Back to Journals » OncoTargets and Therapy » Volume 11
Role of DiGeorge syndrome critical region gene 9, a long noncoding RNA, in gastric cancer
Authors Ni C, Yang P, Guo J , Ye M
Received 11 January 2018
Accepted for publication 13 March 2018
Published 19 April 2018 Volume 2018:11 Pages 2259—2267
DOI https://doi.org/10.2147/OTT.S162253
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr XuYu Yang
Chao Ni,1 Ping Yang,1 Junming Guo,1 Meng Ye2
1Department of Biochemistry and Molecular Biology, Zhejiang Key Laboratory of Pathophysiology, Medical School of Ningbo University, Ningbo, People’s Republic of China; 2Department of Medical Oncology, The Affiliated Hospital of Medical School of Ningbo University, Ningbo, People’s Republic of China
Introduction: Long non-coding RNAs (lncRNAs) regulate and influence cancer cell development and tumor formation. However, the role for lncRNAs in gastric cancer has not been fully established. In this study, DGCR9, a lncRNA, was significantly upregulated in gastric cancer cell lines.
Methods: The expression levels of DGCR9 in each patient between formalin-fixed, paraffin-embedded (FFPE) gastric cancer tissues and adjacent noncancer tissues (NAT) (n=102) were measured by quantitative reverse-transcription polymerase chain reaction (qRT-PCR). The effect of DGCR9 on cellular proliferation, migration, and glucose uptake was investigated in vitro, respectively.
Results: DGCR9 was shown to have increased expression in gastric cancer tissues and in gastric cancer cell lines. Further, DGCR9 was found to be associated with clinicopathological characteristics of patients with gastric cancer. In particular, DGCR9 was positively associated with lymph node invasion and tumor-node-metastasis (TNM) stage in gastric cancer patients. By in vitro functional analysis, knockdown of DGCR9 in gastric cancer cells suppressed cellular proliferation, migration, and glucose uptake. In contrast, overexpression of DGCR9 increased each of these cancer cell characteristics.
Conclusions: DGCR9 was upregulated in gastric cancer tissues and was shown to accelerate cellular proliferation, migration, and glucose metabolism, all of which would promote the formation and development of gastric cancer.
Keywords: long noncoding RNA, DGCR9, gastric cancer, proliferation, migration, glucose metabolism
Introduction
Gastric carcinoma, 1 of the top 5 digestive system neoplasms, is a major cause of cancer mortality worldwide.1,2 In recent years, combined treatments, including surgery, chemotherapy, radiotherapy, and molecular targeted therapy, for gastric cancer have been developed and continuously improved.3–8 Even with these multiple treatments and with early detection, prognosis for patients with advanced gastric carcinoma is still poor.9,10 Hence, understanding the molecular process of cancer development and the involvement of novel molecules in that process is essential for a full appreciation of the molecular mechanisms that result in gastric cancer.
RNA transcripts, longer than 200 nucleotides that lack translated open reading frames, are defined as long noncoding RNAs (lncRNAs).11,12 Many lncRNAs have been associated with tumors and have been shown to influence tumor occurrence by either transcriptional regulation or posttranscriptional processing.13–15 Several lncRNAs have been shown to contribute to hallmark characteristics of cancer cells including proliferation, angiogenesis, and unregulated growth.16 Dysregulation of lncRNAs has been demonstrated in multiple cancer cell types.17,18 For example, the lncRNA MALAT1 is associated with cellular metastasis,19 proliferation, angiogenesis, and apoptosis.20–22 Likewise the lncRNA CASC9, promotes nasopharyngeal carcinoma by regulating HIF-1α.23 The importance of lncRNAs in tumor progression has been demonstrated previously.24
To study the roles of lncRNAs in gastric cancer, we first screened the high-expressed lncRNAs in gastric associated-lncRNA microarray database (GSE96856, www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE96856). Then, we used GO/Pathway analysis to further clarify the possible functions and mechanisms of the lncRNAs. Finally, DiGeorge syndrome critical region gene 9 (DGCR9) was selected as the candidate. DGCR9 gene is located at position 22q11.21.25 In this investigation, the microarray database determination was confirmed. Further, DGCR9 was demonstrated to be upregulated in gastric cancer, to be involved in cell proliferation and migration, and to affect glucose metabolism. These results suggest a key role for DGCR9 in gastric cancer. To the best of our knowledge, this is the first report demonstrating a role for DGCR9 in cancer.
Materials and methods
Clinical specimens
Formalin-fixed, paraffin-embedded (FFPE) gastric cancer tissues and adjacent noncancer tissues (NAT) were obtained from 102 patients who underwent primary gastric cancer resection between November, 2016 and January, 2017 at the Ningbo Pathological Diagnosis Center in People’s Republic of China. The diagnosis of gastric cancer was confirmed by 2 experienced pathologists. Based on the Seventh Edition of the American Joint Committee on Cancer, clinicopathological characteristics, including gender, age, tumor size, lymph node invasion, differentiation, and TNM stage, were collected. The study was approved by the Research Ethics Committee of the Affiliated Hospital of Ningbo University School of Medicine. All patients provided written informed consent documents for this study.
Total RNA extraction
Total RNA was extracted from FFPE gastric cancer and NAT by use of an FFPE RNA Kit (Omega, Norcross, GA, USA), according to the manufacturer’s instructions. TRIzol reagent (Ambion, Carlsbad, CA, USA) was used to extract RNA from cells as described by the manufacturer. RNA concentrations were measured with a DS-11 spectrophotometer (Denovix, Wilmington, DE, USA).
Reverse transcription (RT)
The GoScript Reverse Transcription System (Promega, Madison, WI, USA) was used to synthesize cDNAs. RT reactions in 20 μL were incubated for 60 min at 42°C, 10 min at 75°C, and then stabilized at 4°C.
Real-time quantitative reverse-transcription polymerase chain reaction (qRT-PCR)
For qRT-PCR, 5 μL of diluted RT products were mixed with 12.5 μL of GoTaq qPCR Master Mix kit (Promega, Madison, WI, USA), 1 μL forward and reverse primers, and 5.5 μL of nuclease-free water to a final volume of 25 μL. β-actin was used as a control. PCR primers used were as follows: DGCR9 forward, 5′-TAGCATGGCCAGGTATGCAC-3′, reverse, 5′-TGCGAATCCCAAAGCTGTCA-3′; β-actin forward, 5′-AAGGTGACAGCAGTCGGTTG-3′; reverse, 5′-AAGGTGACAGCAGTCGGTTG-3′. An Mx30005P (Stratagene-Agilent, Santa Clara, CA, USA) was used for measurement. The expression of DGCR5 was calculated using the ΔCt method; with lower ΔCt values indicating higher expression.
Cell culture
Human gastric cancer cell lines, AGS, MGC803, HGC27, and SGC7901, as well as the normal gastric epithelial cell line GES-1, were purchased from the Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (Shanghai, People’s Republic of China). Cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Hyclone, GE Healthcare Life Sciences, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS; PAN-Biotech, Adenbach, Germany) and 1% penicillin–streptomycin in a CO2 incubator at 37°C and 5% CO2.
siRNA transfection
The siRNA targeting DGCR9 (Si-DGCR9) was designed and synthesized by GenePharma, Shanghai, People’s Republic of China. The sequence of DGCR9 SiRNA was as follows: sense: 5′-GCUGGGAGGGACAUACUUUTT-3′ and antisense: 5′-AAAGUAUGUCCCUCCCAGCTT-3′. The negative control missense siRNA (Si-NC) sequence was as follows: sense: 5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense: 5′-ACGUGACACGUUCGGAGAATT-3′. Gastric cancer cells were cultured in 6-well plates to 70%–90% confluence and then transfected with siRNA and Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Cells were harvested 48 h after transfection.
Lentiviral transfection
The overexpressing lentivirus (LV5-DGCR9) and the control virus (LV5-NC) were purchased from Shanghai GenePharma Co., (Shanghai, People’s Republic of China). The MGC803 and AGS cells were transfected by LV5-DGCR9 or LV5-NC for 48 h at a multiplicity of infection of 30. To stabilize transfection, cells were treated with puromycin (0.5 μg/mL) for 1 week. After screening, those cells expressing green fluorescent protein were considered LV-DGCRG and LV-NC. Transfection efficiency was confirmed by qRT-PCR and fluorescence microscopy.
CCK-8 assay
Gastric cancer cells were seeded into 96-well plates at a density of 3×103 cells per well and cultured at 37°C. At 0, 24, 48, 72, and 96 h; 10 μL of CCK-8 (Dojindo Molecular Technologies, Kumamoto, Japan) was added to each well and incubated for 3 h at 37°C. Measurements were made at 450 nm with a microplate reader.
Wound healing assay
Cells were seeded at 5×105 cells/well in 6-well plates and cultured until they reached confluency. The cell monolayer was removed by abrasion with a 200 μL pipette tip. The cells were washed twice with PBS to remove floating cells and then 2 mL of RPMI 1640 medium supplemented with 1% FBS was added to each well. Cells were immediately photographed at 0 h and then again at 48 h. The area into which the cells migrated was measured using Image Pro Plus v6.0 software package (Media Cybernetics Inc., Bethesda, MD, USA).
Colony formation assay
For the colony formation assay, gastric cancer cells were seeded into 6-well plates at a density of 500/well and cultured in RPMI 1640 medium containing 10% FBS for 2 weeks. After washing with PBS 3 times, the colonies were fixed in 4% paraformaldehyde for 30 min and stained with 0.1% crystal violet at room temperature for 30 min.
Transwell migration assay
The migration assay was performed with 24-well Transwell chambers (Costar, Corning, NY, USA). Cells (8×104) were suspended in 200 μL of RPMI 1640 without FBS and seeded into the upper chamber. Then, 500 μL of medium containing 10% FBS was added to the lower chamber. Cells were incubated for 24 h at 37°C and 5% CO2, and then fixed with 4% paraformaldehyde, stained with 0.1% crystal violet, and counted with a light microscope at 100× magnification.
Glucose uptake assay
Glucose uptake was measured with a Glucose Test Kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, People’s Republic of China). Cells were seeded in 6-well plates at a density of 3×105 cells/well in complete culture medium. A day later, the medium was replaced with phenol red-free, Dulbecco’s Modified Eagle’s Medium (1,000 mg/L glucose) supplemented with 10% FBS and antibiotics. The medium at 0 h was collected as background and the medium at 48 h was assessed for glucose concentration. The glucose concentration was normalized to total protein as measured by the BCA assay. Glucose uptake = (background concentration − reading concentration)/protein concentration.
Statistical analysis
Data analysis was performed with SPSS 20.0 software (IBM Corporation, Armonk, NY, USA) and GraphPad Prism 6.0 (GraphPad Software, La Jolla, CA, USA). Association of DGCR5 expression level with FFPE gastric cancer and NAT was calculated by paired t-test. The χ2 test was used to analyze the associations between clinicopathological characteristics and DGCR5 expression. P-values less than 0.05 were considered statistically significant.
Results
Upregulation of DGCR9 in gastric cancer tissues and cell lines
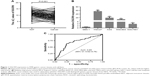
To investigate the association of DGCR9 with gastric cancer, qRT-PCR was used to assess DGCR9 expression in 102 paired FFPE gastric cancer tissues and NAT. DGCR9 expression was significantly increased in gastric cancer tissues compared to NAT (Figure 1A). DGCR9 also showed a higher expression in gastric cancer cell lines (HGC27, AGS, MGC803, and SGC7901) than that in the normal gastric epithelial cell line GES-1 (Figure 1B). Receiver operating characteristic curve analysis was used to evaluate the potential gastric cancer by using DGCR9. DGCR9 showed potential predictive value with an area under curve of 0.691 (Figure 1C). To assess associations among DGCR9 expression and clinicopathological characteristics of patients with gastric cancer, patients were divided into 2 groups, a high expression group (n=77, fold-change ≥2) and a low expression group (n=25, fold-change <2). The clinicopathological characteristics of these 2 groups are summarized in Table 1. DGCR9 was significantly associated with lymph node invasion (P=0.011) and TNM stage (P=0.018). These results suggest that DGCR9 may play a role in gastric cancer progression.
  | Table 1 DGCR9 and the clinicopathological characteristic of patients with gastric cancer |
Knockdown of DGCR9 suppresses cell proliferation and migration
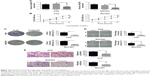
To investigate the biological role of DGCR9, a chemically modified small interfering RNA target for DGCR9 (Si-DGCR9) was used to knockdown the expression of DGCR9 in AGS and MGC803 gastric cancer cells. A significant reduction in DGCR9 expression by Si-DGCR9 was observed by qRT-PCR (Figure 2A). Cell proliferation was significantly suppressed by DGCR9 downregulation as demonstrated by the CCK-8 assay (Figure 2B). Knockdown of DGCR9 in AGS and MGC803 cells dramatically reduced colony formation in comparison to missense-treated cells, Si-NC (Figure 2C). Further, both wound healing and Transwell migration of AGS and MGC803 cells were reduced with Si-DGCR9 when compared to cells treated with Si-NC (Figure 2D and E). These data demonstrate the involvement of DGCR9 in gastric cancer cell proliferation and migration.
Upregulated DGCR9 induces cell proliferation and migration
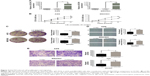
AGS and MGC803 gastric cancer cells were transfected with a lentivirus expressing DGCR9 (LV-DGCR9), and as demonstrated by the results of qRT-PCR, the expression of DGCR9 was remarkably upregulated when compared to cells transfected with a negative control lentivirus, LV-NC (Figure 3A). Similarly, cell proliferation and colony formation (Figure 3B and C) as well as wound healing and cell migration were significantly promoted by LV-DGCR9 when compared to LV-NC (Figure 3D and E). The results demonstrate LV-DGCR9 significantly induced proliferation and migration of gastric cancer cells.
DGCR9 promotes glucose metabolism in gastric cancer cells
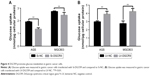
Since glucose metabolism is essential to tumor cell proliferation, the effect of DGCR9 on glucose metabolism was assessed by the glucose uptake assay. Results showed that downregulation of DGCR9 reduced glucose uptake in AGS and MGC803 cells (Figure 4A). In contrast, overexpression increased glucose uptake in gastric cancer cells (Figure 4B). These results demonstrate the regulation of glucose metabolism in gastric cancer cells by DGCR9.
Discussion
With the general implementation of high-throughput sequencing technology, it has become apparent that lncRNAs account for the majority of sequences within the genome. Many lncRNAs regulate biological networks within various tumors.26 Further, lncRNAs have become new and significant diagnostic biomarkers as well as targets for molecular therapies.27
Herein, high expression levels of DGCR9 were found in gastric cancer by microarray analysis. Further, results showed that DGCR9 was upregulated in gastric cancer tissues compared to NAT (Figure 1A). DGCR9 overexpression was also found in gastric cancer cell lines, but not in a normal gastric epithelial cell line (Figure 1A). Moreover, increased expression of DGCR9 was positively associated with lymph node invasion and TNM stage in gastric cancer patients (Table 1). These results suggest that DGCR9 may be involved in gastric cancer progression. For in vitro culture functional studies, interference of DGCR9 resulted in decreased cellular proliferation and migration (Figure 2). In contrast, overexpression of DGCR9 promoted cellular proliferation and migration (Figure 3). These characteristics suggest that aberrant expression of DGCR9 may be associated with gastric cancer.
The underlying mechanism by which DGCR9 regulates proliferation and migration of gastric cancer cells is likely related to glucose uptake (Figure 4). In a previous report, the lncRNA CASC8 was shown to significantly suppress the proliferation of bladder cancer cells by regulating glycolysis.28 ANRIL promotes nasopharnygeal carcinoma progression by reprograming cell glucose metabolism.29 Overexpression of DGCR9 increased glucose uptake, while DGCR9 interference reduced glucose uptake (Figure 4). It is possible that DGCR9 drives progression of gastric cancer through enhanced glucose metabolism. Since the main role of DGCR9 is to regulate glucose uptake, the expression location of DGCR9 should be mainly located in cytoplasm.
DGCR9 is transcripted from DiGeorge syndrome critical region gene 9. And here we found that DGCR9 plays a role in gastric occurrence (Figures 2 and 3). However, to date, there is no evidence to show the correlation between DiGeorge syndrome and gastric cancer.
Although this is the first report of DGCR9 in gastric cancer, there are still some limitations in this study. For instance, DGCR9 expression was significantly upregulated in FFPE gastric cancer tissues compared to NAT. To explore the early diagnostic value of DGCR9, many more samples, such as fresh gastric cancer tissue or gastric juice, should be collected for work in the future. Moreover, the role of related proteins, for example MMP, in regulating proliferation and migration of gastric cancer should be verified.
In conclusion, DGCR9 was significantly expressed in gastric cancer tissues and cell lines. Dysregulated expression of DGCR9 was associated with lymph node invasion and TNM stage. Furthermore, knockdown of DGCR9 suppressed cell proliferation, migration, and glucose uptake, with opposite effects observed upon overexpression. Overall, DGCR9 may represent a novel biomarker for the diagnosis of gastric cancer.
Acknowledgments
This study was supported by grants from Natural Science Foundation of Zhejiang (no LY16H160005), the Applied Research Project on Nonprofit Technology of Zhejiang Province (no 2016C33177), the Scientific Innovation Team Project of Ningbo (no 2017C110019), and the K.C. Wong Magna Fund in Ningbo University.
Disclosure
The authors report no conflicts of interest in this work.
References
Li T, Shao Y, Fu L, et al. Plasma circular RNA profiling of patients with gastric cancer and their droplet digital RT-PCR detection. J Mol Med (Berl). 2018;96(1):85–96. | ||
Tian M, Chen R, Li T, Xiao B. Reduced expression of circRNA hsa_circ_0003159 in gastric cancer and its clinical significance. J Clin Lab Anal. 2018;32(3):e22281. | ||
Hartgrink HH, Jansen EP, van Grieken NC, van de Velde CJ. Gastric cancer. Lancet. 2009;374(9688):477–490. | ||
Wang J, Qu J, Li Z, et al. Pretreatment platelet-to-lymphocyte ratio is associated with the response to first-line chemotherapy and survival in patients with metastatic gastric cancer. J Clin Lab Anal. 2018;32(1):e22185. | ||
Ford HE, Marshall A, Bridgewater JA, et al. Docetaxel versus active symptom control for refractory oesophagogastric adenocarcinoma (COUGAR-02): an open-label, phase 3 randomised controlled trial. Lancet Oncol. 2014;15(1):78–86. | ||
Bagcchi S. Radiotherapy for testicular cancer increases gastric cancer risk. Lancet Oncol. 2014;15(13):e593. | ||
Bang YJ, Van Cutsem E, Feyereislova A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376(9742):687–697. | ||
Wilke H, Muro K, Van Cutsem E, et al. Ramucirumab plus paclitaxel versus placebo plus paclitaxel in patients with previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (RAINBOW): a double-blind, randomised phase 3 trial. Lancet Oncol. 2014;15(11):1224–1235. | ||
Cunningham D, Allum WH, Stenning SP, et al. Perioperative chemotherapy versus surgery alone for resectable gastroesophageal cancer. N Engl J Med. 2006;355(1):11–20. | ||
Smalley SR, Benedetti JK, Haller DG, et al. Updated analysis of SWOG-directed intergroup study 0116: a phase III trial of adjuvant radiochemotherapy versus observation after curative gastric cancer resection. J Clin Oncol. 2012;30(19):2327–2333. | ||
Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. | ||
Li T, Mo X, Fu L, Xiao B, Guo J. Molecular mechanisms of long noncoding RNAs on gastric cancer. Oncotarget. 2016;7(8):8601–8612. | ||
Zhang J, Liu SC, Luo XH, et al. Exosomal long noncoding RNAs are differentially expressed in the cervicovaginal lavage samples of cervical cancer patients. J Clin Lab Anal. 2016;30:1116–1121. | ||
Sun W, Yang Y, Xu C, Guo J. Regulatory mechanisms of long noncoding RNAs on gene expression in cancers. Cancer Genet. 2017;216–217:105–110. | ||
Nie ZL, Wang YS, Mei YP, et al. Prognostic significance of long noncoding RNA Z38 as a candidate biomarker in breast cancer. J Clin Lab Anal. 2018;32(1):e22193. | ||
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. | ||
Sun W, Yang Y, Xu C, Xie Y, Guo J. Roles of long noncoding RNAs in gastric cancer and their clinical applications. J Cancer Res Clin Oncol. 2016;142(11):2231–2237. | ||
Hashad D, Elbanna A, Ibrahim A, Khedr G. Evaluation of the role of circulating long non-coding RNA H19 as a promising novel biomarker in plasma of patients with gastric cancer. J Clin Lab Anal. 2016;30(6):1100–1105. | ||
Puthanveetil P, Chen S, Feng B, Gautam A, Chakrabarti S. Long non-coding RNA MALAT1 regulates hyperglycaemia induced inflammatory process in the endothelial cells. J Cell Mol Med. 2015;19(6):1418–1425. | ||
Michalik KM, You X, Manavski Y, et al. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res. 2014;114(9):1389–1397. | ||
Thum T, Fiedler J. LINCing MALAT1 and angiogenesis. Circ Res. 2014;114(9):1366–1368. | ||
Xin JW, Jiang YG. Long noncoding RNA MALAT1 inhibits apoptosis induced by oxygen-glucose deprivation and reoxygenation in human brain microvascular endothelial cells. Exp Ther Med. 2017;13(4):1225–1234. | ||
Su X, Li G, Liu W. The long noncoding RNA cancer susceptibility candidate 9 promotes nasopharyngeal carcinogenesis via stabilizing HIF1α. DNA Cell Biol. 2017;36(5):394–400. | ||
Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. | ||
Gong W, Emanuel BS, Collins J, et al. A transcription map of the DiGeorge and velo-cardio-facial syndrome minimal critical region on 22q11. Hum Mol Genet. 1996;5(6):789–800. | ||
Sun W, Mo X, Li T, Xie Y, Guo J. Clinical significance of the long noncoding RNA RP11-19P22.6-001 in gastric cancer. Cancer Biomark. 2017;18(4):397–403. | ||
Li Z, Dou P, Liu T, He S. Application of long noncoding RNAs in osteosarcoma: biomarkers and therapeutic targets. Cell Physiol Biochem. 2017;42(4):1407–1419. | ||
Hu R, Zhong P, Xiong L, Duan L. Long noncoding RNA cancer susceptibility candidate 8 suppresses the proliferation of bladder cancer cells via regulating glycolysis. DNA Cell Biol. 2017;36(9):767–774. | ||
Zou ZW, Ma C, Medoro L, et al. LncRNA ANRIL is up-regulated in nasopharyngeal carcinoma and promotes the cancer progression via increasing proliferation, reprograming cell glucose metabolism and inducing side-population stem-like cancer cells. Oncotarget. 2016;7(38):61741–61754. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.