Back to Journals » Cancer Management and Research » Volume 10
Role of intestinal flora in colorectal cancer from the metabolite perspective: a systematic review
Authors Han S , Gao J, Zhou Q , Liu S, Wen C, Yang X
Received 7 October 2017
Accepted for publication 14 November 2017
Published 31 January 2018 Volume 2018:10 Pages 199—206
DOI https://doi.org/10.2147/CMAR.S153482
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Lu-Zhe Sun
Shuwen Han,1 Jianlan Gao,1 Qing Zhou,2 Shanshan Liu,3 Caixia Wen,3 Xi Yang4
1Department of Medical Oncology, Huzhou Central Hospital, 2Department of Critical Care Medicine, Huzhou Central Hospital, 3Medical College of Nursing, Huzhou University, 4Department of Intervention and Radiotherapy, Huzhou Central Hospital, Huzhou, Zhejiang Province, People’s Republic of China
Abstract: Colorectal cancer is one of the most common human malignant tumors. Recent research has shown that colorectal cancer is a dysbacteriosis-induced disease; however, the role of intestinal bacteria in colorectal cancer is unclear. This review explores the role of intestinal flora in colorectal cancer. In total, 57 articles were included after identification and screening. The pertinent literature on floral metabolites in colorectal cancer from three metabolic perspectives – including carbohydrate, lipid, and amino acid metabolism – was analyzed. An association network regarding the role of intestinal flora from a metabolic perspective was constructed by analyzing the previous literature to provide direction and insight for further research on intestinal flora in colorectal cancer.
Keywords: colorectal cancer, bacteria, microbiology, carbohydrate metabolism, lipid metabolism, amino acids, inflammation
Introduction
Colorectal cancer is the third leading cancer in humans and the fourth most common cause of cancer-related death.1 The causes of the occurrence and development of colorectal cancer are unclear, but it is thought to result from a combination of genetic and environmental factors.2 Intestinal flora and their metabolites, as environmental factors, play important roles in colorectal cancer by regulating related genes.3
The main function of the colorectum is to store feces while under siege from complex intestinal flora. Several probiotics,4–6 including Lactobacillus acidophilus, Bifidobacterium, Lactobacillus rhamnosus, and Streptococcus thermophilus, as well as pathogenic bacteria,7–9 including Enterococcus faecalis, Enterotoxigenic bacteroides fragilis, Streptococcus bovis, Salmonella, Clostridia, and Fusobacterium nucleatum, comprise the diverse intestinal flora. This intestinal flora, mucosal epithelial cells, foodborne probiotic components, and small molecules – including hormones, enzymes, mucus, and bile salts – constitute a complex intestinal micro-ecosystem.10 Although individual substances vary, the intestinal micro-ecosystem is relatively stable under physiological conditions. Multiple diseases may result if changes occur beyond the ability of compensatory adjustment.11,12 Studies have shown that the micro-ecosystem equilibrium in patients with colorectal cancer is disrupted.13 Various intestinal flora and metabolites are closely related to colorectal cancer.14
With advanced developments in microbiome and microbial metabolomics, especially rapid advancements in high-throughput sequencing technology, increasing attention has been given to studying intestinal flora and intestinal microecology in recent years.15 Current research has focused on the relationship between intestinal flora and colorectal cancer; however, the specific mechanism of the intestinal flora in causing colorectal cancer is unclear.16,17 Microbial primary metabolites, including amino acids, nucleotides, polysaccharides, lipids, and vitamins, are necessary to sustain intestinal flora growth and reproduction.18 Microbial primary metabolites are similar in most microbial cells. The synthesis of primary metabolites is a constant process, and synthetic obstacles affect normal microbial activities.19,20 Microbial secondary metabolites, including alkaloids, phenols, antibiotics, and pigments, determine the specificity and function of the flora. Microbes are valuable in maintaining the balance of the intestinal microecology;21,22 however, the significance of microbial metabolites in colorectal cancer is unclear. Given that the metabolism of three substances, including carbohydrate, lipid, and amino acid metabolism, is the general metabolic mechanism among all creatures, we tried to build a link between the intestinal flora and colorectal cancer from this angle.
In this review, we comprehensively analyzed and classified the pertinent literature on microflora metabolites in colorectal cancer from three metabolic perspectives, including carbohydrate, lipid, and amino acid metabolism. An association network of intestinal flora, their metabolites, and colorectal cancer was built that may provide direction and insight for further research on intestinal flora in colorectal cancer.
Methods
Literature search
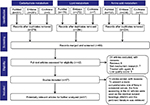
We searched the “PubMed”, “Embase”, and “Cochrane” databases for literature published up to August 11, 2017. To achieve maximum sensitivity of the search strategy and identify all studies, the following terms were combined: (“colorectal or colon or rectal, large intestine or large bowel or intestinum crassum” and “neoplasms or tumor or carcinoma or cancer” and “flora or microflora or microorganism or microbiome or microbiota or microbe or microbiology or germ or bacteria or bacterium or fungus”) and (“glucose or adenosine triphosphate or lactic acid or mitochondria or galactose or sucrase or amylase or hexokinase or glucokinase or pyruvate kinase or glucuronidase” OR “triglyceride or fat or aliphatic acid or lipoprotein or cholesterol or cholesterin or bile acid or lithocholic acid or vitamin d or dehydroxylase” OR “amino acid or ammonia or amine or urea or carbamide or ureophil or mucin or mucoprotein or nitrosamines or nitroguanidine or nitrosourea or aromatic amines or mycotoxin or endotoxin or exotoxin or sulfuretted hydrogen or hydrogen sulfide or hydrothion”). All relevant abstracts were retrieved independently by two authors, and articles with available information for the present systematic review were fully reviewed. In total, 42 articles were included. To present a more comprehensive role of flora in colorectal cancer, flora appearing in the 42 articles were used as the medical subject headings (MeSH) and the pertinent literature was retrieved. Finally, 15 articles were added after identification and screening. Moreover, pertinent literature from the searched studies was analyzed. A detailed search strategy is presented in Figure 1.
Study selection
Studies catering to the following criteria were considered for inclusion: 1) studies that were published in English and 2) studies that involved intestinal flora and intestinal flora metabolism in colorectal cancer, both in vivo and in vitro. Exclusion criteria were as follows: 1) letters, case reports, reviews, or conference reports; 2) predominant studies that were not on intestinal flora metabolism in colorectal cancer; and 3) correlation studies did not involve flora metabolism.
Role of flora metabolites in colorectal cancer
Intestinal flora and carbohydrate metabolism in colorectal cancer
Carbohydrate metabolism refers to a series of complex chemical reactions in vivo. The tricarboxylic acid cycle, as the principal pathway of carbohydrate metabolism, is the final metabolic pathway and metabolic hub of the three major nutrients, including carbohydrates, lipids, and amino acids.23,24 Carbohydrate metabolism is important for intestinal flora and colorectal cancer. First, oxygen plays a decisive role in choosing the carbohydrate metabolic pathway. Both anaerobic and aerobic bacteria coexist in the intestinal tract. Superoxide, oxygen radicals, and oxygen molecules are closely related to the development of colorectal cancer.25,26 Second, carbon dioxide and water are the primary producers in carbohydrate metabolism.27 Various bacteria decompose glucose and lactose and produce acid.28 Intestinal micro-ecology is regulated by maintaining the acid–base balance and regulating osmotic pressure. Third, adenosine triphosphate (ATP) is produced during carbohydrate metabolism and is an important compound that supplies energy to all living cells. Phosphoribose produced during the metabolism of pentose phosphate is necessary to synthesize DNA and RNA, and they are especially important for rapidly reproducing bacteria and infinitely replicating cancer cells.29 Fourth, nicotinamide adenine dinucleotide phosphate (NADPH) is the intermediate metabolite in carbohydrate metabolism, and it participates in phosphorylating proteins and genes. It may be involved in microbial variation and epigenetic regulation of colorectal cancer.30 Finally, mitochondria are the key location for carbohydrate metabolism, and mitochondrial dysfunction is one the most important features in colorectal cancer and intestinal flora imbalance.31,32
Intestinal flora and lipid metabolism in colorectal cancer
Lipids include triglycerides, phospholipids, cholesterol, and glycolipid. Triglycerides provide energy for living organisms by emulsifying bile acid salts and catalyzing lipase in the small intestine.33,34 Phospholipids and sugar esters maintain biomembrane structure and function.35 Cholesterol can transform into vitamins, bile acid, or steroid hormones.36 Many studies indicate that a high-fat diet can induce colorectal cancer, and imply that intestinal flora play irreplaceable roles; however, their specific mechanisms remain unclear.37,38 In this review, we searched for clues on tumorigenesis by summarizing the pertinent literature. High-fat diets can increase bile and bile acid secretion in the colorectum, and some clostridia can accelerate transformation of bile acid into secondary bile acid by participating in the synthesis of various enzymes during fatty acid metabolism.39,40 Secondary bile acid, as a carcinogenic substance, promotes colorectal cancer by multiple molecular mechanisms – synthesizing oxygen free radicals, fracturing DNA strands, making chromosomes unstable, and forming cancer stem cells.41,42 Interactions between fatty acids, bile acids, and intestinal flora can produce diacylglycerol, prostaglandin, and leukotriene, leading to tumorigenesis by activating immune or inflammatory responses.43–45
Intestinal flora and amino acid metabolism in colorectal cancer
Amino acid metabolism involves two parts. Amino acids can be used to synthesize proteins, peptides, and other nitrogenous substances and, moreover, they can be decomposed into α-ketonic acid, amines, and carbon dioxide through deamination, transamination, and decarboxylation. Many toxic substances such as sulfur, nitrates, hydrogen sulfide, ammonia, and amines are involved in the metabolic process, and these toxic substances can lead to colorectal cancer.46 Food residue with high protein content can stimulate sulfate-reducing bacterial growth. Hydrogen sulfide is a product of sulfate-reducing bacteria as well as an intermediate product of amino acid metabolism.47 Hydrogen sulfide elicits several pathogenic events, including cell proliferation, differentiation, apoptosis, and inflammation – ultimately leading to malignant enterocyte transformation.48,49 Nitrate is not toxic, but easily reduces to nitrite due to the intestinal flora. Nitrite combines with nitrogenous compounds such as amines, amino compounds, and methyl urea to form carcinogenic nitroso compounds.50,51 Furthermore, mucin as an intermediate product of amino acid metabolism is a mutagenic agent with cooperation from the intestinal flora.52 Many enzymes, peptides, and other nitrogenous substances secreted by the intestinal flora are involved in activating and regulating important signal molecules and signaling pathways in tumorigenesis.53,54
Results and discussion
The intestinal flora and host maintain a dynamic balance under physiological conditions. When this balance is disrupted, the entire micro-ecological system is significantly altered.55,56 The synergistic effect among intestinal flora, metabolites, and the host plays a pivotal role in the occurrence and development of colorectal cancer. First, microbes are the initial factors in colorectal cancer. Changes to the intestinal flora distribution and abundance contributes to inflammatory and immunological responses and induces malignant transformation of the intestinal mucosal cells.57 Second, epidemiological surveys have indicated that the balance of intestinal flora in patients with precancerous lesions, including inflammatory bowel disease (IBD) and intestinal polyps, were altered significantly.58,59 Third, various metabolic products of the intestinal flora can, directly or indirectly, promote development and progression of colorectal cancer.60 Fourth, micro-ecology helps to prevent tumorigenesis by reestablishing the intestinal micro-ecological balance.61,62 In conclusion, colorectal cancer is a dysbacteriosis-induced disease, and the understanding of this disease has changed in the molecular age.
Researchers have increasingly focused on determining the specific bacteria or microbial community structural changes in colorectal cancer by sequencing 16S rRNA and bioinformatics analysis in recent years. Many researchers support that Streptococcus bovis63 and Streptococcus gallolyticus64 are the specific bacteria involved in colorectal cancer; however, there are some lacunae in this research. Sequencing of the 16s rRNA variable region only identifies the bacterial species, and the intra-individual variability of the bacteria was not considered. In addition, a better scientific method for studying the intestinal flora in colorectal cancer is to explore its relationship with the intestinal micro-ecological system. The intestinal micro-ecological system is complex and integral, with individual differences. Establishing an association network for the intestinal micro-ecological system in colorectal cancer may offer an approach to solving this dilemma. As shown in Figure 2, the association network for the intestinal flora and microbial metabolites in colorectal cancer, from a metabolic perspective, was constructed by analyzing the previous literature. Although we tried to search all pertinent literature, mistakes of omission inevitably occurred because of the complexity of microbial metabolism and the many compounds involved in it. Research on the relationship between microbial metabolites and colorectal cancer were relatively insufficient.
Future directions
There appears to be a complex relationship between colorectal cancer and intestinal flora. Microbial metabolites may play vital roles in balancing the intestinal micro-ecology and in developing colorectal cancer. The intestinal flora is insufficiently understood, intestinal micro-ecology is complex, and intestinal flora show significant intra-individual variability; thus, evaluating all of these interactions is challenging. With the great progress of integrated systems, molecular biology, and bioinformatics, we urgently call for a synthesis of the existing research to establish a comprehensive database that focuses on individual relationships among the intestinal flora, microbial metabolites, and colorectal cancer.
It is plausible that the intestinal flora and microbial metabolites in colorectal cancer are related to the immune system and inflammatory abnormalities. Although much effort has been expended, many bottlenecks must be addressed before stepping from the imbalanced intestinal micro-ecological system to immune system and inflammatory abnormalities to genesis and development of colorectal cancer. The clinical correlation of the intestinal flora, microbial metabolites, and colorectal cancer remains unknown; thus, it is essential to conduct further functional assays on pathogenesis such as the microbiome, microbial metabolomics, and peptidome assays. Multiple probiotics have been applied clinically for some time, and preclinical trials involving intestinal flora transfusion are also underway. Prospective and retrospective studies on the incidence of colorectal cancer after clinical interventions with microbial preparations should be scheduled.
Several avenues are available to pursue translational applications. First, microchip arrays or metabonomic technologies can assess the risk and monitor the curative effects of a bacterial species or specific microbial metabolite in colorectal cancer. Second, probiotics, microbial metabolites, and fecal microbial transfusions can all be used to recover the intestinal micro-ecological system to prevent colorectal cancer; however, this requires further clinical testing. Third, intestinal micro-ecology is influenced by many factors, including the endocrine system, diet, sleep, and stress. Testing the intestinal flora and microbial metabolites in feces can guide the adjustment of dietary structure or living habits to prevent colorectal cancer.
Disclosure
The authors report no conflicts of interest in this work.
References
Mármol I, Sánchez-de-Diego C, Pradilla Dieste A, Cerrada E, Rodriguez Yoldi MJ. Colorectal Carcinoma: a general overview and future perspectives in colorectal cancer. Int J Mol Sci. 2017;18(1). pii:E197. | ||
Ibáñez-Sanz G, Díez-Villanueva A, Alonso MH, et al. Risk model for colorectal cancer in Spanish population using environmental and genetic factors: results from the MCC-Spain study. Sci Rep. 2017;7:43263. | ||
Manzat-Saplacan RM, Mircea PA, Balacescu L, Chira RI, Berindan-Neagoe I, Balacescu O. Can we change our microbiome to prevent colorectal cancer development? Acta Oncol. 2015;54(8):1085–1095. | ||
Dodoo CC, Wang J, Basit AW, Stapleton P, Gaisford S. Targeted delivery of probiotics to enhance gastrointestinal stability and intestinal colonisation. Int J Pharm. 2017;530(1–2):224–229. | ||
Quagliariello A, Aloisio I, Bozzi Cionci N, et al. Effect of Bifidobacterium breve on the intestinal microbiota of coeliac children on a gluten free diet: a pilot study. Nutrients. 2016;8(10). pii:E660. | ||
Yoon JS, Sohn W, Lee OY, et al. Effect of multispecies probiotics on irritable bowel syndrome: a randomized, double-blind, placebo-controlled trial. J Gastroenterol Hepatol. 2014;29(1):52–59. | ||
Adesida SA, Ezenta CC, Adagbada AO, Aladesokan AA, Coker AO. Carriage of multidrug resistant Enterococcus faecium and Enterococcus faecalis among apparently healthy humans. Afr J Infect Dis. 2017;11(2):83–89. | ||
Fukugaiti MH, Ignacio A, Fernandes MR, Ribeiro Júnior U, Nakano V, Avila-Campos MJ. High occurrence of Fusobacterium nucleatum and Clostridium difficile in the intestinal microbiota of colorectal carcinoma patients. Braz J Microbiol. 2015;46(4):1135–1140. | ||
Paritsky M, Pastukh N, Brodsky D, Isakovich N, Peretz A. Association of Streptococcus bovis presence in colonic content with advanced colonic lesion. World J Gastroenterol. 2015;21(18):5663–5667. | ||
Gomez A, Petrzelkova K, Yeoman CJ, et al. Gut microbiome composition and metabolomic profiles of wild western lowland gorillas (Gorilla gorilla gorilla) reflect host ecology. Mol Ecol. 2015;24(10):2551–2565. | ||
Ni J, Wu GD, Albenberg L, Tomov VT. Gut microbiota and IBD: causation or correlation? Nat Rev Gastroenterol Hepatol. 2017;14(10):573–584. | ||
Sommer F, Anderson JM, Bharti R, Raes J, Rosenstiel P. The resilience of the intestinal microbiota influences health and disease. Nat Rev Microbiol. 2017;15(10):630–638. | ||
Sheflin AM, Whitney AK, Weir TL. Cancer-promoting effects of microbial dysbiosis. Curr Oncol Rep. 2014;16(10):406. | ||
Wang X, Wang J, Rao B, Deng L. Gut flora profiling and fecal metabolite composition of colorectal cancer patients and healthy individuals. Exp Ther Med. 2017;13(6):2848–2854. | ||
Shaffer M, Armstrong AJS, Phelan VV, Reisdorph N, Lozupone CA. Microbiome and metabolome data integration provides insight into health and disease. Transl Res. 2017;189:51–64. | ||
Calistri D, Rengucci C, Casadei Gardini A, et al. Fecal DNA for noninvasive diagnosis of colorectal cancer in immunochemical fecal occult blood test-positive individuals. Cancer Epidemiol Biomarkers Prev. 2010;19(10):2647–2654. | ||
Sinha R, Ahn J, Sampson JN, et al. Fecal microbiota, fecal metabolome, and colorectal cancer interrelations. PLoS One. 2016;11(3):e0152126. | ||
Singh R, Kumar M, Mittal A, Mehta PK. Microbial metabolites in nutrition, healthcare and agriculture. 3 Biotech. 2017;7(1):15. | ||
Yap IK, Li JV, Saric J, et al. Metabonomic and microbiological analysis of the dynamic effect of vancomycin-induced gut microbiota modification in the mouse. J Proteome Res. 2008;7(9):3718–3728. | ||
Patterson E, Cryan JF, Fitzgerald GF, Ross RP, Dinan TG, Stanton C. Gut microbiota, the pharmabiotics they produce and host health. Proc Nutr Soc. 2014;73(4):477–489. | ||
Shi Y, Pan C, Wang K, et al. Synthetic multispecies microbial communities reveals shifts in secondary metabolism and facilitates cryptic natural product discovery. Environ Microbiol. 2017;19(9):3606–3618. | ||
Narsing Rao MP, Xiao M, Li WJ. Fungal and bacterial pigments: secondary metabolites with wide applications. Front Microbiol. 2017;8:1113. | ||
Ribel-Madsen A, Ribel-Madsen R, Brøns C, Newgard CB, Vaag AA, Hellgren LI. Plasma acylcarnitine profiling indicates increased fatty acid oxidation relative to tricarboxylic acid cycle capacity in young, healthy low birth weight men. Physiol Rep. 2016;4(19). pii:e12977. | ||
Patkova A, Joskova V, Havel E, et al. Energy, protein, carbohydrate, and lipid intakes and their effects on morbidity and mortality in critically ill adult patients: a systematic review. Adv Nutr. 2017;8(4):624–634. | ||
Azzolin VF, Cadoná FC, Machado AK, et al. Superoxide-hydrogen peroxide imbalance interferes with colorectal cancer cells viability, proliferation and oxaliplatin response. Toxicol In Vitro. 2016;32:8–15. | ||
Tsuruya A, Kuwahara A, Saito Y, et al. Major anaerobic bacteria responsible for the production of carcinogenic acetaldehyde from ethanol in the colon and rectum. Alcohol Alcohol. 2016;51(4):395–401. | ||
Vinke PC, El Aidy S, van Dijk G. The role of supplemental complex dietary carbohydrates and gut microbiota in promoting cardiometabolic and immunological health in obesity: lessons from healthy non-obese individuals. Front Nutr. 2017;4:34. | ||
Liu Z, Liu T. Production of acrylic acid and propionic acid by constructing a portion of the 3-hydroxypropionate/4-hydroxybutyrate cycle from Metallosphaera sedula in Escherichia coli. J Ind Microbiol Biotechnol. 2016;43(12):1659–1670. | ||
Dean BM, Perrett D. Studies on adenine and adenosine metabolism by intact human erythrocytes using high performance liquid chromatography. Biochim Biophys Acta. 1976;437(1):1–5. | ||
Li Y, Kundu P, Seow SW, et al. Gut microbiota accelerate tumor growth via c-jun and STAT3 phosphorylation in APCMin/+ mice. Carcinogenesis. 2012;33(6):1231–1238. | ||
Shuwen H, Xi Y, Yuefen P. Can mitochondria DNA provide a novel biomarker for evaluating the risk and prognosis of colorectal cancer? Dis Markers. 2017;2017:5189803. | ||
Saint-Georges-Chaumet Y, Edeas M. Microbiota-mitochondria inter-talk: consequence for microbiota-host interaction. Pathog Dis. 2016;74(1):ftv096. | ||
Ryan M, Kochunov P, Rowland LM, et al. Lipid metabolism, abdominal adiposity, and cerebral health in the Amish. Obesity (Silver Spring). 2017;25(11):1876–1880. | ||
Giang TM, Gaucel S, Brestaz P, et al. Dynamic modeling of in vitro lipid digestion: individual fatty acid release and bioaccessibility kinetics. Food Chem. 2016;194:1180–1188. | ||
Shen C, Xue M, Qiu H, Guo W. Insertion of neurotransmitters into a lipid bilayer membrane and its implication on membrane stability: a molecular dynamics study. Chemphyschem. 2017;18(6):626–633. | ||
Buitenwerf E, Dullaart RPF, Muller Kobold AC, et al. Cholesterol delivery to the adrenal glands estimated by adrenal venous sampling: an in vivo model to determine the contribution of circulating lipoproteins to steroidogenesis in humans. J Clin Lipidol. 2017;11(3):733–738. | ||
Wang CZ, Huang WH, Zhang CF, et al. Role of intestinal microbiome in American ginseng-mediated colon cancer protection in high fat diet-fed AOM/DSS mice. Clin Transl Oncol. Epub 2017 Aug 14. | ||
Weber C. Nutrition. Diet change alters microbiota and might affect cancer risk. Nat Rev Gastroenterol Hepatol. 2015;12(6):314. | ||
Ridlon JM, Wolf PG, Gaskins HR. Taurocholic acid metabolism by gut microbes and colon cancer. Gut Microbes. 2016;7(3):201–215. | ||
O’Keefe SJ, Li JV, Lahti L, et al. Fat, fibre and cancer risk in African Americans and rural Africans. Nat Commun. 2015;6:6342. | ||
Farhana L, Nangia-Makker P, Arbit E, et al. Bile acid: a potential inducer of colon cancer stem cells. Stem Cell Res Ther. 2016;7(1):181. | ||
Ajouz H, Mukherji D, Shamseddine A. Secondary bile acids: an underrecognized cause of colon cancer. World J Surg Oncol. 2014;12:164. | ||
Savari S, Vinnakota K, Zhang Y, Sjölander A. Cysteinyl leukotrienes and their receptors: bridging inflammation and colorectal cancer. World J Gastroenterol. 2014;20(4):968–977. | ||
Wang D, DuBois RN. An inflammatory mediator, prostaglandin E2, in colorectal cancer. Cancer J. 2013;19(6):502–510. | ||
Kai M, Yamamoto E, Sato A, et al. Epigenetic silencing of diacylglycerol kinase gamma in colorectal cancer. Mol Carcinog. 2017;56(7):1743–1752. | ||
Ma N, Tian Y, Wu Y, Ma X. Contributions of the interaction between dietary protein and gut microbiota to intestinal health. Curr Protein Pept Sci. 2017;18(8):795–808. | ||
Carbonero F, Benefiel AC, Alizadeh-Ghamsari AH, Gaskins HR. Microbial pathways in colonic sulfur metabolism and links with health and disease. Front Physiol. 2012;3:448. | ||
Attene-Ramos MS, Nava GM, Muellner MG, Wagner ED, Plewa MJ, Gaskins HR. DNA damage and toxicogenomic analyses of hydrogen sulfide in human intestinal epithelial FHs 74 Int cells. Environ Mol Mutagen. 2010;51(4):304–314. | ||
Yazici C, Wolf PG, Kim H, et al. Race-dependent association of sulfidogenic bacteria with colorectal cancer. Gut. 2017;66(11):1983–1994. | ||
Espejo-Herrera N, Gràcia-Lavedan E, Boldo E, et al. Colorectal cancer risk and nitrate exposure through drinking water and diet. Int J Cancer. 2016;139(2):334–346. | ||
Bingham SA, Pignatelli B, Pollock JR, et al. Does increased endogenous formation of N-nitroso compounds in the human colon explain the association between red meat and colon cancer? Carcinogenesis. 1996;17(3):515–523. | ||
Rokhsefat S, Lin A, Comelli EM. Mucin-microbiota interaction during postnatal maturation of the intestinal ecosystem: clinical implications. Dig Dis Sci. 2016;61(6):1473–1486. | ||
Yang T, Owen JL, Lightfoot YL, Kladde MP, Mohamadzadeh M. Microbiota impact on the epigenetic regulation of colorectal cancer. Trends Mol Med. 2013;19(12):714–725. | ||
Kumar M, Nagpal R, Verma V, et al. Probiotic metabolites as epigenetic targets in the prevention of colon cancer. Nutr Rev. 2013;71(1):23–34. | ||
Oke S, Martin A. Insights into the role of the intestinal microbiota in colon cancer. Therap Adv Gastroenterol. 2017;10(5):417–428. | ||
Gagnière J, Raisch J, Veziant J, et al. Gut microbiota imbalance and colorectal cancer. World J Gastroenterol. 2016;22(2):501–518. | ||
García-Castillo V, Sanhueza E, McNerney E, Onate SA, García A. Microbiota dysbiosis: a new piece in the understanding of the carcinogenesis puzzle. J Med Microbiol. 2016;65(12):1347–1362. | ||
Pascal V, Pozuelo M, Borruel N, et al. A microbial signature for Crohn’s disease. Gut. 2017;66(5):813–822. | ||
Brim H, Yooseph S, Zoetendal EG, et al. Microbiome analysis of stool samples from African Americans with colon polyps. PLoS One. 2013;8(12):e81352. | ||
Weir TL, Manter DK, Sheflin AM, Barnett BA, Heuberger AL, Ryan EP. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS One. 2013;8(8) :e70803. | ||
Gao Z, Guo B, Gao R, Zhu Q, Wu W, Qin H. Probiotics modify human intestinal mucosa-associated microbiota in patients with colorectal cancer. Mol Med Rep. 2015;12(4):6119–6127. | ||
Dos Reis SA, da Conceiçãoo LL, Siqueira NP, Rosa DD, da Silva LL, Peluzio MD. Review of the mechanisms of probiotic actions in the prevention of colorectal cancer. Nutr Res. 2017;37:1–19. | ||
Tsai CE, Chiu CT, Rayner CK, et al. Associated factors in Streptococcus bovis bacteremia and colorectal cancer. Kaohsiung J Med Sci. 2016;32(4):196–200. | ||
Andres-Franch M, Galiana A, Sanchez-Hellin V, et al. Streptococcus gallolyticus infection in colorectal cancer and association with biological and clinical factors. PLoS One. 2017;12(3):e0174305. | ||
Nava GM, Ou J, O;Keefe SJ, Gaskins HR. Diversity of colonic Archaea and sulfate reducing bacteria populations in native Africans differs from those in Caucasian and African Americans. Gastroenterology. 2009;136(5):A410. | ||
Balamurugan R, Rajendiran E, George S, Samuel GV, Ramakrishna BS. Real-time polymerase chain reaction quantification of specific butyrate-producing bacteria, Desulfovibrio and Enterococcus faecalis in the feces of patients with colorectal cancer. J Gastroenterol Hepatol. 2008;23(8 Pt 1):1298–1303. | ||
Attene-Ramos MS, Wagner ED, Gaskins HR, Plewa MJ. Hydrogen sulfide induces direct radical-associated DNA damage. Mol Cancer Res. 2007;5(5):455–459. | ||
Moore DR, Kotake Y, Huycke MM. Effects of iron and phytic acid on production of extracellular radicals by Enterococcus faecalis. Exp Biol Med (Maywood). 2004;229(11):1186–1195. | ||
Deplancke B, Gaskins HR. Hydrogen sulfide induces serum-independent cell cycle entry in nontransformed rat intestinal epithelial cells. FASEB J. 2003;17(10):1310–1312. | ||
Weir T, Marschke RF, Brown RJ, et al. Fecal metabolome and microflora differences between colorectal cancer patients and healthy adults. J Clin Oncol. 2013;31(15 Suppl):11050. | ||
Chmelař D, Hájek M, Janečková J, Vobejdová J, Martineková P, Kašíková A. [The effect of oxygen on endotoxin production in bacteria of the Bacteroides fragilis group isolated from patients with colorectal carcinoma]. Epidemiol Mikrobiol Imunol. 2016;65(2):129–135. Czech. | ||
Purcell RV, Pearson J, Aitchison A, Dixon L, Frizelle FA, Keenan JI. Colonization with enterotoxigenic Bacteroides fragilis is associated with early-stage colorectal neoplasia. PLoS One. 2017;12(2):e0171602. | ||
Thiele Orberg E, Fan H, Tam AJ, et al. The myeloid immune signature of enterotoxigenic Bacteroides fragilis-induced murine colon tumorigenesis. Mucosal Immunol. 2017;10(2):421–433. | ||
Irrazabal T, Martin A. T regulatory cells gone bad: an oncogenic immune response against Enterotoxigenic B. fragilis infection leads to colon cancer. Cancer Discov. 2015;5(10):1021–1023. | ||
Tsoi H, Chu ESH, Zhang X, et al. Peptostreptococcus anaerobius induces intracellular cholesterol biosynthesis in colon cells to induce proliferation and causes dysplasia in mice. Gastroenterology. 2017;152(6):1419–1433.e5. | ||
Meeker SM, Paik J, Hsu CC, et al. The gut microbiome is modulated by dietary vitamin d. J Am Assoc Lab Anim Sci. 2015;54(5):643–644. | ||
Wu WT, Cheng HC, Chen HL. Ameliorative effects of konjac glucomannan on human faecal β-glucuronidase activity, secondary bile acid levels and faecal water toxicity towards Caco-2 cells. Br J Nutr. 2011;105(4):593–600. | ||
Appleyard CB, Cruz ML, Isidro AA, Arthur JC, Jobin C, De Simone C. Pretreatment with the probiotic VSL#3 delays transition from inflammation to dysplasia in a rat model of colitis-associated cancer. Am J Physiol Gastrointest Liver Physiol. 2011;301(6):G1004–G1013. | ||
Xu MQ, Cao HL, Wang S, Cao XC, Yan F, Wang BM. The influence of intestinal microbiota on the secondary bile acid-induced colorectal carcinogenesis. J Dig Dis. 2014;15:130. | ||
Minamida K, Kaneko M, Ohashi M, et al. Effects of difructose anhydride III (DFA III) administration on bile acids and growth of DFA III-assimilating bacterium Ruminococcus productus on rat intestine. J Biosci Bioeng. 2005;99(6):548–554. | ||
Zampa A, Silvi S, Fabiani R, Morozzi G, Orpianesi C, Cresci A. Effects of different digestible carbohydrates on bile acid metabolism and SCFA production by human gut micro-flora grown in an in vitro semi-continuous culture. Anaerobe. 2004;10(1):19–26. | ||
Wells JE, Williams KB, Whitehead TR, Heuman DM, Hylemon PB. Development and application of a polymerase chain reaction assay for the detection and enumeration of bile acid 7alpha-dehydroxylating bacteria in human feces. Clin Chim Acta. 2003;331(1–2):127–134. | ||
Fadden K, Owen RW. Faecal steroids and colorectal cancer: the effect of lactulose on faecal bacterial metabolism in a continuous culture model of the large intestine. Eur J Cancer Prev. 1992;1(2):113–127. | ||
Zheng ZY, Bernstein C. Bile salt/acid induction of DNA damage in bacterial cells: effect of taurine conjugation. Nutr Cancer. 1992;18(2):157–164. | ||
Murray WR, Blackwood A, Trotter JM, Calman KC, MacKay C. Faecal bile acids and clostridia in the aetiology of colorectal cancer. Br J Cancer. 1980;41(6):923–928. | ||
Pool-Zobel BL, Neudecker C, Domizlaff I, et al. Lactobacillus- and bifidobacterium-mediated antigenotoxicity in the colon of rats. Nutr Cancer. 1996;26(3):365–380. | ||
Cholewa K, Wȩglarz L, Parfiniewicz B, Lodowska J, Jaworska-Kik M. The influence of desulfovibrio desulfuricans endotoxin on IL-6 and IL-6 receptor genes expression in colon cancer Caco-2 cells. Farmaceutyczny Przeglad Naukowy. 2010;7(3):27–32. | ||
Kostic AD, Chun E, Robertson L, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14(2):207–215. | ||
Chen Y, Peng Y, Yu J, et al. Invasive Fusobacterium nucleatum activates beta-catenin signaling in colorectal cancer via a TLR4/P-PAK1 cascade. Oncotarget. 2017;8(19):31802–31814. | ||
Becker S, Oelschlaeger TA, Wullaert A, et al. Bacteria regulate intestinal epithelial cell differentiation factors both in vitro and in vivo. PLoS One. 2013;8(2):e55620. | ||
Burger-van Paassen N, Bouma J, Boehm G, van Goudoever H, Van Seuningen I, Renes IB. Lactobacillus GG stimulates mucin MUC2 synthesis in the human colon cancer cell line LS174T. Gastroenterology. 2009;136(5):A267. | ||
Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A. 2008;105(39):15064–15069. | ||
Femia AP, Giannini A, Fazi M, et al. Identification of mucin depleted foci in the human colon. Cancer Prev Res (Phila). 2008;1(7):562–567. | ||
Akiba T, Abe Y, Kitada S, et al. Crystal structure of the parasporin-2 Bacillus thuringiensis toxin that recognizes cancer cells. J Mol Biol. 2009;386(1):121–133. | ||
Witthöft T, Eckmann L, Kim JM, Kagnoff MF. Enteroinvasive bacteria directly activate expression of iNOS and NO production in human colon epithelial cells. Am J Physiol. 1998;275(3 Pt 1):G564–G571. | ||
Yoshimura K, Laird LS, Chia CY, et al. Live attenuated Listeria monocytogenes effectively treats hepatic colorectal cancer metastases and is strongly enhanced by depletion of regulatory T cells. Cancer Res. 2007;67(20):10058–10066. | ||
Oliveira MJ, Lauwaet T, De Bruyne G, Mareel M, Leroy A. Listeria monocytogenes produces a pro-invasive factor that signals via ErbB2/ErbB3 heterodimers. J Cancer Res Clin Oncol. 2005;131(1):49–59. | ||
Boleij A, Muytjens CM, Bukhari SI, et al. Novel clues on the specific association of Streptococcus gallolyticus subsp gallolyticus with colorectal cancer. J Infect Dis. 2011;203(8):1101–1109. | ||
Tsai TL, Li AC, Chen YC, Liao YS, Lin TH. Antimicrobial peptide m2163 or m2386 identified from Lactobacillus casei ATCC 334 can trigger apoptosis in the human colorectal cancer cell line SW480. Tumor Biol. 2015;36(5):3775–3789. | ||
de Moreno de LeBlanc A, Perdigón G. Reduction of beta-glucuronidase and nitroreductase activity by yoghurt in a murine colon cancer model. Biocell. 2005;29(1):15–24. | ||
de Moreno de LeBlanc A, Perdigón G. Mechanisms involved in the antitumor activity exerted by yoghurt in an experimental colon cancer model. Int J Cancer Prev. 2005;2(3):181–193. | ||
Lim D, Kim KS, Kim H, et al. Anti-tumor activity of an immunotoxin (TGFα-PE38) delivered by attenuated Salmonella typhimurium. Oncotarget. 2017;8(23):37550–37560. | ||
Lenoir M, Del Carmen S, Cortes-Perez NG, et al. Lactobacillus casei BL23 regulates Treg and Th17 T-cell populations and reduces DMH-associated colorectal cancer. J Gastroenterol. 2016;51(9):862–873. | ||
Szemes T, Vlkova B, Minarik G, et al. On the origin of reactive oxygen species and antioxidative mechanisms in Enterococcus faecalis. Redox Rep. 2010;15(5):202–206. | ||
Nepelska M, Cultrone A, Béguet-Crespel F, et al. Butyrate produced by commensal bacteria potentiates phorbol esters induced AP-1 response in human intestinal epithelial cells. PLoS One. 2012;7(12):e52869. | ||
Lv Y, Ye T, Wang HP, et al. Suppression of colorectal tumorigenesis by recombinant Bacteroides fragilis enterotoxin-2 in vivo. World J Gastroenterol. 2017;23(4):603–613. | ||
DeStefano Shields CE, Van Meerbeke SW, Housseau F, et al. Reduction of murine colon tumorigenesis driven by enterotoxigenic Bacteroides fragilis using cefoxitin treatment. J Infect Dis. 2016;214(1):122–129. | ||
Geis AL, Fan H, Wu X, et al. Regulatory T-cell response to enterotoxigenic Bacteroides fragilis colonization triggers IL17-dependent colon carcinogenesis. Cancer Discov. 2015;5(10):1098–1109. | ||
Han YW. Fusobacterium nucleatum: a commensal-turned pathogen. Curr Opin Microbiol. 2015;23:141–147. | ||
Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14(2):195–206. | ||
Choi HJ, Kim J, Do KH, Park SH, Moon Y. Enteropathogenic Escherichia coli-induced macrophage inhibitory cytokine 1 mediates cancer cell survival: an in vitro implication of infection-linked tumor dissemination. Oncogene. 2013;32(41):4960–4969. | ||
Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, Nougayrède JP. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc Natl Acad Sci U S A. 2010;107(25):11537–11542. | ||
Maddocks OD, Short AJ, Donnenberg MS, Bader S, Harrison DJ. Attaching and effacing Escherichia coli downregulate DNA mismatch repair protein in vitro and are associated with colorectal adenocarcinomas in humans. PLoS One. 2009;4(5):e5517. | ||
Olino K, Wada S, Edil BH, et al. Tumor-associated antigen expressing Listeria monocytogenes induces effective primary and memory T-cell responses against hepatic colorectal cancer metastases. Ann Surg Oncol. 2012;19(Suppl 3):S597–S607. | ||
Pan ZK, Ikonomidis G, Lazenby A, Pardoll D, Paterson Y. A recombinant Listeria monocytogenes vaccine expressing a model tumour antigen protects mice against lethal tumour cell challenge and causes regression of established tumours. Nat Med. 1995;1(5):471–477. | ||
Biarc J, Nguyen IS, Pini A, et al. Carcinogenic properties of proteins with pro-inflammatory activity from Streptococcus infantarius (formerly S.bovis). Carcinogenesis. 2004;25(8):1477–1484. | ||
Ellmerich S, Schöller M, Duranton B, et al. Promotion of intestinal carcinogenesis by Streptococcus bovis. Carcinogenesis. 2000;21(4):753–756. | ||
Lecuit M. Understanding how Listeria monocytogenes targets and crosses host barriers. Clin Microbiol Infect. 2005;11(6):430–436. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.