84, member B antisense RNA; GHET1, Gastric carcinoma high expressed transcript 1; Bax: Bcl-2-associated X protein; Bcl-2, B-cell lymphoma 2; HOTAIR, HOX antisense intergenic RNA; HOTTIP, HOXA distal transcript antisense RNA; HULC, Highly up-regulated in liver cancer; MALAT1, Metastasis-associated lung adenocarcinoma transcript 1; ATG12: Autophagy related 12; PCAT-1: Prostate cancer–associated transcript 1; PTEN: Phosphatase and tensin homolog; EZH2: Enhancer of zeste homolog 2; ZEB1: Zinc finger E-box binding homeobox 1; PVT1: Plasmacytoma variant translocation 1; TBL1XR1: Transducin beta like 1 x-linked receptor 1; ROR: Regulator of reprogramming; SNHG5: Small nucleolar RNA host gene 5; UCA1: Urothelial cancer associated 1; XIST: X inactive specific transcript; ZFAS1: ZNFX1 antisense RNA 1; BLACAT1: Bladder cancer associated transcript 1; ABCB1: ATP-binding cassette subfamily B member 1; DDX11-AS1: DDX11 antisense RNA 1; HCP5: HLA complex P5; MACC1-AS1: MACC1 antisense RNA 1; ZFP91: Zinc finger protein 91; CASC9: Cancer susceptibility candidate 9; GACAT1: Gastric cancer associated transcript 1; MRUL: MDR-related and up-regulated lncRNA; NEAT1: Nuclear-enriched abundant transcript 1; TRIM37: Tripartite motif containing 37; HMGA1: High mobility group AT-hook 1; ATG5: Autophagy related 5; HMGA2: High mobility group AT-hook 2; ZFP91: Zinc finger protein 91; CDH1: Cadherin 1; ATG12: Autophagy related 12; MEF2D: Myocyte enhancer factor 2D; GPC5: Glypican 5; PTPN14: Protein tyrosine phosphatase non-receptor type 14.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81672458, 30672058).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
2. Engreitz JM, Ollikainen N, Guttman M. Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat Rev Mol Cell Biol. 2016;17:756–770.
3. Kretz M, Siprashvili Z, Chu C, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–235.
4. Uthaya Kumar DB, Williams A. Long non-coding RNAs in immune regulation and their potential as therapeutic targets. Int Immunopharmacol. 2020;81:106279.
5. Zhang X, Liu H, Chen S. Mechanisms of long non-coding RNAs in cancers and their dynamic regulations. Cancers. 2020;12:1245.
6. Jiang W, Kai J, Li D, Wei Z, Wang Y, Wang W. lncRNA HOXB-AS3 exacerbates proliferation, migration, and invasion of lung cancer via activating the PI3K-AKT pathway. J Cell Physiol. 2020.
7. Li Y, Guo X, Wei Y. LncRNA GAS5 affects epithelial-mesenchymal transition and invasion of breast cancer cells by regulating miR-216b. Eur Rev Med Pharmacol Sci. 2020;24:4873.
8. Lan Y, Xiao X, He Z, et al. Long noncoding RNA OCC-1 suppresses cell growth through destabilizing HuR protein in colorectal cancer. Nucleic Acids Res. 2018;46:5809–5821.
9. Liu K, Gao L, Ma X, et al. Long non-coding RNAs regulate drug resistance in cancer. Mol Cancer. 2020;19:13–54.
10. Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6:e1000097.
11. Ren N, Jiang T, Wang C, et al. LncRNA ADAMTS9-AS2 inhibits gastric cancer (GC) development and sensitizes chemoresistant GC cells to cisplatin by regulating miR-223-3p/NLRP3 axis. Aging. 2020;12:11025–11041.
12. Lan WG, Xu DH, Xu C, et al. Silencing of long non-coding RNA ANRIL inhibits the development of multidrug resistance in gastric cancer cells. Oncol Rep. 2016;36:263–270.
13. Zhu L, Zhu Y, Han S, et al. Impaired autophagic degradation of lncRNA ARHGAP5-AS1 promotes chemoresistance in gastric cancer. Cell Death Dis. 2019;10.
14. Hang Q, Sun R, Jiang C, Li Y. Notch 1 promotes cisplatin-resistant gastric cancer formation by upregulating lncRNA AK022798 expression. Anti-Cancer Drug. 2015;1.
15. Fu T, Ji K, Jin L, et al. ASB16-AS1 up-regulated and phosphorylated TRIM37 to activate NF-κB pathway and promote proliferation, stemness, and cisplatin resistance of gastric cancer. Gastric Cancer. 2020.
16. Wang L, Chunyan Q, Zhou Y, et al. BCAR4 increase cisplatin resistance and predicted poor survival in gastric cancer patients. Eur Rev Med Pharmacol Sci. 2017;21:4064–4070.
17. Li Y, Lv S, Ning H, et al. Down-regulation of CASC2 contributes to cisplatin resistance in gastric cancer by sponging miR-19a. Biomed Pharmacother. 2018;108:1775–1782.
18. Wang Z, Wang Q, Xu G, et al. The long noncoding RNA CRAL reverses cisplatin resistance via the miR-505/CYLD/AKT axis in human gastric cancer cells. RNA Biol. 2020;1–14.
19. Xu YD, Shang J, Li M, Zhang YY. LncRNA DANCR accelerates the development of multidrug resistance of gastric cancer. Eur Rev Med Pharmacol Sci. 2019;23:2794–2802.
20. Zhang Y, Li Q, Yu S, et al. Long non-coding RNA FAM84B-AS promotes resistance of gastric cancer to platinum drugs through inhibition of FAM84B expression. Biochem Bioph Res Co. 2019;509:753–762.
21. Zhang X, Bo P, Liu L, Zhang X, Li J. Overexpression of long non-coding RNA GHET1 promotes the development of multidrug resistance in gastric cancer cells. Biomed Pharmacother. 2017;92:580–585.
22. Cheng C, Qin Y, Zhi Q, Wang J, Qin C. Knockdown of long non-coding RNA HOTAIR inhibits cisplatin resistance of gastric cancer cells through inhibiting the PI3K/Akt and Wnt/beta-catenin signaling pathways by up-regulating miR-34a. Int J Biol Macromol. 2018;107:2620–2629.
23. Yan J, Dang Y, Liu S, Zhang Y, Zhang G. LncRNA HOTAIR promotes cisplatin resistance in gastric cancer by targeting miR-126 to activate the PI3K/AKT/MRP1 genes. Tumor Biology. 2016;37:16345–16355.
24. Wang J, Lv B, Su Y, Wang X, Bu J, Yao L. Exosome-mediated transfer of lncRNA HOTTIP promotes cisplatin resistance in gastric cancer cells by regulating HMGA1/miR-218 axis. Onco Targets Ther. 2019;12:11325–11338.
25. Mao Z, Wu Y, Zhou J, Xing C. Salinomycin reduces epithelial-mesenchymal transition-mediated multidrug resistance by modifying long noncoding RNA HOTTIP expression in gastric cancer cells. Anticancer Drugs. 2019;30:892–899.
26. Xin L, Zhou Q, Yuan YW, et al. METase/lncRNA HULC/FoxM1 reduced cisplatin resistance in gastric cancer by suppressing autophagy. J Cancer Res Clin Oncol. 2019;145:2507–2517.
27. Zhang Y, Song X, Wang X, Hu J, Jiang L. Silencing of LncRNA HULC enhances chemotherapy induced apoptosis in human gastric cancer. J Med Biochem. 2016;35:137–143.
28. YiRen H, YingCong Y, Sunwu Y, et al. Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR-23b-3p sequestration in gastric cancer. Mol Cancer. 2017;16.
29. Dai Q, Zhang T, Li C. LncRNA MALAT1 regulates the cell proliferation and cisplatin resistance in gastric cancer via PI3K/AKT pathway. Cancer Manag Res. 2020;12:1929–1939.
30. Xi Z, Si J, Nan J. LncRNA MALAT1 potentiates autophagy-associated cisplatin resistance by regulating the microRNA30b/autophagy-related gene 5 axis in gastric cancer. Int J Oncol. 2019;54:239–248.
31. Zhang YF, Li CS, Zhou Y, Lu XH. Propofol facilitates cisplatin sensitivity via lncRNA MALAT1/miR-30e/ATG5 axis through suppressing autophagy in gastric cancer. Life Sci. 2020;244:117280.
32. Li H, Ma X, Yang D, Suo Z, Dai R, Liu C. PCAT-1 contributes to cisplatin resistance in gastric cancer through epigenetically silencing PTEN via recruiting EZH2. J Cell Biochem. 2019;121:1353–1361.
33. Guo Y, Yue P, Wang Y, Chen G, Li Y. PCAT-1 contributes to cisplatin resistance in gastric cancer through miR-128/ZEB1 axis. Biomed Pharmacother. 2019;118:109255.
34. Zhang X, Bu P, Liu L, Zhang X, Li J. Overexpression of long non-coding RNA PVT1 in gastric cancer cells promotes the development of multidrug resistance. Biochem Bioph Res Co. 2015;462:227–232.
35. Wu C, Hu Y, Ning Y, Zhao A, Zhang G, Yan L. Long noncoding RNA plasmacytoma variant translocation 1 regulates cisplatin resistance via miR-3619-5p/TBL1XR1 axis in gastric cancer. Cancer Biother Radio. 2020.
36. Jin W, Zhang H, Li M, Lin S. Long noncoding RNA regulator of reprogramming regulates cell growth, metastasis, and cisplatin resistance in gastric cancer via miR-519d-3p/HMGA2 Axis. Cancer Biother Radio. 2020.
37. Li M, Zhang YY, Shang J, Xu YD. LncRNA SNHG5 promotes cisplatin resistance in gastric cancer via inhibiting cell apoptosis. Eur Rev Med Pharmacol Sci. 2019;23:4185–4191.
38. Dai Q, Zhang T, Pan J, Li C. LncRNA UCA1 promotes cisplatin resistance in gastric cancer via recruiting EZH2 and activating PI3K/AKT pathway. J Cancer. 2020;11:3882–3892.
39. Fang Q, Chen X, Zhi X. Long non-coding RNA (LncRNA) Urothelial Carcinoma Associated 1 (UCA1) increases multi-drug resistance of gastric cancer via downregulating miR-27b. Med Sci Monitor. 2016;22:3506–3513.
40. Li Y, Zhang Q, Tang X. Long non-coding RNA XIST contributes into drug resistance of gastric cancer cell. Minerva Med. 2019;110:270–272.
41. Zeng L, Liao Q, Zou Z, et al. Long non-coding RNA XLOC_006753 promotes the development of multidrug resistance in gastric cancer cells through the PI3K/AKT/mTOR signaling pathway. Cell Physiol Biochem. 2018;51:1221–1236.
42. Xu W, He L, Li Y, Tan Y, Zhang F, Xu H. Silencing of lncRNA ZFAS1 inhibits malignancies by blocking Wnt/beta-catenin signaling in gastric cancer cells. Biosci Biotechnol Biochem. 2018;82:456–465.
43. Wu X, Zheng Y, Han B, Dong X. Long noncoding RNA BLACAT1 modulates ABCB1 to promote oxaliplatin resistance of gastric cancer via sponging miR-361. Biomed Pharmacother. 2018;99:832–838.
44. Song W, Qian Y, Zhang MH, et al. The long non-coding RNA DDX11-AS1 facilitates cell progression and oxaliplatin resistance via regulating miR-326/IRS1 axis in gastric cancer. Eur Rev Med Pharmacol Sci. 2020;24:3049–3061.
45. Wu H, Liu B, Chen Z, Li G, Zhang Z. MSC-induced lncRNA HCP5 drove fatty acid oxidation through miR-3619-5p/AMPK/PGC1α/CEBPB axis to promote stemness and chemo-resistance of gastric cancer. Cell Death Dis. 2020;11.
46. He W, Liang B, Wang C, et al. MSC-regulated lncRNA MACC1-AS1 promotes stemness and chemoresistance through fatty acid oxidation in gastric cancer. Oncogene. 2019;38:4637–4654.
47. Zhang Z, Li M, Zhang Z. lncRNA MALAT1 modulates oxaliplatin resistance of gastric cancer via sponging miR-22-3p. Onco Targets Ther. 2020;13:1343–1354.
48. Han Y, Ye J, Wu D, et al. LEIGC long non-coding RNA acts as a tumor suppressor in gastric carcinoma by inhibiting the epithelial-to-mesenchymal transition. Bmc Cancer. 2014;14:932.
49. Du P, Hu C, Qin Y, et al. LncRNA PVT1 mediates antiapoptosis and 5-fluorouracil resistance via increasing Bcl2 expression in gastric cancer. J Oncol. 2019;2019:9325407.
50. Shang C, Sun L, Zhang J, et al. Silence of cancer susceptibility candidate 9 inhibits gastric cancer and reverses chemoresistance. Oncotarget. 2017;8:15393–15398.
51. Zhou Z, Lin Z, He Y, et al. The long noncoding RNA D63785 regulates chemotherapy sensitivity in human gastric cancer by targeting miR-422a. Molecular Therapy - Nucleic Acids. 2018;12:405–419.
52. Ding H, Sun J, Li R, Wang G. Long non-coding RNA GACAT1 alleviates doxorubicin and vincristine resistance through a PTEN/AKT/mTOR/S6K1 regulatory pathway in gastric cancer. RSC Adv. 2019;9:8048–8055.
53. Wang H, Qin R, Guan A, et al. HOTAIR enhanced paclitaxel and doxorubicin resistance in gastric cancer cells partly through inhibiting miR-217 expression. J Cell Biochem. 2018;119:7226–7234.
54. Wang Y, Zhang D, Wu K, Zhao Q, Nie Y, Fan D. Long noncoding RNA MRUL promotes ABCB1 expression in multidrug-resistant gastric cancer cell sublines. Mol Cell Biol. 2014;34:3182–3193.
55. Zhang J, Zhao B, Chen X, Wang Z, Xu H, Huang B. Silence of long noncoding RNA NEAT1 inhibits malignant biological behaviors and chemotherapy resistance in gastric cancer. Pathology Oncol Res. 2017;24:109–113.
56. Wang S, Chen W, Yu H, et al. lncRNA ROR promotes gastric cancer drug resistance. Cancer Control. 2020;27:544330725.
57. Shang C, Guo Y, Zhang J, Huang B. Silence of long noncoding RNA UCA1 inhibits malignant proliferation and chemotherapy resistance to adriamycin in gastric cancer. Cancer Chemother Pharm. 2016;77:1061–1067.
58. Ding J, Li D, Gong M, et al. Expression and clinical significance of the long non-coding RNA PVT1 in human gastric cancer. Onco Targets Ther. 2014;7:1625–1630.
59. Arnold M, Park JY, Camargo MC, Lunet N, Forman D, Soerjomataram I. Is gastric cancer becoming a rare disease? A global assessment of predicted incidence trends to 2035. Gut. 2020;69:823–829.
60. Li ZL, Jiang HW, Song M, Xu L, Xia D, Liu Q. Secondary sentinel lymph node tracing technique: a new method for tracing lymph nodes in radical gastrectomy for advanced gastric cancer. J Zhejiang Univ Sci B. 2015;16:897–903.
61. Poursheikhani A, Bahmanpour Z, Razmara E, et al. Non-coding RNAs underlying chemoresistance in gastric cancer. Cell Oncol. 2020.
62. Ruan T, Liu W, Tao K, Wu C. A review of research progress in multidrug-resistance mechanisms in gastric cancer. Onco Targets Ther. 2020;13:1797–1807.
63. Mirzaei H, Hamblin MR. Regulation of glycolysis by non-coding RNAs in cancer: switching on the warburg effect. Mol Ther Oncolytics. 2020;19:218–239.
64. Marchese FP, Raimondi I, Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017;18:206.
65. Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77:3965–3981.
66. Wang Z, Qin B. Prognostic and clinicopathological significance of long noncoding RNA CTD-2510F5.4 in gastric cancer. Gastric Cancer. 2019;22:692–704.
67. Chong DQ, Shan JL, Yang CS, Wang R, Du ZM. Clinical prognostic value of A FOXM1 related long non-coding RNA expression in gastric cancer. Eur Rev Med Pharmacol Sci. 2018;22:417–421.
68. Wei L, Sun J, Zhang N, et al. Noncoding RNAs in gastric cancer: implications for drug resistance. Mol Cancer. 2020;19:62.
69. Zhang X, Xie K, Zhou H, et al. Role of non-coding RNAs and RNA modifiers in cancer therapy resistance. Mol Cancer. 2020;19:47.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
