Back to Journals » OncoTargets and Therapy » Volume 14
Role of Antizyme Inhibitor Proteins in Cancers and Beyond
Authors Tulluri V, Nemmara VV
Received 10 September 2020
Accepted for publication 5 November 2020
Published 25 January 2021 Volume 2021:14 Pages 667—682
DOI https://doi.org/10.2147/OTT.S281157
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Leo Jen-Liang Su
Vennela Tulluri, Venkatesh V Nemmara
Department of Chemistry and Biochemistry, Rowan University, Glassboro, NJ 08028, USA
Correspondence: Venkatesh V Nemmara
Department of Chemistry and Biochemistry, Rowan University, 201 Mullica Hill Road, Glassboro, NJ 08028, USA
Tel +1 856-256-5460
Email [email protected]
Abstract: Polyamines are multivalent organic cations essential for many cellular functions, including cell growth, differentiation, and proliferation. However, elevated polyamine levels are associated with a slew of pathological conditions, including multiple cancers. Intracellular polyamine levels are primarily controlled by the autoregulatory circuit comprising two different protein types, Antizymes (OAZ) and Antizyme Inhibitors (AZIN), which regulate the activity of the polyamine biosynthetic enzyme ornithine decarboxylase (ODC). While OAZ functions to decrease the intracellular polyamine levels by inhibiting ODC activity and exerting a negative control of polyamine uptake, AZIN operates to increase intracellular polyamine levels by binding and sequestering OAZ to relieve ODC inhibition and to increase polyamine uptake. Interestingly, OAZ and AZIN exhibit autoregulatory functions on polyamine independent pathways as well. A growing body of evidence demonstrates the dysregulation of AZIN expression in multiple cancers. Additionally, RNA editing of the Azin1 transcript results in a “gain-of-function” phenotype, which is shown to drive aggressive tumor types. This review will discuss the recent advances in AZIN’s role in cancers via aberrant polyamine upregulation and its polyamine-independent protein regulation. This report will also highlight AZIN interaction with proteins outside the polyamine biosynthetic pathway and its potential implication to cancer pathogenesis. Finally, this review will reveal the protein interaction network of AZIN isoforms by analyzing three different interactome databases.
Keywords: polyamine, ornithine decarboxylase, antizyme inhibitor, antizyme, putrescine, spermine, spermidine, mRNA editing, 26S proteasome, degradation, ubiquitin-independent, protein interactome
Introduction
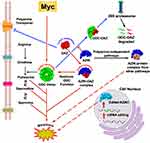
Reprogramming of the metabolic landscape is a hallmark of cancer cells.1 Transformed cells modulate metabolic pathways and nutrient uptake using various adaptation mechanisms to support tumor progression.2 In this regard, the polyamine metabolism is a central pathway hijacked by cancer cells.3,4 Polyamines are small aliphatic polycations biosynthesized from amino acids such as methionine and ornithine and are tightly regulated by de novo synthesis and diet.5 Polyamines play a crucial role in key cellular processes such as cell growth, cell proliferation, apoptosis, and gene regulation.6,7 Notably, polyamine levels are frequently dysregulated in multiple cancer types.3,4 Besides malignant transformation, elevated polyamines are essential for the developmental and compensatory growth of cancer cells in response to systemic stimuli like hormones.8 Elevated polyamine levels contribute to cancer progression by binding to nucleic acids, thereby controlling DNA replication, transcription, translation, and cell cycle progression.9–12 Dysregulation of polyamine metabolism is directly controlled by oncogenic signaling pathways, including RAS, PI3K, mTORC, and MYC genes.13 Specifically, MYC oncogenes upregulate the expression of ornithine decarboxylase (ODC), a pyridoxal phosphate-dependent enzyme. ODC converts ornithine to putrescene, which is the first rate-limiting step in polyamine biosynthesis.14 Putrescine is an important polyamine that is further converted to the other two polyamines, spermidine and spermine in cells (Figure 1). Spermidine is further utilized for the post-translational modification of a lysine residue on the eukaryotic initiation factor 5A isoform 1 (eIF5A).15 This process, termed as hypusination, is essential for eIF5A to influence neoplastic transformation.16
ODC is catalytically active as a homodimer, and its expression is tightly regulated.17 The monomeric form of ODC is short-lived and gets degraded by the proteasome machinery.18 Notably, ODC levels are elevated at low and repressed at high polyamine levels, respectively. The post-translational regulation of ODC is mediated by two different antagonistic protein isoforms, Ornithine Decarboxylase Antizymes (OAZs) and Antizyme Inhibitors (AZINs). OAZ targets ODC for proteasomal degradation via ubiquitin independent pathways.19 Specifically, OAZ binds the carboxylic end of monomeric ODC and presents it to the 26S proteasome, thereby decreasing the half-life of ODC to minutes.18 Interestingly, ODC is stabilized by its non-catalytic homolog AZIN, which forms a tighter complex with OAZ, thereby disrupting ODC-OAZ interaction. This rescues ODC activity resulting in increased polyamine synthesis.18
Polyamines regulate the expression of functional OAZ at the transcriptional level. OAZ is encoded by two open reading frames ORF1 and ORF2. The ORF1 with a premature stop codon leads to the expression of a truncated OAZ, which lacks the ability to target ODC for proteasomal degradation. Interestingly, high polyamine levels manipulate the ribosome to switch to a +1 reading frame, which then reads ORF2 and skips the premature stop codon in ORF1. This results in the expression of functional OAZ.20,21 Four different isoforms of antizymes have been reported in mammals – OAZ1, OAZ2, OAZ3, and OAZ4.22,23 OAZ1 is commonly found in almost all tissues and functions to inhibit and degrade ODC. OAZ2 is functionally similar to OAZ1 and can inhibit ODC and target it for proteasomal degradation. OAZ3 is specific to the testis and is restricted to the late stage of sperm production. The putative fourth member OAZ4 isolated from the human brain has been demonstrated to bind to ODC, but the ability to promote ODC degradation has not yet been examined.24–30 In addition to modulating ODC activity, OAZ also regulates polyamine levels by inhibiting polyamine transport.31–33 However, the regulatory mechanism of OAZ-mediated polyamine transport is not clear.
AZINs play a pivotal role in polyamine homeostasis by modulating OAZ activity. Two isoforms of antizyme inhibitors are reported - AZIN1 and AZIN2. Both isoforms can affect intracellular polyamine levels by binding to OAZ isoforms, thereby rescuing ODC activity.34,35 Notably, AZINs can also increase the uptake of extracellular polyamines, likely by binding to and sequestering OAZ, thereby preventing the negative regulation of polyamine transport by OAZ.36 AZIN’s role in mammalian pathophysiology is slowly emerging as researchers focus on understanding its function outside the polyamine biosynthetic pathway. Moreover, both AZIN1 and AZIN2 have been implicated in a variety of mammalian disease . Notable research advancements involve understanding the role of AZIN1 in promoting multiple cancers. The current review focuses on structural and functional aspects of AZIN isoforms in polyamine dependent and independent processes. Furthermore, it also highlights the contribution of AZIN isoforms in human cancers.
Regulation of Antizyme Inhibitors
AZIN1 Cellular Distribution and Regulation
AZIN1 is ubiquitously expressed in all proliferating cell types, as evidenced by its mRNA levels.37,38 AZIN1 mRNA in mice is reported to undergo alternative splicing to generate multiple forms of Azin1 transcripts.39 AZIN1 expression is regulated by various factors, including nutritional stimuli and other growth factors.40 Moreover, increased polyamine levels can affect Azin1 transcription and splicing patterns.39 Cellular AZIN1 levels is also controlled by its proteasomal degradation. Unlike ODC, AZIN1 is degraded by the proteasome machinery via the ubiquitin-dependent pathway.41 Notably, AZIN1 is stabilized by its interaction with OAZ1, likely by interfering with the ubiquitin-mediated degradation.29,42
AZIN2 Cellular Distribution and Regulation
AZIN2 is primarily expressed in the testis and brain. Because of its tissue-specific distribution, AZIN2 is thought to play a major role in terminal differentiation.43 As a short-lived protein, AZIN2 has a lower half-life than ODC but is interestingly more stable than AZIN1.43,44 Like AZIN1, its interaction with OAZ isoforms increases the stability of AZIN2. Furthermore, cellular studies indicate that while AZIN2 is primarily degraded via the proteasomal pathway, it may undergo degradation via alternative mechanisms as well.44
Structural and Functional Features of Antizyme Inhibitors
Structural Features of AZIN1 Interaction with OAZ1
Originally discovered in rat liver extracts as an antizyme scavenger, AZIN1 was thought to be an ODC derivative due to the similarities in protein sequences.40,45,46 While it shares a 49% sequence identity with ODC, AZIN1 lacks ornithine decarboxylating activity. Besides, AZIN1 is 41% structurally identical to Gm853, an ODC homolog in mice. Interestingly, Gm853 exhibits the function of both AZIN1 and ODC.47
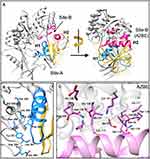
Structural studies indicate that the binding mode of AZIN-OAZ1 interaction is similar to ODC-OAZ1 interaction.48 OAZ1 interacts with AZIN1 predominantly at two different sites, site-A, and site-B, as seen from a co-crystal structure of OAZ1 bound to AZIN1 (Figure 2).48 Site-B in AZIN1 includes the following residues – S91, K92, N93, C114, Q116, V117, S118, Q119, D134, N135, E136, I137, E138, K140, R144. These residues in AZIN1 interact with the helical region H2 (shown in pink) of OAZ1, including the N-terminal α-helix (residues 152–165) and residues D99, T123, and D124. Residues in site B collectively form the Antizyme-Binding Element (AZBE). Furthermore, Site-A in AZIN1 harbors important residues that interact with the helical region H1 (shown in blue) of OAZ1. Residues in site-A includes F170, Y321, A325, S329, L328, D359, E360, L361, H390, S393, F395, N396, and D397. The helical region H1 in OAZ1 includes the c-terminal α-helix (residues 178–192) and the adjoining loop (residues193-200) in OAZ1.
Interestingly AZIN1-OAZ1 interaction (Kd = 20 nM) is 10-fold stronger than ODC-OAZ1 interaction (Kd = 200 nM).49,50 Mutational studies suggest that the differences in specific amino acid residues in the Antizyme-Binding Element (AZBE) of ODC and AZIN1 may contribute towards differential OAZ-binding affinities.49 Notably, the corresponding mutations K125N and K140M in AZIN1 decrease its binding to OAZ1 by 10-fold. Conversely, mutating N125 and M140 in ODC to lysine residues markedly increases ODC’s binding towards OAZ1. However, it is unclear as to whether these residues contribute to the increased affinity of AZIN1-OAZ1 interaction. Biochemical studies on truncated OAZ1 variants reveal that residues from 95–228 are sufficient for optimal AZIN1 binding.51
Structural comparison of AZIN1-OAZ1 and ODC-OAZ1 complexes reveal that amino acid residues A325 and S329 in AZIN1 are conserved in vertebrates. N327 and Y331 substitute these residues in ODC.48 Ghalali et al. in a recent study demonstrated that ODC mutations N327A and Y331S displayed increased binding affinity (Kd = 192 nM and 277 nM respectively) towards OAZ1. This suggests that these two residues may likely play a key role in the decreased binding affinity of ODC-OAZ1 interaction. Furthermore, Ghalali et al. performed an alanine scanning mutagenesis experiment on AZIN1 wherein 39 alanine point mutations were introduced on the protein, and the effect on OAZ1 binding was evaluated.50 Results from this experiment indicate that while no single residue in AZIN1 contributes to OAZ1 binding, a combination of small contributions from several single residues may likely be responsible.50 Additionally, results from the experiment indicate that individual mutations at site A of AZIN1 decrease OAZ1 binding (increased Kd from 20 nM for WT to ~80 nM for F395A) by up to 4-fold. Contrarily, individual mutations at site B of AZIN1 (AZBE) did not decrease OAZ1 binding. This suggests that the AZBE site in AZIN1 may not necessarily contribute to the increased affinity of AZIN1-OAZ1 interaction. More importantly, biophysical and structural characterization of AZIN1 interactions with other OAZ isoforms is not yet reported.
AZIN2 Interaction with OAZ Isoforms
The Human AZIN2 isoform shares 49% sequence identity with human ODC. Not surprisingly, AZIN2 lacks ornithine decarboxylase activity, likely due to its inability to bind PLP.52,53 However, AZIN2 binds efficiently to three OAZ isoforms (OAZ1, OAZ2, and OAZ3), thereby inhibiting OAZ-mediated degradation of ODC.36,53 Although biochemical interactions of AZIN2 with OAZ isoforms have not been quantitated at the protein level, the magnitude of interaction has been compared with AZIN1 in mammalian cell lines. Interestingly, AZIN2 binds less efficiently to both OAZ1 and OAZ3 than AZIN1.43 Notably, investigations on the molecular basis of AZIN2 interaction with three OAZ isoforms via mutational analysis indicate that amino acid residues in the antizyme-binding element (AZBE) are essential for AZIN2-OAZ binding.44
To date, no structural studies on AZIN2-OAZ1 interaction have been reported, except for an AZIN2 homology model.44 While AZIN2 shares only 42% sequence identity with AZIN1, the AZBE in AZIN2 is 62% identical to the AZBE in AZIN1 (). A quick analysis of the sequence alignments of AZIN1, AZIN2, and ODC reveal differences in amino acid residues that could explain the differential binding of these proteins to OAZ1. AZIN2 has six different amino acid substitutions, three of which happen to be in the Antizyme Binding Element region of the protein. Residues N93, S118, and K140 in AZIN1 are replaced by A94, A119, and A141 in AZIN2. Additionally, residues A325, S329, E360, H390, and D397 in AZIN1 are substituted by the following residues, N328, F332, G363, G393, and G400 in AZIN2. These are notable substitutions as these residues in AZIN1 interact with residues in the H1 and H2 regions of OAZ1. Particularly, H390 and D397 in AZIN1 make ionic interactions with E196 and R199 from the loop next to the H2 helical region in OAZ. Both ODC and AZIN2 likely lack these interactions due to alanine substitutions at these sites. Additionally, like ODC, residues N328 and F332 in AZIN2 may likely contribute to a decreased binding towards OAZ1 compared to AZIN1. Therefore, to further understand the functional features of AZIN2, it is imperative to elucidate the biophysical and structural aspects of AZIN2 binding to individual OAZ isoforms.
Physiological Role of Antizyme Inhibitors
Physiological Role of AZIN1
Early reports on the in vivo role of AZIN1 suggest that the Azin1 gene is essential for the survival of Azin1 knockout transgenic mice.54 Decreased polyamine levels found in transgenic mice reinforce the fact that AZIN1 is a positive driver of intracellular polyamine levels. Moreover, increased AZIN1 expression is correlated to elevated polyamine levels leading to increased cell proliferation.29,55 Notably, different roles of AZIN1 in polyamine independent pathways are also reported. AZIN1 plays an active role in cell proliferation by preventing cyclin D1 degradation, thereby improving the half-life of cyclin D1.56 However, structural details of AZIN1 interaction with cyclin D1 are not clear. The subcellular localization of AZIN1 changes at various phases of the cell cycle.55 AZIN1 expression increases during the early cell cycle at G1 and G2/M phases.57 Also, AZIN1 colocalizes with OAZ1 at centrosomes during the period from prophase to late anaphase. Interestingly, AZIN1 colocalization with OAZ1 was not observed during interphase, when ODC activity increased.55,57 AZIN1 is believed to stabilize ODC activity during interphase. However, the stabilization mechanism is not clear.
AZIN1 expression pattern has been correlated to genes involved in the rat oestrous cycle and avian ovarian follicles, suggesting a possible role of AZIN1 in reproductive physiology.58,59 Additionally, AZIN1 is highly expressed in vasopressinergic and oxytocinergic neurons of the male rat hypothalamus. AZIN expression in hypothalamic nuclei is directly correlated to increased polyamines resulting in transcriptional regulation of the neuropeptide hormone arginine vasopressin.60 Furthermore, transcriptomic studies reveal that glucocorticoids consistently upregulate the Azin1 gene in brain cells.61
Physiological Role of AZIN2
Although AZIN2 expression stimulates ODC activity and polyamine uptake, not much is known about its effects on polyamine levels in vivo.36,52 Contrary to Azin1 knockout mice, transgenic mice with deleted Azin2 gene were found to be viable.54 AZIN2 is widely expressed in differentiated cells. However, it does not seem to be involved in cell proliferation, unlike AZIN1.62 Specifically, AZIN2 is expressed in neuronal cells of the frontal cortex and co-localizes with N-methyl-d-aspartate (NMDA) glutamate receptor, suggesting a possible role in the central nervous system.63
AZIN2 is highly expressed in the testes of adult humans and mice, suggesting a possible role in spermiogenesis.64 Strikingly, the spatial and temporal expression pattern of AZIN2 is similar to OAZ3, the testes-specific OAZ.27 However, unlike Oaz3 knockout mice, Azin2 knockout mice are fertile and exhibit decreased polyamine levels.63,65,66 Interestingly, OAZ3 interacts with testicular proteins that are unrelated to polyamine metabolism.67,68 Given the correlation in the expression patterns of AZIN2 and OAZ3, it is plausible that AZIN2 may regulate OAZ3-mediated polyamine independent pathways. A recent study showed decreased testosterone levels in plasma and testis and decreased sperm motility in Azin2 knockout mice, suggesting a possible role of AZIN2 in testes physiology and reproductive biology.66
Furthermore, AZIN2 is expressed in the pancreas and adrenal glands, suggesting a possible role in secretory processes.62 AZIN2 also localizes to post-Golgi vesicles of the secretory pathway and regulates intracellular vesicle transport.69 Recent studies show that AZIN2 is expressed in serotonin containing mast cell granules, thereby demonstrating AZIN2 as a regulator of biogenic amines like serotonin and histamines.70,71
Antizyme Inhibitors in Mammalian Disease
Role of AZIN1 in Cancers
AZIN1 is overexpressed in different types of human malignancies, including gastric, lung, prostate, liver, and ovarian cancers.72–75 Specifically, AZIN1 overexpressing NIH3T3 fibroblast cells proliferated much faster than control cells and generated tumors when injected into nude mice.35 RAS transformed cell lines had increased AZIN1 expression and were more sensitive to increased polyamine uptake. Additionally, silencing AZIN1 expression in A549 lung cancer cells decreased polyamine levels and reduced cell proliferation due to increased OAZ1 mediated ODC degradation.76 More importantly, shRNA mediated knockdown of Azin1 in both human and rat prostate cancer diminished tumor volume in vivo following subcutaneous injection into nude mice.77 These studies demonstrate the therapeutic potential of AZIN1 in multiple cancers.
Recent studies reveal that the Azin1 transcript undergoes mRNA editing in certain cancers, including hepatocellular carcinoma (HCC), esophageal squamous cell carcinoma, Non-Small Cell Lung Cancer (NSCLC), colorectal cancers, and prostate cancers. mRNA editing involves the conversion of Adenosine to Inosine in primary mRNA transcripts, a reaction catalyzed by the enzyme Adenosine Deaminases Acting on RNA (ADAR), leading to diversification of the transcriptome in human cells.78 mRNA editing of the Azin1 transcript, a target of ADAR, leads to a “gain-of-function” phenotype. This phenotype leads to the substitution of serine residue to glycine (S367G) on the translated protein.79 Such a phenotype was shown to augment tumor-initiating potential leading to a more aggressive tumor behavior in HCC and esophageal squamous cell carcinoma.79,80 mRNA editing of Azin1 and subsequent translation of the mutant AZIN1 protein was also observed in NSCLC patient samples.81 In these NSCLC samples, AZIN1 protein expression was higher in tumors with edited Azin1 than in those with the non-edited transcript. In parallel, edited AZIN1 at the protein level induced proliferation, invasion, and migration of lung cancer cell lines.81 Furthermore, analysis of tissue samples from multiple independent colorectal cancer patient cohorts revealed Azin1 mRNA as one of the most frequently edited transcripts.82 Additionally, these edited transcripts enhanced the invasive potential of cancer-associated fibroblasts within the tumor microenvironment in the colon.83 Specifically, edited Azin1 enhances spheroid formation, with a corresponding increase in stemness markers in colorectal cancers.82 A recent study unveiled a correlation between Azin1 mRNA editing levels and lymph node metastasis in gastric cancer patients, thereby demonstrating the potential of measuring Azin1 mRNA editing status as a promising prognostic biomarker for this cancer type.84 Another recent study analyzed the expression and localization of AZIN1 in prostate cancer specimens and demonstrated that nuclear localization of the mRNA-edited AZIN1 protein is associated with a decreased survival rate.85 mRNA-edited Azin1 transcripts are thought to express a mutant protein localized in the cell nucleus with an increased binding affinity towards OAZ1. However, recombinantly expressed S367G mutant did not display any increased affinity towards OAZ1, suggesting that nuclear localization of the mutant AZIN1 may contribute to oncogenesis via mechanisms independent of AZIN1-OAZ1 interactions.50,85
AZIN1 plays a major role in modulating cell proliferation and oncogenesis via polyamine independent mechanisms. Notably, AZIN1 regulates Cyclin D1 (CCND1) levels by preventing its degradation. CCND1 serves as an active switch in regulating cell-cycle progression through its association with Cyclin-Dependent Kinases (CDKs). Normal cell-cycle progression involves constant degradation of CCND1. Dysregulation of CCND1 activity by preventing its degradation can lead to aberrant cell proliferation in various cancer types.86,87 In this regard, Azin1 silencing leads to diminished CCND1 levels resulting in reduced cell proliferation.56 Interestingly, OAZ1 was shown to prevent cell cycle progression by binding to and targeting CCND1 for proteasomal degradation.88 Strikingly, OAZ1 binds to CCND1 with Kd of 810 nM, 40 fold higher than its interaction with AZIN1 (Kd = 20 nM).89 AZIN1 overexpression can thus lead to OAZ-sequestration, thereby preventing CCND1degradation.
AZIN1 also contributes to cancer progression by neutralizing the tumor-suppressor functions of OAZ1 in polyamine independent pathways. Notably, OAZ1 is a potent tumor suppressor, the expression of which can prevent cell growth and tumorigenesis.29,90 For example, OAZ1 expression levels are downregulated in cisplatin-resistant Non-Small Cell Lung Cancer Cells (NSCLC), thereby attributing a major role in drug-resistant cancers.91 OAZ1 also mediates the proteasomal degradation of DNP73 in a c-Jun dependent manner.92 The anti-apoptotic DNP73 is a trans-activation deficient isoform of P73, which is elevated in multiple cancers.92–94 Interestingly, co-expression of AZIN1 with either DNP73 or c-Jun prevented OAZ-mediated degradation of DNP73, suggesting a possible role of AZIN1 as a tumor-proliferator. Additionally, the OAZ1/AZIN1 system is implicated to play a role in centrosome amplification.95 Centrosome amplification is a common feature of numerous solid tumors and is an early event in tumorigenesis.96 While OAZ1 overexpression reduces numerical centrosome abnormalities, depletion of OAZ1 leads to centrosome overamplification.95 Contrarily, AZIN1 overexpression leads to centrosome overamplification, whereas silencing the Azin1 gene decreases numerical centrosome abnormalities.95 Interestingly, OAZ1 also mediates degradation of other proteins associated with cell-cycle progression, including SMAD1,97 AuroraA,98 MPs I,99 and loricrin.100 Therefore, AZIN1-mediated sequestering of OAZ may likely regulate proteasomal degradation of these proteins, thereby controlling cell-cycle progression.
Role of AZIN2 in Cancers
AZIN2 is mainly expressed in terminally differentiated cells, such as neural cells,63 Leydig cells,101 and mast cells.70 Elevated expression of AZIN2 is observed in Purkinje cells and the hippocampus of brains affected by Alzheimer’s disease.63 However, the pathological relevance of this observation is not clear. Although AZIN2 stimulates ODC activity and polyamine uptake, it is not involved in cell proliferation and is not found to be upregulated in many cancer types. Strikingly, a recent study identified elevated expression of AZIN2 in colorectal cancer (CRC) samples from a cohort of 840 patients.102 Immunohistochemical staining of CRC samples revealed strong AZIN2 expression in invasive cells of the tumor front. Also, they displayed morphological features of epithelial-mesenchymal transition (EMT). Additionally, AZIN2 overexpression in T84 CRC cells induced the accumulation of cell-derived CD63+ exosomes in the culture medium, indicating a secretory phenotype.102 Notably, CD63 expression is associated with advanced-stage CRC with mucinous histology103 Taken together, these findings attribute a pathological role for AZIN2 in CRC progression.
Cellular Protein-Interactome of Antizyme Inhibitors
Besides polyamine homeostasis, AZINs are thought to function in various cellular processes. While some AZIN-regulated cellular processes are implicated in diseases such as cancers, most AZIN-mediated functions are unknown. A likely reason is the lack of information about the cellular protein interactome of AZINs. To date, no study has focused exclusively on identifying AZIN-interacting protein networks in tissue-specific cell lines. To bridge this gap, the current review will examine the identities of AZIN-interacting proteins from three different databases.
Protein Interaction Network of AZIN1
The AZIN1 protein-interaction network is obtained from three-different databases – STRING,104 BioGRID,105 and IntAct.106 All three databases provide information regarding AZIN1 interactors based on predicted and experimental evidence. To avoid algorithmic discrepancies among these databases, only AZIN1 interactors with experimental evidence is examined. A total of 18 different proteins are identified as AZIN1 interactors with eight proteins found exclusively on BioGRID, whereas only four proteins are found to be common to all three databases (Figure 4A and Table 1). Experimental evidence comes mainly from two experimental techniques, High-throughput Affinity Capture Mass Spectrometry and Yeast-two Hybrid methodology employed in various studies to identify tissue, and cell line-specific global proteome-wide interaction networks.
 |
Table 1 AZIN1-Interacting Proteins and Their Description |
Interestingly, of the 18 protein interactors, 15 are unrelated to polyamine-dependent pathways. Five different studies utilized the Yeast-two Hybrid methodology to identify proteome-wide interaction networks. Of these, eight different proteins were identified as AZIN1 interactors. ABCC2, ADSSL1, ARNTL, and KHDRBS1 were identified as AZIN1 interactors in a comprehensive study to map the human liver protein interaction network using yeast two-hybrid methodology.107 The functional relevance of these interactions is unclear. However, It is worth noting that increased expression of ABCC2 (MRP2)108,109 and KHDRBS1 (Sam68)110,111 is associated with multiple cancers.
A systematic study on acute lymphoblastic leukemia cancer gene products revealed CDKN2A as a gene set with 29 distinct mutations from 1545 patient samples examined.112 A closer look at the functional associations between ALL-gene products and their partners in the human proteome using yeast two-hybrid detection assay revealed Azin1 as an interactor of CDKN2A.112 The CDKN2A gene encodes two tumor-suppressor proteins p16 and p14ARF, both involved in cell cycle regulation.113 Notably, the downregulation of CDKN2A is common in multiple cancers due to somatic mutations. Therefore, it would be interesting to examine whether AZIN1 interaction with CDKN2A might contribute to the AZIN1 function (at protein level) in cell-cycle regulation via preventing CCND1 degradation.
Another yeast two-hybrid screen identified AZIN1 as a potential interactor of Fanconi Anemia complementation group A (FANCA) and group C (FANC) proteins from a list of 69 proteins belonging to various cellular pathways.114 This study provided evidence for Fanconi Anemia proteins’ involvement in several pathways linked to the clinical phenotype. However, it did not examine the role of AZIN1-FANCA/FANC interactions in Fanconi Anemia.114
Exploring of the human protein interactome using high-throughput affinity-purification mass spectrometry in HEK292T cells identified 10 different protein interactors of AZIN1 from three separate studies. Not surprisingly, the three different OAZ isoforms, OAZ1,43,115–117 OAZ2,115,116,118 and OAZ3,116,119,120 were identified as AZIN1 interactors. These studies also revealed the interaction of AZIN1 with selective heavy chain (KIF5B, KIF5C) and light chain (KLC1, KLC2, and KLC4) Kinesin isoforms.115,116 Both KIF5B and KIF5C isoforms are expressed due to gene duplication of the Kinesis heavy chain 5 (KIF5) gene belonging to the kinesin 1 family.121 KIF5 protein is a tetramer comprising two heavy chains and two light chains and exhibits motor activity to transport various cargo along microtubules. On the other hand, KLCs (KLC1-3) associates with KIF5 through an N-terminal domain and are involved in binding various cargos such as vesicles and Golgi complexes.121 Furthermore, Kif5b knockdown in mitotic cells results in centrosome amplification and chromosomal segregation defect, suggesting a crucial role for KIF5B in meiotic cell development and mitotic cell division.122 Given these observations, AZIN1 might play a likely role in centrosome regulation via kinesin interaction. However, there is no evidence for the physiological relevance of AZIN1-Kinesin interaction, and it remains to be seen as to whether such an interaction has relevance in pathological cell lines.
High-throughput affinity-purification mass spectrometry experiments in HEK292T cells also revealed the interaction of AZIN1 with two different enzymes, MSRB3 and TDP2.115,116 While MSRB3 decreases oxidative stress by catalyzing the reduction of methionine sulfoxide in proteins, TDP2 functions as a DNA repair enzyme. The functional relevance of AZIN1 association with these enzymes remains to be determined. AZIN1 also interreacts with ELAV1 (Human antigen R), an RNA-binding protein that binds and stabilizes mRNA to regulate gene expression.123 The functional consequence of AZIN1-ELAV1 interaction has not yet been explored.
Protein Interaction Network of AZIN2
Given the experimental evidence, AZIN2 interacts with 12 different proteins, of which 9 of them are common to BioGRID105 and IntAct,106 whereas only 2 are common to all three databases. Although the STRING104 database lists 10 unique AZIN2 interactors, 8 lack experimental evidence and is purely based on text mining (Figure 4B). AZIN2 Interactors with experimental evidence are listed in Table 2.
 |
Table 2 AZIN2-Interacting Proteins and Their Description |
AZIN2 was shown to interact with OAZ1,116 and OAZ2,116 from a study that explored human interactome using high-throughput affinity-purification mass spectrometry. Additionally, AZIN2 interaction with OAZ3119,120 was established from two different yeast two-hybrid screens. Although there is precedent in the literature (as discussed above) for the physiological basis of AZIN2-OAZ interactions, their structural and biophysical aspects remain unclear.
In addition to OAZ1 and OAZ2, seven other protein interactors of AZIN2 are identified from the high-throughput affinity-purification mass spectrometry study (Table 2).116 AZIN2 interacts with CORO7, a ubiquitous mammalian protein involved in maintaining Golgi structure and function.124 Interestingly, AZIN2 localizes in the ER-Golgi intermediate compartment and the cis-Golgi network125 suggesting a likely physiological relevance for AZIN2-CORO7 interaction. AZIN2 also interacts with DDX11 and SMARCAL1, two different proteins involved in DNA replication and chromatin regulation. DDX11 is a DNA-dependent ATPase and a helicase that is involved in DNA replication and sister chromatid cohesion.126 SMARCAL1, on the other hand, is a mediator of nucleosome restructuring and chromatin remodeling during gene regulation and DNA repair.127 Besides, AZIN2 also interacts with LRF1, a nuclear receptor protein that regulates chromosome segregation during mitosis through its interaction with the Heterochromatin protein 1-α.128 Although there is precedent for polyamine-dependent chromatin regulation, the functional relevance of AZIN2 interaction with DDX11, SMARCL1, or LRF1 is unclear.
SMG8 is a protein cofactor that interacts with SMG1 kinase and is involved in non-sense mediated mRNA decay (NMD) pathway,129 a surveillance process that targets and degrades defective mRNA containing premature translation termination codons.130 Interestingly, polyamines are known to modulate alternative splicing of SSAT pre-mRNA via the NMD pathway.131 However, the role of AZIN2 in regulating the NMD pathway via SMG8 interaction is unknown. Strikingly, AZIN2 interacts with tumor-associated proteins PRAME132 and TPD52L1,133,134 which are overexpressed in a variety of malignancies including breast cancers. Particularly, TPD52L1 is shown to function as a cell-cycle regulator protein involved in the G2-M transition in breast cancer cell lines.133,134 However, AZIN2 is shown to have no involvement in cell cycle regulation.62 Therefore, the functional relevance of AZIN2 interaction with PRAME and TPD52L1 in the context of human malignancies remains to be determined.
On a different note, a recent study demonstrated the interaction of TLN-1 and PSMC5 with a long non-coding splice variant RNA of AZIN2 (lncRNA-AZIN2-sv), through RNA-pulldown and RNA-immunoprecipitation experiments.135 This study was aimed to identify the role of AZIN2-splice variant mRNA in cardiomyocyte regeneration via endothelial activation and angiogenesis.135 Notably, Tln1, a cytoskeletal protein, was shown to regulate integrin activity along with its involvement in cell spreading and flattening during angiogenesis.136 AZIN2-splice variant mRNA was further shown to promote ubiquitin-dependent Tln1 degradation via 26S proteasome subunit ATPase 5 (PSMC5), leading to decreased β-integrin expression, an essential factor for endothelial development and vessel formation. Taken together, AZIN2-sv plays an active role in angiogenesis via Tln1 downregulation, thereby blocking the miR-214/PTEN/Akt pathway.135
Conclusions and Future Perspectives
This review summarizes the recent efforts in understanding the multi-faceted role of AZIN isoforms in mammalian pathophysiology and sheds light on the cellular interactome of both AZIN isoforms. In short, AZINs play a crucial role in polyamine homeostasis via their interaction with OAZ isoforms and regulate polyamine-independent cellular processes through their interaction with multiple proteins. Despite our understanding of AZINs, many questions remain unanswered due to a lack of structural and biophysical evidence, which forms the backbone of protein-protein interactions. For example, while the structural and biophysical basis of the AZIN1-OAZ1 interaction is reported,48–51 no such information exists for AZIN1 interactions with OAZ2 or OAZ3 and AZIN2 interactions with OAZ1, OAZ2, or OAZ3. Albeit identifying novel protein interactors of AZIN isoforms outside the polyamine pathway, their biochemical and biophysical aspects remain unknown. Additionally, to understand and correlate AZIN’s role in disease progression, it is imperative to identify AZIN interactors at the protein level in various pathogenic states.
AZIN1 is upregulated in multiple cancers and is thought to drive cancer progression via dysregulation of polyamine synthesis and cell-cycle control. Although AZIN2 was originally thought to have no involvement in cancer progression, a recent study demonstrated otherwise.102 Given its role in cancers, therapeutic targeting of AZIN1-OAZ1 interactions using small molecules, peptides, or proteins, may prove to be a viable approach for cancer treatment, provided it is validated rigorously.50,77 The rationale for the preceding statement can be provided in two ways. 1) Because of its tight control over the polyamine biosynthetic pathway, AZIN1 regulates polyamine levels, thereby controlling cell-proliferation by sequestering OAZ1. Although targeting ODC using DFMO (an established ODC inhibitor) prevents intracellular polyamine synthesis, it does not prevent polyamine uptake from tumor microenvironments. On the contrary, inhibiting AZIN-OAZ1 interaction may likely free OAZ1, resulting in ODC degradation and decreased polyamine uptake. 2) AZIN1 also controls cell cycle progression by preventing OAZ1-mediated degradation of proteins associated with cell-cycle regulation. Therefore, inhibiting AZIN1-OAZ1 interaction may likely prevent AZIN1-mediated cell-proliferation.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi:10.1126/science.1160809
2. Vander Heiden MG, DeBerardinis RJ. Understanding the intersections between metabolism and cancer biology. Cell. 2017;168(4):657–669. doi:10.1016/j.cell.2016.12.039
3. Casero RA
4. Gerner EW, Meyskens FL
5. Dever TE, Ivanov IP. Roles of polyamines in translation. J Biol Chem. 2018;293(48):18719–18729. doi:10.1074/jbc.TM118.003338
6. Ivanov IP, Shin BS, Loughran G, et al. Polyamine control of translation elongation regulates start site selection on antizyme inhibitor mRNA via ribosome queuing. Mol Cell. 2018;70(2):254–264.e256. doi:10.1016/j.molcel.2018.03.015
7. Sánchez-Jiménez F, Medina M, Villalobos-Rueda L, Urdiales JL. Polyamines in mammalian pathophysiology. Cell Mol Life Sci. 2019;76(20):3987–4008.
8. Damiani E, Wallace HM. Polyamines and cancer. In: Alcázar R, Tiburcio AF, editors. Polyamines: Methods and Protocols. New York: Springer New York; 2018:469–488.
9. Celano P, Baylin SB, Casero RA
10. Frugier M, Florentz C, Hosseini MW, Lehn JM, Giege R. Synthetic polyamines stimulate in vitro transcription by T7 RNA polymerase. Nucleic Acids Res. 1994;22(14):2784–2790. doi:10.1093/nar/22.14.2784
11. Gerner EW, Russell DH. The relationship between polyamine accumulation and DNA replication in synchronized Chinese hamster ovary cells after heat shock. Cancer Res. 1977;37(2):482–489.
12. Yamashita T, Nishimura K, Saiki R, et al. Role of polyamines at the G1/S boundary and G2/M phase of the cell cycle. Int J Biochem Cell Biol. 2013;45(6):1042–1050. doi:10.1016/j.biocel.2013.02.021
13. Zabala-Letona A, Arruabarrena-Aristorena A, Martín-Martín N, et al. mTORC1-dependent AMD1 regulation sustains polyamine metabolism in prostate cancer. Nature. 2017;547(7661):109–113. doi:10.1038/nature22964
14. Bello-Fernandez C, Packham G, Cleveland JL. The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc Natl Acad Sci U S A. 1993;90(16):7804–7808. doi:10.1073/pnas.90.16.7804
15. Shin BS, Katoh T, Gutierrez E, Kim JR, Suga H, Dever TE. Amino acid substrates impose polyamine, eIF5A, or hypusine requirement for peptide synthesis. Nucleic Acids Res. 2017;45(14):8392–8402. doi:10.1093/nar/gkx532
16. Mathews MB, Hershey JW. The translation factor eIF5A and human cancer. Biochim Biophys Acta. 2015;1849(7):836–844. doi:10.1016/j.bbagrm.2015.05.002
17. Pegg AE. Regulation of ornithine decarboxylase. J Biol Chem. 2006;281(21):14529–14532. doi:10.1074/jbc.R500031200
18. Coffino P. Regulation of cellular polyamines by antizyme. Nat Rev Mol Cell Biol. 2001;2(3):188–194. doi:10.1038/35056508
19. Miller-Fleming L, Olin-Sandoval V, Campbell K, Ralser M. Remaining mysteries of molecular biology: the role of polyamines in the cell. J Mol Biol. 2015;427(21):3389–3406. doi:10.1016/j.jmb.2015.06.020
20. Rom E, Kahana C. Polyamines regulate the expression of ornithine decarboxylase antizyme in vitro by inducing ribosomal frame-shifting. Proc Natl Acad Sci U S A. 1994;91(9):3959–3963. doi:10.1073/pnas.91.9.3959
21. Matsufuji S, Matsufuji T, Miyazaki Y, et al. Autoregulatory frameshifting in decoding mammalian ornithine decarboxylase antizyme. Cell. 1995;80(1):51–60. doi:10.1016/0092-8674(95)90450-6
22. Mangold U. The antizyme family: polyamines and beyond. IUBMB Life. 2005;57(10):671–676. doi:10.1080/15216540500307031
23. Kahana C. The antizyme family for regulating polyamines. J Biol Chem. 2018;293(48):18730–18735. doi:10.1074/jbc.TM118.003339
24. Zhu C, Lang DW, Coffino P. Antizyme2 is a negative regulator of ornithine decarboxylase and polyamine transport. J Biol Chem. 1999;274(37):26425–26430. doi:10.1074/jbc.274.37.26425
25. Murai N, Murakami Y, Matsufuji S. Identification of nuclear export signals in antizyme-1. J Biol Chem. 2003;278(45):44791–44798. doi:10.1074/jbc.M308059200
26. Murai N, Shimizu A, Murakami Y, Matsufuji S. Subcellular localization and phosphorylation of antizyme 2. J Cell Biochem. 2009;108(4):1012–1021. doi:10.1002/jcb.22334
27. Ivanov IP, Rohrwasser A, Terreros DA, Gesteland RF, Atkins JF. Discovery of a spermatogenesis stage-specific ornithine decarboxylase antizyme: antizyme 3. Proc Natl Acad Sci U S A. 2000;97(9):4808–4813. doi:10.1073/pnas.070055897
28. Tosaka Y, Tanaka H, Yano Y, et al. Identification and characterization of testis specific ornithine decarboxylase antizyme (OAZ-t) gene: expression in haploid germ cells and polyamine-induced frameshifting. Genes Cells. 2000;5(4):265–276. doi:10.1046/j.1365-2443.2000.00324.x
29. Olsen RR, Zetter BR. Evidence of a role for antizyme and antizyme inhibitor as regulators of human cancer. Mol Cancer Res. 2011;9(10):1285–1293. doi:10.1158/1541-7786.MCR-11-0178
30. Mangold U, Leberer E. Regulation of all members of the antizyme family by antizyme inhibitor. Biochem J. 2005;385(Pt 1):21–28. doi:10.1042/BJ20040547
31. Mitchell JL, Judd GG, Bareyal-Leyser A, Ling SY. Feedback repression of polyamine transport is mediated by antizyme in mammalian tissue-culture cells. Biochem J. 1994;299(Pt 1):19–22. doi:10.1042/bj2990019
32. Hoshino K, Momiyama E, Yoshida K, et al. Polyamine transport by mammalian cells and mitochondria: role of antizyme and glycosaminoglycans. J Biol Chem. 2005;280(52):42801–42808. doi:10.1074/jbc.M505445200
33. Ramos-Molina B, Lopez-Contreras AJ, Lambertos A, Dardonville C, Cremades A, Penafiel R. Influence of ornithine decarboxylase antizymes and antizyme inhibitors on agmatine uptake by mammalian cells. Amino Acids. 2015;47(5):1025–1034. doi:10.1007/s00726-015-1931-3
34. Mitchell JL, Thane TK, Sequeira JM, Marton LJ, Thokala R. Antizyme and antizyme inhibitor activities influence cellular responses to polyamine analogs. Amino Acids. 2007;33(2):291–297. doi:10.1007/s00726-007-0523-2
35. Keren-Paz A, Bercovich Z, Porat Z, Erez O, Brener O, Kahana C. Overexpression of antizyme-inhibitor in NIH3T3 fibroblasts provides growth advantage through neutralization of antizyme functions. Oncogene. 2006;25(37):5163–5172. doi:10.1038/sj.onc.1209521
36. Lopez-Contreras AJ, Ramos-Molina B, Cremades A, Penafiel R. Antizyme inhibitor 2 (AZIN2/ODCp) stimulates polyamine uptake in mammalian cells. J Biol Chem. 2008;283(30):20761–20769. doi:10.1074/jbc.M801024200
37. Murakami Y, Ichiba T, Matsufuji S, Hayashi S. Cloning of antizyme inhibitor, a highly homologous protein to ornithine decarboxylase. J Biol Chem. 1996;271(7):3340–3342. doi:10.1074/jbc.271.7.3340
38. Ramos-Molina B, López-Contreras AJ, Cremades A, Peñafiel R. Differential expression of ornithine decarboxylase antizyme inhibitors and antizymes in rodent tissues and human cell lines. Amino Acids. 2012;42(2–3):539–547. doi:10.1007/s00726-011-1031-y
39. Murakami Y, Ohkido M, Takizawa H, Murai N, Matsufuji S. Multiple forms of mouse antizyme inhibitor 1 mRNA differentially regulated by polyamines. Amino Acids. 2014;46(3):575–583. doi:10.1007/s00726-013-1598-6
40. Murakami Y, Matsufuji S, Nishiyama M, Hayashi S. Properties and fluctuations in vivo of rat liver antizyme inhibitor. Biochem J. 1989;259(3):839–845. doi:10.1042/bj2590839
41. Bercovich Z, Kahana C. Degradation of antizyme inhibitor, an ornithine decarboxylase homologous protein, is ubiquitin-dependent and is inhibited by antizyme. J Biol Chem. 2004;279(52):54097–54102. doi:10.1074/jbc.M410234200
42. Li X, Coffino P. Degradation of ornithine decarboxylase: exposure of the C-terminal target by a polyamine-inducible inhibitory protein. Mol Cell Biol. 1993;13(4):2377–2383. doi:10.1128/MCB.13.4.2377
43. Snapir Z, Keren-Paz A, Bercovich Z, Kahana C. ODCp, a brain- and testis-specific ornithine decarboxylase paralogue, functions as an antizyme inhibitor, although less efficiently than AzI1. Biochem J. 2008;410(3):613–619. doi:10.1042/BJ20071423
44. Ramos-Molina B, Lambertos A, Lopez-Contreras AJ, et al. Structural and degradative aspects of ornithine decarboxylase antizyme inhibitor 2. FEBS Open Bio. 2014;4:510–521. doi:10.1016/j.fob.2014.05.004
45. Fujita K, Murakami Y, Hayashi S. A macromolecular inhibitor of the antizyme to ornithine decarboxylase. Biochem J. 1982;204(3):647–652. doi:10.1042/bj2040647
46. Kitani T, Fujisawa H. Purification and characterization of antizyme inhibitor of ornithine decarboxylase from rat liver. Biochim Biophys Acta. 1989;991(1):44–49. doi:10.1016/0304-4165(89)90026-3
47. Lambertos A, Ramos-Molina B, Cerezo D, López-Contreras AJ, Peñafiel R. The mouse Gm853 gene encodes a novel enzyme: leucine decarboxylase. Biochim Biophys Acta Gen Subj. 2018;1862(3):365–376. doi:10.1016/j.bbagen.2017.11.007
48. Wu HY, Chen SF, Hsieh JY, et al. Structural basis of antizyme-mediated regulation of polyamine homeostasis. Proc Natl Acad Sci U S A. 2015;112(36):11229–11234. doi:10.1073/pnas.1508187112
49. Liu YC, Liu YL, Su JY, Liu GY, Hung HC. Critical factors governing the difference in antizyme-binding affinities between human ornithine decarboxylase and antizyme inhibitor. PLoS One. 2011;6(4):e19253. doi:10.1371/journal.pone.0019253
50. Ghalali A, Rice JM, Kusztos A, Jernigan FE, Zetter BR, Rogers MS. Developing a novel FRET assay, targeting the binding between Antizyme-AZIN. Sci Rep. 2019;9(1):4632. doi:10.1038/s41598-019-40929-4
51. Hsieh JY, Yang JY, Lin CL, Liu GY, Hung HC. Minimal antizyme peptide fully functioning in the binding and inhibition of ornithine decarboxylase and antizyme inhibitor. PLoS One. 2011;6(9):e24366. doi:10.1371/journal.pone.0024366
52. López-Contreras AJ, López-Garcia C, Jiménez-Cervantes C, Cremades A, Peñafiel R. Mouse ornithine decarboxylase-like gene encodes an antizyme inhibitor devoid of ornithine and arginine decarboxylating activity. J Biol Chem. 2006;281(41):30896–30906. doi:10.1074/jbc.M602840200
53. Kanerva K, Mäkitie LT, Pelander A, Heiskala M, Andersson LC. Human ornithine decarboxylase paralogue (ODCp) is an antizyme inhibitor but not an arginine decarboxylase. Biochem J. 2008;409(1):187–192. doi:10.1042/BJ20071004
54. Tang H, Ariki K, Ohkido M, et al. Role of ornithine decarboxylase antizyme inhibitor in vivo. Genes Cells. 2009;14(1):79–87. doi:10.1111/j.1365-2443.2008.01249.x
55. Silva TM, Cirenajwis H, Wallace HM, Oredsson S, Persson L. A role for antizyme inhibitor in cell proliferation. Amino Acids. 2015;47(7):1341–1352. doi:10.1007/s00726-015-1957-6
56. Kim SW, Mangold U, Waghorne C, et al. Regulation of cell proliferation by the antizyme inhibitor: evidence for an antizyme-independent mechanism. J Cell Sci. 2006;119(Pt 12):2583–2591. doi:10.1242/jcs.02966
57. Murakami Y, Suzuki J, Samejima K, et al. The change of antizyme inhibitor expression and its possible role during mammalian cell cycle. Exp Cell Res. 2009;315(13):2301–2311. doi:10.1016/j.yexcr.2009.04.024
58. Ma R, Jiang D, Kang B, et al. Molecular cloning and mRNA expression analysis of antizyme inhibitor 1 in the ovarian follicles of the Sichuan white goose. Gene. 2015;568(1):55–60. doi:10.1016/j.gene.2015.05.014
59. Fernandes JRD, Jain S, Banerjee A. Expression of ODC1, SPD, SPM and AZIN1 in the hypothalamus, ovary and uterus during rat estrous cycle. Gen Comp Endocrinol. 2017;246:9–22. doi:10.1016/j.ygcen.2017.03.005
60. Greenwood MP, Greenwood M, Paton JF, Murphy D. Control of polyamine biosynthesis by antizyme inhibitor 1 is important for transcriptional regulation of arginine vasopressin in the male rat hypothalamus. Endocrinology. 2015;156(8):2905–2917. doi:10.1210/en.2015-1074
61. Juszczak GR, Stankiewicz AM. Glucocorticoids, genes and brain function. Prog Neuropsychopharmacol Biol Psychiatry. 2018;82:136–168. doi:10.1016/j.pnpbp.2017.11.020
62. López-Garcia C, Ramos-Molina B, Lambertos A, López-Contreras AJ, Cremades A, Peñafiel R. Antizyme inhibitor 2 hypomorphic mice. New patterns of expression in pancreas and adrenal glands suggest a role in secretory processes. PLoS One. 2013;8(7):e69188. doi:10.1371/journal.pone.0069188
63. Mäkitie LT, Kanerva K, Polvikoski T, Paetau A, Andersson LC. Brain neurons express ornithine decarboxylase-activating antizyme inhibitor 2 with accumulation in Alzheimer’s disease. Brain Pathol. 2010;20(3):571–580. doi:10.1111/j.1750-3639.2009.00334.x
64. López-Contreras AJ, Ramos-Molina B, Martínez-de-la-Torre M, et al. Expression of antizyme inhibitor 2 in male haploid germinal cells suggests a role in spermiogenesis. Int J Biochem Cell Biol. 2009;41(5):1070–1078. doi:10.1016/j.biocel.2008.09.029
65. Tokuhiro K, Isotani A, Yokota S, et al. OAZ-t/OAZ3 is essential for rigid connection of sperm tails to heads in mouse. PLoS Genet. 2009;5(11):e1000712. doi:10.1371/journal.pgen.1000712
66. Lambertos A, Ramos-Molina B, López-Contreras AJ, Cremades A, Peñafiel R. New insights of polyamine metabolism in testicular physiology: a role of ornithine decarboxylase antizyme inhibitor 2 (AZIN2) in the modulation of testosterone levels and sperm motility. PLoS One. 2018;13(12):e0209202. doi:10.1371/journal.pone.0209202
67. Zhang J, Wang Y, Zhou Y, Cao Z, Huang P, Lu B. Yeast two-hybrid screens imply that GGNBP1, GGNBP2 and OAZ3 are potential interaction partners of testicular germ cell-specific protein GGN1. FEBS Lett. 2005;579(2):559–566. doi:10.1016/j.febslet.2004.10.112
68. Ruan Y, Cheng M, Ou Y, Oko R, van der Hoorn FA. Ornithine decarboxylase antizyme Oaz3 modulates protein phosphatase activity. J Biol Chem. 2011;286(33):29417–29427. doi:10.1074/jbc.M111.274647
69. Kanerva K, Makitie LT, Back N, Andersson LC. Ornithine decarboxylase antizyme inhibitor 2 regulates intracellular vesicle trafficking. Exp Cell Res. 2010;316(11):1896–1906. doi:10.1016/j.yexcr.2010.02.021
70. Kanerva K, Lappalainen J, Mäkitie LT, Virolainen S, Kovanen PT, Andersson LC. Expression of antizyme inhibitor 2 in mast cells and role of polyamines as selective regulators of serotonin secretion. PLoS One. 2009;4(8):e6858. doi:10.1371/journal.pone.0006858
71. Acosta-Andrade C, Lambertos A, Urdiales JL, Sanchez-Jimenez F, Penafiel R, Fajardo I. A novel role for antizyme inhibitor 2 as a regulator of serotonin and histamine biosynthesis and content in mouse mast cells. Amino Acids. 2016;48(10):2411–2421. doi:10.1007/s00726-016-2230-3
72. Jung MH, Kim SC, Jeon GA, et al. Identification of differentially expressed genes in normal and tumor human gastric tissue. Genomics. 2000;69(3):281–286. doi:10.1006/geno.2000.6338
73. Schaner ME, Ross DT, Ciaravino G, et al. Gene expression patterns in ovarian carcinomas. Mol Biol Cell. 2003;14(11):4376–4386. doi:10.1091/mbc.e03-05-0279
74. van Duin M, van Marion R, Vissers K, et al. High-resolution array comparative genomic hybridization of chromosome arm 8q: evaluation of genetic progression markers for prostate cancer. Genes Chromosomes Cancer. 2005;44(4):438–449. doi:10.1002/gcc.20259
75. Peng L, Guo J, Zhang Z, et al. A candidate gene study for the association of host single nucleotide polymorphisms with liver cirrhosis risk in chinese hepatitis B patients. Genet Test Mol Biomarkers. 2013;17(9):681–686. doi:10.1089/gtmb.2013.0058
76. Choi KS, Suh YH, Kim WH, Lee TH, Jung MH. Stable siRNA-mediated silencing of antizyme inhibitor: regulation of ornithine decarboxylase activity. Biochem Biophys Res Commun. 2005;328(1):206–212. doi:10.1016/j.bbrc.2004.11.172
77. Olsen RR, Chung I, Zetter BR. Knockdown of antizyme inhibitor decreases prostate tumor growth in vivo. Amino Acids. 2012;42(2–3):549–558. doi:10.1007/s00726-011-1032-x
78. Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817–846. doi:10.1146/annurev.biochem.71.110601.135501
79. Chen L, Li Y, Lin CH, et al. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat Med. 2013;19(2):209–216. doi:10.1038/nm.3043
80. Qin YR, Qiao JJ, Chan TH, et al. Adenosine-to-inosine RNA editing mediated by ADARs in esophageal squamous cell carcinoma. Cancer Res. 2014;74(3):840–851. doi:10.1158/0008-5472.CAN-13-2545
81. Hu X, Chen J, Shi X, et al. RNA editing of AZIN1 induces the malignant progression of non-small-cell lung cancers. Tumour Biol. 2017;39(8):1010428317700001. doi:10.1177/1010428317700001
82. Shigeyasu K, Okugawa Y, Toden S, et al. AZIN1 RNA editing confers cancer stemness and enhances oncogenic potential in colorectal cancer. JCI Insight. 2018;3(12). doi:10.1172/jci.insight.99976.
83. Takeda S, Shigeyasu K, Okugawa Y, et al. Activation of AZIN1 RNA editing is a novel mechanism that promotes invasive potential of cancer-associated fibroblasts in colorectal cancer. Cancer Lett. 2019;444:127–135. doi:10.1016/j.canlet.2018.12.009
84. Okugawa Y, Toiyama Y, Shigeyasu K, et al. Enhanced AZIN1 RNA editing and overexpression of its regulatory enzyme ADAR1 are important prognostic biomarkers in gastric cancer. J Transl Med. 2018;16(1):366. doi:10.1186/s12967-018-1740-z
85. Ghalali A, Rice JM, Wang L, Wu CL, Rogers MS, Zetter B Nuclear localization of antizyme inhibitor may be a marker for aggressiveness of prostate cancer. AACR; 2019.
86. Casimiro MC, Velasco-Velázquez M, Aguirre-Alvarado C, Pestell RG. Overview of cyclins D1 function in cancer and the CDK inhibitor landscape: past and present. Expert Opin Investig Drugs. 2014;23(3):295–304. doi:10.1517/13543784.2014.867017
87. Finn RS, Aleshin A, Slamon DJ. Targeting the cyclin-dependent kinases (CDK) 4/6 in estrogen receptor-positive breast cancers. Breast Cancer Res. 2016;18(1):17. doi:10.1186/s13058-015-0661-5
88. Newman RM, Mobascher A, Mangold U, et al. Antizyme targets cyclin D1 for degradation. A novel mechanism for cell growth repression. J Biol Chem. 2004;279(40):41504–41511. doi:10.1074/jbc.M407349200
89. Liu YC, Lee CY, Lin CL, Chen HY, Liu GY, Hung HC. Multifaceted interactions and regulation between antizyme and its interacting proteins cyclin D1, ornithine decarboxylase and antizyme inhibitor. Oncotarget. 2015;6(27):23917–23929. doi:10.18632/oncotarget.4469
90. Fong LY, Feith DJ, Pegg AE. Antizyme overexpression in transgenic mice reduces cell proliferation, increases apoptosis, and reduces N-nitrosomethylbenzylamine-induced forestomach carcinogenesis. Cancer Res. 2003;63(14):3945–3954.
91. Sun Y, Bao X, Ren Y, et al. Targeting HDAC/OAZ1 axis with a novel inhibitor effectively reverses cisplatin resistance in non-small cell lung cancer. Cell Death Dis. 2019;10(6):400. doi:10.1038/s41419-019-1597-y
92. Dulloo I, Gopalan G, Melino G, Sabapathy K. The antiapoptotic DeltaNp73 is degraded in a c-Jun-dependent manner upon genotoxic stress through the antizyme-mediated pathway. Proc Natl Acad Sci U S A. 2010;107(11):4902–4907. doi:10.1073/pnas.0906782107
93. Ishimoto O, Kawahara C, Enjo K, Obinata M, Nukiwa T, Ikawa S. Possible oncogenic potential of DeltaNp73: a newly identified isoform of human p73. Cancer Res. 2002;62(3):636–641.
94. Belloni L, Moretti F, Merlo P, et al. DNp73alpha protects myogenic cells from apoptosis. Oncogene. 2006;25(25):3606–3612. doi:10.1038/sj.onc.1209321
95. Mangold U, Hayakawa H, Coughlin M, Münger K, Zetter BR. Antizyme, a mediator of ubiquitin-independent proteasomal degradation and its inhibitor localize to centrosomes and modulate centriole amplification. Oncogene. 2008;27(5):604–613. doi:10.1038/sj.onc.1210685
96. Meraldi P, Nigg EA. The centrosome cycle. FEBS Lett. 2002;521(1–3):9–13. doi:10.1016/S0014-5793(02)02865-X
97. Gruendler C, Lin Y, Farley J, Wang T. Proteasomal degradation of Smad1 induced by bone morphogenetic proteins. J Biol Chem. 2001;276(49):46533–46543. doi:10.1074/jbc.M105500200
98. Lim SK, Gopalan G. Antizyme1 mediates AURKAIP1-dependent degradation of Aurora-A. Oncogene. 2007;26(46):6593–6603. doi:10.1038/sj.onc.1210482
99. Kasbek C, Yang CH, Fisk HA. Antizyme restrains centrosome amplification by regulating the accumulation of Mps1 at centrosomes. Mol Biol Cell. 2010;21(22):3878–3889. doi:10.1091/mbc.e10-04-0281
100. Xiong A, Zhang Y, Chen X, et al. Ornithine decarboxylase antizyme 1 upregulate LOR to promote differentiation of SCC15 cells by binding CBP/p300 in promoter region. Int J Clin Exp Med. 2016;9(2):2359–2366.
101. Mäkitie LT, Kanerva K, Sankila A, Andersson LC. High expression of antizyme inhibitor 2, an activator of ornithine decarboxylase in steroidogenic cells of human gonads. Histochem Cell Biol. 2009;132(6):633–638. doi:10.1007/s00418-009-0636-7
102. Kaprio T, Rasila T, Hagström J, et al. Ornithine decarboxylase antizyme inhibitor 2 (AZIN2) is a signature of secretory phenotype and independent predictor of adverse prognosis in colorectal cancer. PLoS One. 2019;14(2):e0211564. doi:10.1371/journal.pone.0211564
103. Kaprio T, Hagström J, Andersson LC, Haglund C. Tetraspanin CD63 independently predicts poor prognosis in colorectal cancer. Histol Histopathol. 2020;35:18209. doi:10.14670/HH-18-209
104. Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–D368. doi:10.1093/nar/gkw937
105. Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34(Databaseissue):D535–D539. doi:10.1093/nar/gkj109
106. Orchard S, Ammari M, Aranda B, et al. The MIntAct project–IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014;42(Databaseissue):D358–D363. doi:10.1093/nar/gkt1115
107. Wang J, Huo K, Ma L, et al. Toward an understanding of the protein interaction network of the human liver. Mol Syst Biol. 2011;7:536. doi:10.1038/msb.2011.67
108. Cervenkova L, Vycital O, Bruha J, et al. Protein expression of ABCC2 and SLC22A3 associates with prognosis of pancreatic adenocarcinoma. Sci Rep. 2019;9(1):19782. doi:10.1038/s41598-019-56059-w
109. Li Z, Xing X, Shan F, et al. ABCC2-24C > T polymorphism is associated with the response to platinum/5-Fu-based neoadjuvant chemotherapy and better clinical outcomes in advanced gastric cancer patients. Oncotarget. 2016;7(34):55449–55457. doi:10.18632/oncotarget.10961
110. Benoit YD, Mitchell RR, Risueño RM, et al. Sam68 allows selective targeting of human cancer stem cells. Cell Chem Biol. 2017;24(7):833–844.e839. doi:10.1016/j.chembiol.2017.05.026
111. Sumithra B, Saxena U, Das AB. A comprehensive study on genome-wide coexpression network of KHDRBS1/Sam68 reveals its cancer and patient-specific association. Sci Rep. 2019;9(1):11083. doi:10.1038/s41598-019-47558-x
112. Daakour S, Hajingabo LJ, Kerselidou D, et al. Systematic interactome mapping of acute lymphoblastic leukemia cancer gene products reveals EXT-1 tumor suppressor as a Notch1 and FBWX7 common interactor. BMC Cancer. 2016;16:335. doi:10.1186/s12885-016-2374-2
113. Monzon J, Liu L, Brill H, et al. CDKN2A mutations in multiple primary melanomas. N Engl J Med. 1998;338(13):879–887. doi:10.1056/NEJM199803263381305
114. Reuter TY, Medhurst AL, Waisfisz Q, et al. Yeast two-hybrid screens imply involvement of Fanconi anemia proteins in transcription regulation, cell signaling, oxidative metabolism, and cellular transport. Exp Cell Res. 2003;289(2):211–221. doi:10.1016/S0014-4827(03)00261-1
115. Huttlin EL, Ting L, Bruckner RJ, et al. The BioPlex network: a systematic exploration of the human interactome. Cell. 2015;162(2):425–440. doi:10.1016/j.cell.2015.06.043
116. Huttlin EL, Bruckner RJ, Paulo JA, et al. Architecture of the human interactome defines protein communities and disease networks. Nature. 2017;545(7655):505–509. doi:10.1038/nature22366
117. Hein MY, Hubner NC, Poser I, et al. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell. 2015;163(3):712–723. doi:10.1016/j.cell.2015.09.053
118. Ewing RM, Chu P, Elisma F, et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol. 2007;3:89. doi:10.1038/msb4100134
119. Rolland T, Taşan M, Charloteaux B, et al. A proteome-scale map of the human interactome network. Cell. 2014;159(5):1212–1226. doi:10.1016/j.cell.2014.10.050
120. Luck K, Kim DK, Lambourne L, et al. A reference map of the human binary protein interactome. Nature. 2020;580(7803):402–408. doi:10.1038/s41586-020-2188-x
121. Hirokawa N, Noda Y, Tanaka Y, Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol. 2009;10(10):682–696. doi:10.1038/nrm2774
122. Kidane D, Sakkas D, Nottoli T, McGrath J, Sweasy JB. Kinesin 5B (KIF5B) is required for progression through female meiosis and proper chromosomal segregation in mitotic cells. PLoS One. 2013;8(4):e58585. doi:10.1371/journal.pone.0058585
123. Abdelmohsen K, Srikantan S, Yang X, et al. Ubiquitin-mediated proteolysis of HuR by heat shock. EMBO J. 2009;28(9):1271–1282. doi:10.1038/emboj.2009.67
124. Rybakin V, Rastetter RH, Stumpf M, et al. Molecular mechanism underlying the association of Coronin-7 with Golgi membranes. Cell Mol Life Sci. 2008;65(15):2419–2430. doi:10.1007/s00018-008-8278-9
125. López-Contreras AJ, Sánchez-Laorden BL, Ramos-Molina B, de la Morena ME, Cremades A, Peñafiel R. Subcellular localization of antizyme inhibitor 2 in mammalian cells: influence of intrinsic sequences and interaction with antizymes. J Cell Biochem. 2009;107(4):732–740. doi:10.1002/jcb.22168
126. Marchese FP, Grossi E, Marín-Béjar O, et al. A long noncoding RNA regulates sister chromatid cohesion. Mol Cell. 2016;63(3):397–407. doi:10.1016/j.molcel.2016.06.031
127. Boerkoel CF, Takashima H, John J, et al. Mutant chromatin remodeling protein SMARCAL1 causes Schimke immuno-osseous dysplasia. Nat Genet. 2002;30(2):215–220. doi:10.1038/ng821
128. Akram S, Yang F, Li J, et al. LRIF1 interacts with HP1α to coordinate accurate chromosome segregation during mitosis. J Mol Cell Biol. 2018;10(6):527–538. doi:10.1093/jmcb/mjy040
129. Kurosaki T, Maquat LE. Nonsense-mediated mRNA decay in humans at a glance. J Cell Sci. 2016;129(3):461–467. doi:10.1242/jcs.181008
130. Yamashita A, Izumi N, Kashima I, et al. SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev. 2009;23(9):1091–1105. doi:10.1101/gad.1767209
131. Hyvönen MT, Uimari A, Keinänen TA, et al. Polyamine-regulated unproductive splicing and translation of spermidine/spermine N1-acetyltransferase. Rna. 2006;12(8):1569–1582. doi:10.1261/rna.39806
132. Epping MT, Hart AA, Glas AM, Krijgsman O, Bernards R. PRAME expression and clinical outcome of breast cancer. Br J Cancer. 2008;99(3):398–403. doi:10.1038/sj.bjc.6604494
133. Zhu VW, Upadhyay D, Schrock AB, Gowen K, Ali SM, Ou SH. TPD52L1-ROS1, a new ROS1 fusion variant in lung adenosquamous cell carcinoma identified by comprehensive genomic profiling. Lung Cancer. 2016;97:48–50. doi:10.1016/j.lungcan.2016.04.013
134. Boutros R, Byrne JA. D53 (TPD52L1) is a cell cycle-regulated protein maximally expressed at the G2-M transition in breast cancer cells. Exp Cell Res. 2005;310(1):152–165. doi:10.1016/j.yexcr.2005.07.009
135. Li X, Sun Y, Huang S, et al. Inhibition of AZIN2-sv induces neovascularization and improves prognosis after myocardial infarction by blocking ubiquitin-dependent talin1 degradation and activating the Akt pathway. EBioMedicine. 2019;39:69–82. doi:10.1016/j.ebiom.2018.12.001
136. Monkley SJ, Kostourou V, Spence L, et al. Endothelial cell talin1 is essential for embryonic angiogenesis. Dev Biol. 2011;349(2):494–502. doi:10.1016/j.ydbio.2010.11.010
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.