Back to Journals » International Journal of General Medicine » Volume 14
Role of CD5L and SRD5A2 as Prognostic Biomarkers for Hepatocellular Carcinoma
Authors Luo Y, Huang X , Zhan J, Zhang S
Received 20 September 2021
Accepted for publication 18 November 2021
Published 1 December 2021 Volume 2021:14 Pages 9247—9260
DOI https://doi.org/10.2147/IJGM.S337769
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Yunxiu Luo,1,* Xiaopeng Huang,2,* Jiabin Zhan,3 Shuai Zhang2
1Department of Radiation Oncology, Hainan Cancer Hospital, Affiliated Cancer Hospital of Hainan Medical University, Haikou, 570311, Hainan Province, People’s Republic of China; 2Department of Radiation Oncology, Hainan General Hospital, Hainan Affiliated Hospital of Hainan Medical University, Haikou, 570311, Hainan Province, People’s Republic of China; 3Department of Otolaryngology, Hainan General Hospital, Hainan Affiliated Hospital of Hainan Medical University, Haikou, 570311, Hainan Province, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jiabin Zhan
Department of Otolaryngology, Hainan General Hospital, Hainan Affiliated Hospital of Hainan Medical University, Haikou, 570311, Hainan Province, People’s Republic of China
Tel +86-18907669339
Email [email protected]
Shuai Zhang Tel +86-1387642896
Email [email protected]
Purpose: Due to the limitations of currently available biomarkers, new biomarkers are needed to accurately predict the prognosis of patients with hepatocellular carcinoma (HCC) patients.
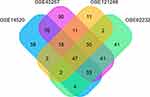
Methods: In this study, we screened for differentially expressed genes (DEGs) in the tumor and the adjacent tissues using the four gene expression array (GSE14520, GSE45267, GSE121248, GSE62232) of the Gene Express Omnibus (GEO) database.
Results: Subsequently, 47 overlapping DEGs were identified in four GEO datasets, which were mostly located on chromosomes 5q and 6q, distributed in the liver and CD105-positive endothelial cells, and closely related to HCC. Function enrichment revealed 47 DEGs were related to HCC, and involved in steroid /lipid /retinol metabolism, bile secretion and p53 signalling pathway. The Kaplan–Meier plotter analysis (http://www.kmplot.com/) identified 26 and 40 genes associated with the 5-year overall survival (OS) and relapse-free survival (RFS). We found that CD5L and SRD5A2 were independent prognostic factors for 5-year OS (P=0.036) and RFS (P=0.044) in HCC patients from GSE14520, respectively. Clinicopathological features including BCLC stage, cirrhosis, and risk signature for predicted metastasis were used to construct and validate a nomogram for 5-year OS with C-index of 0.732 and 0.717 in the training and validation cohort, respectively. SRD5A2, BCLC stage and gender was independent prognostic factors for RFS which were used to build a nomogram with the C-index of 0.666 and 0.682 in the training and validation cohort, respectively.
Conclusion: CD5L can facilitate individualized, targeted therapy for HCC patients.
Keywords: hepatocellular carcinoma, CD5L, prognostic, nomogram, SRD5A2
Introduction
Hepatocellular carcinoma (HCC) is the sixth most common malignant disease and the second leading cause of cancer-related mortality in males.1 Despite the improvements in the clinical diagnosis and treatment strategies, the 5-year overall survival (OS) rate of HCC patients remains less than 40%.1,2 Intrahepatic recurrence and extrahepatic metastasis are the major causes of death.2 The identification of specific biomarkers and responsive therapeutic targets could significantly help to predict early recurrence and subsequently improve the prognosis of HCC patients.3 High throughput gene expression profiling allows identification of potential biomarkers for constructing prognosis model and prediction of latent molecular mechanisms. In the last decade, there have been few reports of the combined use of multiple genes for prognostication of HCC.3–5 However, there is a clear need to identify a more sensitive and specific prognostic biomarker for clinical practice.
CD5L is highly expressed in macrophages and acts as regulator of lipid synthesis, immune homeostasis and inflammatory response.6–10 CD5L could inhibit chronic liver injury by controlling SMAD7 expression and TGFß signal pathway. But, representative analysis found CD5L protein had higher expression in HCC tissues than that in adjacent liver tissues, and the patients with high expression of CD5L had terrible prognosis comparing to those with under expression.11 In vitro study indicated CD5L prompted the proliferation and colony formation of HCC cells and helped them to escape from cisplatin induced apoptosis.11 The aim of the current study was to investigate the expression and clinical significance of CD5L and other differentially expressed genes (DEGs) in HCC using public databases.
Materials and Methods
Patients and Gene Expression Data
Gene expression and clinical data were obtained from the gene expression omnibus (GEO) database of the National Center for Biotechnology Information (NCBI) (accession numbers GSE45267, 12 GSE62232,4 GSE121248,13 and GSE14520-GPL571).14 The data on the effect of gene expression on prognosis in liver cancer were acquired from the Kaplan-Meier plotter (KM plotter, http://www.kmplot.com/). Kaplan-Meier plotter is a public database containing 54,675 genes of 18,674 cancer patients, including 364 liver cancer cases with relapse-free and overall survival data derived from TCGA database.
Analysis of Differentially Expressed Genes (DEGs)
DEGs in HCC and precancerous samples were screened using the GEO2R online tool (https://www.ncbi.nlm.nih.gov/geo/geo2r/) through which we compared two or more groups of samples in a GEO dataset in order to identify genes that are differentially expressed across experimental conditions. The GEO2R online analysis tool was used to screen the DEGs in four datasets which detected the expression profile of paired samples from HCC and adjacent non-cancerous tissues. To take care of false-positive results, an adjusted p-value was calculated using the Benjamini and Hochberg false discovery rate. An adjusted P-value <0.01, P-value <0.01, and a log fold change (log FC) ≥2 were considered as the thresholds for DEGs screening. Gene with more than one probe set was average. The common DEGs among the four datasets were analyzed using a Venn diagram. The data processing has been shown in Figure S1.
Gene Function Annotation and PPI Network Assay
The database for annotation, visualization and integrated discovery (DAVID) (https://david.ncifcrf.gov/), Omicsbean (http://www.omicsbean.cn/) and KEGG Orthology-Based Annotation System (KOBAS) (http://kobas.cbi.pku.edu.cn/kobas3/) were used to determine the function enrichment for biological process (BP), molecular function (MF), and cellular component (CC), and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway of DEGs. P<0.05 was considered to be statistically significant. The protein-protein interaction network analysis of DEGs was conducted using the String online database (http://www.string-db.org/, version 11.1). A combined score of more than 0.4 was set as the cut-off value.
Screening for Survival-Related DEGs
The univariate survival analysis of overlapped DEGs in four datasets was performed according to the K-M method using the KM plotter (http://kmplot.com/). The screening filters for gene selection were as follows: multiple genes (gene symbols), split patients by median value, threshold overall survival (OS) of 60 months, median survival, censored at threshold, the pathology and clinical features of the patients were set as default all (stage, grade, AJCC T, vascular invasion, gender, race, sorafenib treatment, alcohol consumption, hepatitis virus were set all). P< 0.05 was used for the purpose of analyzing significantly different. The data processing has been shown in Figure S1.
Statistical Analysis
Statistical analysis was performed using the EmpowerStats, Graph Pad Prism 7.0, IBM SPSS 23.0, and R 3.5.3 software packages. Continuous variables were assayed by two-sided Student’s t-test, or one-way analysis of variance (ANOVA), while nonparametric or χ2 test (Fisher exact probability method) were used for categorical variables. Normally distributed data are presented as means ± standard deviation, and the data with skewed distribution are presented as median. Factors that appeared statistically significant on univariate analysis were subjected to multivariate Cox regression analysis. P <0.05 was considered to be statistically significant. The R 3.5.3 software was used to construct the nomogram and the calibration curve.
Results
Identification of DEGs with GEO2R and Analysis of Genetic Characteristics
On analysis, 241, 346, 200 and 124 DEGs were identified in GSE14520, GSE45267, GSE62232, and GSE121248 datasets, respectively (Figure 1). The four datasets contained 47 overlapped DEGs. Out of these, 81% of DEGs (38/47) were downregulated, and 19% of DEGs (9/47) were upregulated. The 47 DEGs were subsequently analyzed. The TOP2A, CDKN3, and IGFBP3 genes were over-expressed, and CXCR12, CXCR14, CD5L, LCAT were under-expressed (Table 1). The analysis of genetic features of the DEGs indicated that most of them were located in the chromosome regions 5q, 6p26, 11p, and 5p (Figure 2A) and distributed in the liver and CD105-positive endothelial cells (Figure 2B). Downregulated DEGs were strongly linked to HCC (Figure 2C), while the upregulated DEGs were likely associated with the pathogenesis of non-alcoholic steatohepatitis (NASH) and breast cancer (Figure 2D).
 |
Table 1 Forty-Seven DEGs |
Functional Classification of DEGs and Protein-Protein Interaction Network
Using the Gene Ontology (GO) annotation, the number of DEGs in each GO term, including CC, BP, MF and corresponding KEGG pathways were also identified. The DEGs were predominantly involved in protein binding, monooxygenase activity, oxidoreductase activity, and the reduction of molecular oxygen (Figure S2). The BP included steroid metabolism, lipid metabolism, monocarboxylic acid metabolism, and response to chemicals (Figure S2). The KEGG pathways included retinol metabolism, bile secretion, tryptophan metabolism, cytokine-cytokine receptor interaction, and the p53 signaling pathway, as shown in Table S1. To understand the interactions involving DEGs, PPI, (Figure 3), network was generated using String 11.1. The detected interactions indicated that TOP2A and CDKN3 were the key nodes in the network. They participated in the regulation of cell cycle and were related to carcinogenesis. TOP2A was also involved in metabolism of protein and CDKN3 was associated with HCC.15–18
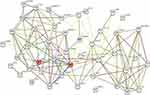 |
Figure 3 Analysis of PPI network of the 47 identified DEGs using Sting 11.1. Abbreviations: DEG, differentially expressed gene; PPI, protein-protein interaction. |
Identification of the Prognosis-Related DEGs Through KM Plotter Analysis (TCGA Cohort)
The K-M method was used in the present data to gain insight into the association between the DEGs and the prognosis (5-year OS, RFS) of patients with HCC. A total of 47 DEGs were used to analyze their association with 5-year OS and RFS in 364 and 313 patients from TCGA database, respectively. Twenty-six genes (BBOX1, ZG16, ASPM, CD5L, TOP2A, DTL, ESR1, FCN2, NAT2, LCAT, GLS2, GHR, as seen in Table 2) were associated with 5-year OS, and 40 genes were linked to RFS in HCC patients (Table 2). Twenty-two genes related to both OS and RFS were analyzed further using a regression model, as detailed below.
 |
Table 2 Validation of DEGs Correlated to RFS and 5-Year OS with K-M Plotter |
Validation of Candidate Genes and Identification of Independent Prognostic Factors (GSE14520 Cohort)
To confirm the relationship between the DEGs and prognosis, validation EGs was performed using different cases from GSE14520 cohort. The clinicopathologic features and 22 genes linked to 5-year OS were analyzed by univariate Cox regression, and the results are reported in Table 3. Tumor stage (AJCC TNM stage, BCLC stage, CLIP stage), risk signature for predicted metastasis (RSPM), tumor size, multiple nodular, cirrhosis, AFP, ZG16, ASPM, ACSM3, CD5L, PRC1, DNASE1L3, NAT2, RACGAP1, TOP2A, GLYAT, SRD5A2, SLC22A1, LCAT, ESR1, and GHR were associated with 5-year OS. Moreover, tumor stage (AJCC TNM stage, BCLC stage), RSPM, tumor size, gender, TOP2A, SLC22A1, LCAT, SRD5A2, CRHBP, GLYAT, NAT2, DNASE1L3, ZG16, and LPA were linked to RFS.
 |
Table 3 Clinicopathologic Features and Their Correlation with 5-Year OS and RFS on Univariate Analysis |
Based on statistical significance, the BCLC stage was selected for OS and RFS analysis. The multivariate COX regression hazard analysis demonstrated that RSPM, cirrhosis, BCLC stage, and CD5L were independent prognostic factors for OS, while gender, BCLC stage, and SRD5A2 were risk factors for RFS, as shown in Figure 4.
Construction and Validation of a DEGs-Based Predictive Nomogram Model Using GSE14520 Cohort
The cases of GSE14520 were divided into the training (n=144) and validation cohort (n=98). The validation cohort was used for the external validation of the nomogram model. The stage of cirrhosis (hazard ratio [HR] 4.97, 95% confidence interval [95% CI] 1.17–21.18, P=0.028), RSPM (HR 11.73, 95% CI 1.63–84.58, P=0.015), BCLC (HR 0.55, 95% CI 0.27–1.13, P<0.001), and CD5L (HR 0.712748, 95% CI 0.54–0.93, P=0.036) were found to be independent risk factors for 5-year OS (Figure 4A). BCLC stage (HR 4.48, 95% CI 1.66–7.04, P=0.005), gender (HR 0.42, 95% CI 0.22–0.81, P=0.031), and SRD5A2 (HR 0.87, 95% CI 0.77–0.97, P=0.044) were risk factors for RFS (Figure 4B). The independent prognostic factors were incorporated into a nomogram to estimate the OS and RFS, respectively (Figure 5A and D). The performance of the nomogram was assessed by the index of concordance (C-index). The calibration plot displays ideal consistency for 5-y OS using bootstrap sampling, with a C-index of 0.73 (95% CI 0.71–0.90) in the training cohort and 0.72 (95% CI:0.70–0.89) in the validation cohort, respectively (Figure 5B and C). The C-index for the nomogram predicting 1-yr RFS was 0.67 (95% CI 0.68–0.87) in the training cohort and 0.68 (95% CI 0.68–0.89) in the validation cohort, respectively (Figures 5E and F).
Correlations Between CD5L, SRD5A2 and Clinicopathological Features (GSE14520)
The correlation between CD5L and SRD5A2 with the clinicopathological features was investigated, and the results indicated that CD5L was related to the risk index for predicted metastasis (RIPM, P=0.015), TNM stage (P=0.05), but not to gender (P=0.336), cirrhosis (P=0.811), multiple nodularity (P=0.06), ALT (P=0.296), HBV (P=0.329), tumor size (P=0.827), BCLC stage (P=0.186), CLIP stage (P=0.633), and AFP (P=0.197). On the other hand, SRD5A2 expression was associated with RIPM (P<0.001), TNM stage (P=0.012), BCLC stage (P=0.011), CLIP stage (P<0.001), and AFP (P<0.001), but not with gender (P=0.178), cirrhosis (P=0.473), multiple nodularity (P=0.118), ALT (P=0.117), HBV (P=0.308), and tumor size (P=0.068). Lastly, all 22 genes correlated with CD5L and SRD5A2 (Table 4).
 |
Table 4 Correlation Between CD5L and SRD5A2 Expression and Clinicopathologic Features |
Discussion
The pathogenesis of HCC is related to the dysregulation of oncogenes and tumor suppressor genes, which are also responsible for the tumor progression and metastasis. In the current study, we identified 47 DEGs in HCC tissues, most of which were located on chromosomes 5q and 6q, and expressed mostly in the liver and CD105-positive endothelial cells. Previous studies also had similar observations with the tumor suppressor gene of non-cirrhotic HCC located on chromosome 5q and the gene related to recurrence located on chromosome 6q.19–22 In this study, most of DEGs with low expression were located on chromosome 5q, suggesting that the decreased expression of these genes promoted hepatocarcinogenesis.21 The upregulated genes were related to NASH and breast cancer, while the downregulated genes were closely associated with HCC. The GO and KEGG pathway analysis revealed that the 47 aberrant genes were involved in pathways of steroid, lipid, and retinol metabolism, bile secretion, cytokine-cytokine interaction, and p53 signalling, which are implicated in the cell cycle progression, proliferation, and invasion of cancer cells. The above biological phenotypes were in conformity to the role of the hub genes (TOP2A, ESR1, CDKN3, and PRC1).
In this study, 26 and 40 genes related to 5-year OS and RFS in HCC were identified using K-M plotter through the third-party database (TCGA cohort database), respectively. Subsequent analysis demonstrated that CD5L and SRD5A2 were independent risk factors for 5-year OS and RFS, respectively. Importantly, all independent prognostic factors were combined to build a predictive nomogram model, which was effective in predicting the prognosis of HCC patients.
CD5L is mostly expressed in the macrophages, lymphoid and inflamed tissues and regulates inflammatory responses and lipid synthesis.11,23,26 It is also known as the inhibitor of apoptosis in macrophages. It promotes macrophage survival by protecting them from the apoptotic effects of oxidized lipids in atherosclerosis.10 Moreover, CD5L is involved in the early response to the infection by bacteria and other pathogens, where it acts as a pattern recognition receptor and activates autophagy.10,24 CD5L also controls the metabolic switch in T-helper Th17 cells and regulates their expression of pro-inflammatory genes.9,23 CD5L accumulation on the hepatic surface could inhibit chronic liver injury by attenuating CCL4-induced injury and fibrosis, repressing TGF-ß signal and immune cell infiltration.6 Circulating CD5L potentially protects from the development of fatty liver and HCC.23 The current assay also identified CD5L was associated with improved OS in HCC. Contrastingly, Aran et al observed that CD5L was upregulated in HCC and it enhanced HCC cell growth and antiapoptotic responses by binding to HSPA5 (GRP78).11 It was also argued that CD5L was more suitable for HCC or cirrhosis accompanied by a viral infection than in the absence of an inflammatory response because CD5L itself was a factor associated with immune regulation.9,23
Previous studies pointed to the CD5L-related modulation of immune responses in malignancies, such as lung adenocarcinoma and HCC. Interestingly, CD5L has opposite effects on these tumors, promoting lung cancer but inhibiting liver cancer. It was established that CD5L accumulates on the surface of transformed hepatocytes and induces necrosis of the tumor. CD5L-deficient mice were susceptible to HCC and formed multiple liver tumors after feeding with a high-fat diet for one year. It was found that mouse CD5L was internalized together with CD36 by normal hepatocytes and modulated intracellular lipid metabolism.24 Therefore, CD5L may serve as a potential target for the treatment of HCC. In humans, CD5L protein is present at a high concentration in the serum, especially in women. However, CD5L peaks in women in their 20s and decreases with age. Several proteomic assays pointed to the CD5L protein as a putative biomarker for inflammatory conditions as well as liver diseases.25 The continuous inflammation caused by liver damage due to hepatitis virus infection, alcohol abuse, and NASH leads to hepatic fibrosis, which frequently triggers cirrhosis and, ultimately leads to HCC. Based on these findings, hCD5L can also be considered as a plasma biomarker for early detection of liver fibrosis and HCC, and the ratio of hCD5L-to- liver marker score might discriminate between HCC and non-HCC patients. HCC. Thus, CD5L may serve as a potential target in the development of HCC treatment. Future studies are required to verify the role of CD5L in HCC and develop strategies to alter its expression.
SRD5A2 is expressed in androgen-dependent tissues and responsible for converting testosterone to the more metabolically active dihydrotestosterone. Therefore, SRD5A2 V89L gene polymorphism has been associated with breast cancer and prostate cancer in previous studies.26–28 In the present study, we found that SRD5A2 was an independent risk factor for RFS in HCC. Liver regulates the metabolism and activity of sex hormones. Hence, SRD5A2 might be aberrantly expressed in HCC and serve as an useful biomarker for early diagnosis of HCC.29–31 However, in the current study, the C-index of SRD5A2 was 0.67 and 0.68 indicating that SRD5A2 may not have an adequate performance for the prognostic predictive nomogram. Future studies are required to delineate the role of SRD5A2 in predicting the recurrence and survival of HCC patients.
Conclusion
In this study, 47 genes associated with HCC were identified, with most of them being located on chromosomes 5q and 6q. The potential pathways involving these genes were steroid metabolism, lipid metabolism, retinol metabolism, bile secretion, and p53 signalling pathway. There were 26 and 40 genes associated with the 5-year OS and RFS of HCC patients, respectively. Among them, CD5L and SRD5A2 were independent risk factors for 5-year OS and RFS. A nomogram model combining CD5L, cirrhosis, RSPM and BCLC stage was constructed for accurate prognostication of patients with HCC. CD5L might be useful as a potential biomarker for HCC.
Abbreviations
AFP, Alpha-feto protein; AJCC, American Joint Committee on Cancer; ALT, Alanine aminotransferase; AST, Aspartate aminotransferase; BCLC, Barcelona Clinic Liver Cancer; CLIP, Cancer of the Liver Italian Program; DEG, differentially expressed gene; GEO, Gene Express Omnibus; GO, Gene Ontology; HCC, hepatocellular carcinoma; KEGG, Kyoto Encyclopedia of Genes and Genomes; NASH, Non-alcoholic steatohepatitis; OS, overall survival; RFS, relapse-free survival; RSPM, risk signature for predicted metastasis; CD5L, CD5 Antigen-Like; SMAD7, SMAD Family Member 7; TGFβ, Transforming Growth Factor Beta; TCGA, The Cancer Genome Atlas; TOP2A, DNA Topoisomerase II Alpha; CDKN3, Cyclin-Dependent Kinase Inhibitor 3; IGFBP3, Insulin-Like Growth Factor Binding Protein 3; CXCR12, C-X-C Motif Chemokine Ligand 12; CXCL14, C-X-C Motif Chemokine Ligand 14; LCAT, Lecithin-Cholesterol Acyltransferase; BBOX1, Gamma-Butyrobetaine Hydroxylase 1.
Disclosure
Yunxiu Luo and Xiaopeng Huang are co-first authors for this study. The authors have no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391(10127):1301–1314. doi:10.1016/S0140-6736(18)30010-2
3. Takeda H, Nishikawa H, Osaki Y. The new era of precision medicine in hepatocellular carcinoma: the urgent need for promising biomarkers. Hepatobiliary Surg Nutr. 2018;7(6):490–491. doi:10.21037/hbsn.2018.08.06
4. Schulze K, Imbeaud S, Letouzé E, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47(5):505–511. doi:10.1038/ng.3252
5. Grinchuk OV, Yenamandra SP, Iyer R, et al. Tumor-adjacent tissue co-expression profile analysis reveals pro-oncogenic ribosomal gene signature for prognosis of resectable hepatocellular carcinoma. Mol Oncol. 2018;12(1):89–113. doi:10.1002/1878-0261.12153
6. Bárcena C, Aran G, Perea L, et al. CD5L is a pleiotropic player in liver fibrosis controlling damage, fibrosis and immune cell content. EBioMedicine. 2019;43:513–524. doi:10.1016/j.ebiom.2019.04.052
7. Gebe JA, Kiener PA, Ring HZ, Li X, Francke U, Aruffo A. Molecular cloning, mapping to human chromosome 1 q21-q23, and cell binding characteristics of Spalpha, a new member of the scavenger receptor cysteine-rich (SRCR) family of proteins. J Biol Chem. 1997;272(10):6151–6158. doi:10.1074/jbc.272.10.6151
8. Sanchez-Moral L, Ràfols N, Martori C, Paul T, Téllez É, Sarrias M-R. Multifaceted roles of CD5L in infectious and sterile inflammation. Int J Mol Sci. 2021;22(8):4076. doi:10.3390/ijms22084076
9. Sanjurjo L, Aran G, Roher N, Valledor AF, Sarrias M-R. AIM/CD5L: a key protein in the control of immune homeostasis and inflammatory disease. J Leukoc Biol. 2015;98(2):173–184. doi:10.1189/jlb.3RU0215-074R
10. Sanjurjo L, Aran G, Téllez É, et al. CD5L promotes M2 macrophage polarization through autophagy-mediated upregulation of ID3. Front Immunol. 2018;9:480. doi:10.3389/fimmu.2018.00480
11. Aran G, Sanjurjo L, Bárcena C, et al. CD5L is upregulated in hepatocellular carcinoma and promotes liver cancer cell proliferation and antiapoptotic responses by binding to HSPA5 (GRP78). FASEB J. 2018;32(7):3878–3891. doi:10.1096/fj.201700941RR
12. Wang H-W, Hsieh T-H, Huang S-Y, et al. Forfeited hepatogenesis program and increased embryonic stem cell traits in young hepatocellular carcinoma (HCC) comparing to elderly HCC. BMC Genomics. 2013;14(1):736. doi:10.1186/1471-2164-14-736
13. Wang SM, Ooi LLPJ, Hui KM. Identification and validation of a novel gene signature associated with the recurrence of human hepatocellular carcinoma. Clin Cancer Res. 2007;13(21):6275–6283. doi:10.1158/1078-0432.CCR-06-2236
14. Roessler S, Jia H-L, Budhu A, et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70(24):10202–10212. doi:10.1158/0008-5472.CAN-10-2607
15. Chromosomal mapping of the genes for the human cell cycle proteins cyclin C (CCNC), cyclin E (CCNE), p21 (CDKN1) and KAP (CDKN3) - PubMed [Internet]; [cited August 5 2020]. Available from: https://pubmed.ncbi.nlm.nih.gov/7698009/.
16. Wang L, Sun L, Huang J, Jiang M. Cyclin-dependent kinase inhibitor 3 (CDKN3) novel cell cycle computational network between human non-malignancy associated hepatitis/cirrhosis and hepatocellular carcinoma (HCC) transformation. Cell Prolif. 2011;44(3):291–299. doi:10.1111/j.1365-2184.2011.00752.x
17. Labbé DP, Sweeney CJ, Brown M, et al. TOP2A and EZH2 provide early detection of an aggressive prostate cancer subgroup. Clin Cancer Res. 2017;23(22):7072–7083. doi:10.1158/1078-0432.CCR-17-0413
18. Wong N, Yeo W, Wong W-L, et al. TOP2A overexpression in hepatocellular carcinoma correlates with early age onset, shorter patients survival and chemoresistance. Int J Cancer. 2009;124(3):644–652. doi:10.1002/ijc.23968
19. Schaefer I-M, Schweyer S, Kuhlgatz J. Chromosomal imbalances in primary hepatic carcinosarcoma. Hum Pathol. 2012;43(8):1328–1333. doi:10.1016/j.humpath.2011.11.007
20. Ding SF, Habib NA, Dooley J, Wood C, Bowles L, Delhanty JD. Loss of constitutional heterozygosity on chromosome 5q in hepatocellular carcinoma without cirrhosis. Br J Cancer. 1991;64(6):1083–1087. doi:10.1038/bjc.1991.468
21. Raidl M, Pirker C, Schulte-Hermann R, et al. Multiple chromosomal abnormalities in human liver (pre)neoplasia. J Hepatol. 2004;40(4):660–668. doi:10.1016/j.jhep.2003.12.020
22. Nishida N, Nishimura T, Ito T, Komeda T, Fukuda Y, Nakao K. Chromosomal instability and human hepatocarcinogenesis. Histol Histopathol. 2003;18(3):897–909. doi:10.14670/HH-18.897
23. Maehara N, Arai S, Mori M, et al. Circulating AIM prevents hepatocellular carcinoma through complement activation. Cell Rep. 2014;9:61–74.
24. Sanjurjo L, Amézaga N, Aran G, et al. The human CD5L/AIM-CD36 axis: a novel autophagy inducer in macrophages that modulates inflammatory responses. Autophagy. 2015;11(3):487–502. doi:10.1080/15548627.2015.1017183
25. Gray J, Chattopadhyay D, Beale GS, et al. A proteomic strategy to identify novel serum biomarkers for liver cirrhosis and hepatocellular cancer in individuals with fatty liver disease. BMC Cancer. 2009;9(1):271. doi:10.1186/1471-2407-9-271
26. Zhang D, Li Q, Qu H-C, Yu T, Liu Y-R. Associations between the SRD5A2 gene V89L and TA repeat polymorphisms and breast cancer risk: a meta-analysis. Genet Mol Res. 2015;14(3):9004–9012. doi:10.4238/2015.August.7.9
27. Dušenka R, Tomaškin R, Kliment J, et al. Polymorphism of the SRD5A2 gene and the risk of prostate cancer. Mol Med Rep. 2014;10(6):3151–3156. doi:10.3892/mmr.2014.2621
28. Francis A, Sarkar S, Pooja S, et al. SRD5A2 gene polymorphisms affect the risk of breast cancer. Breast. 2014;23(2):137–141. doi:10.1016/j.breast.2013.11.010
29. Moribe T, Iizuka N, Miura T, et al. Identification of novel aberrant methylation of BASP1 and SRD5A2 for early diagnosis of hepatocellular carcinoma by genome-wide search. Int J Oncol. 2008;33(5):949–958.
30. Iizuka N, Oka M, Sakaida I, et al. Efficient detection of hepatocellular carcinoma by a hybrid blood test of epigenetic and classical protein markers. Clin Chim Acta. 2011;412(1–2):152–158. doi:10.1016/j.cca.2010.09.028
31. Tsunedomi R, Ogawa Y, Iizuka N, et al. The assessment of methylated BASP1 and SRD5A2 levels in the detection of early hepatocellular carcinoma. Int J Oncol. 2010;36(1):205–212.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.