Back to Journals » OncoTargets and Therapy » Volume 13
RNF128 Promotes Malignant Behaviors via EGFR/MEK/ERK Pathway in Hepatocellular Carcinoma
Authors Bai XS, Zhang C, Peng R, Jiang GQ , Jin SJ, Wang Q, Ke AW , Bai DS
Received 6 July 2020
Accepted for publication 28 August 2020
Published 9 October 2020 Volume 2020:13 Pages 10129—10141
DOI https://doi.org/10.2147/OTT.S269606
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Arseniy Yuzhalin
Xue-Song Bai,1,* Chi Zhang,2,* Rui Peng,2,* Guo-Qing Jiang,2 Sheng-Jie Jin,2 Qian Wang,2 Ai-Wu Ke,3 Dou-Sheng Bai2
1The First Clinical Medical College, Dalian Medical University, Dalian, Liaoning 116044, People’s Republic of China; 2Department of Hepatobiliary Surgery, Clinical Medical College, Yangzhou University, Yangzhou 225009, Jiangsu, People’s Republic of China; 3Liver Cancer Institute, Zhongshan Hospital, Fudan University, Shanghai 200032, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Dou-Sheng Bai
Clinical Medical College, Yangzhou University, 98 West Nantong Road, Yangzhou, Jiangsu 225000, People’s Republic of China
Tel +86-514-87373385
Fax +86-514-87990188
Email [email protected]
Background: The ubiquitin-proteasome system participates in the pathogenesis and progression of hepatocellular carcinoma (HCC). As an E3 ubiquitin ligase, RNF128 has been proved vital in carcinogenesis, whereas, little is known about the oncogenic mechanisms of RNF128 in HCC.
Materials and Methods: Through tissue microarray from HCC patients, we analyzed RNF128 expression and its relationship with clinical outcomes in HCC. Western blot and quantitative realtime polymerase chain reaction (qRT-PCR) were performed to examine expression levels of RNF128 in HCC tissues and cell lines. Effects of RNF128 on HCC cellular biological functions and the potential mechanism were evaluated through knockdown and overexpression assays in vitro and in vivo methods.
Results: RNF128 expression was found to be remarkably elevated in HCC tissues compared with adjacent normal tissues. Furthermore, the overexpression of RNF128 enhanced hepatoma cells proliferation, colony formation, migration, invasion, and apoptotic resistance both in vitro and in vivo. Mechanistically, RNF128 activated EGFR/MEK/ERK signaling pathway and the EGFR inhibitor, gefitinib partially reversed RNF128-enhanced proliferation, invasion, and migration in hepatoma cells.
Conclusion: RNF128 promotes HCC progression by activating EGFR/MEK/ERK signaling pathway, which might function as a novel prognostic molecular signature with the potential to be a candidate therapeutic target for HCC patients.
Keywords: hepatocellular carcinoma, RNF128, ubiquitination, EGFR/MEK/ERK signaling pathway, prognosis
Introduction
As a predominant type of primary liver cancer, hepatocellular carcinoma (HCC) mainly originates from liver injury and inflammation triggered by various risk factors, including hepatitis virus infection, obesity, chronic alcohol consumption, and exposure to aflatoxin.1,2 Due to the high recurrence, early invasion and metastasis, the number of deaths attributed to HCC are the third highest among all cancers worldwide.3,4 Currently, the molecular mechanisms underlying HCC have not been elucidated and clinical methods for early diagnosis and treatments are lacking. Despite recent advances in targeted therapies for HCC, the benefits for HCC patients are still limited.5 Therefore, profound comprehension of the underlying molecular mechanism linked to HCC development is urgently needed for the identification of diagnosis and treatment targets of HCC.
The ubiquitin-proteasome system (UPS) plays a pivotal function in protein post-translational modification and is a modulator of the physiological, as well as pathological processes in eukaryotes. E3 ubiquitin ligases are the core components of the system because of their specific recognition for substrates.6,7 The RING finger (RNF) protein family is a complex set of E3 ubiquitin ligase that possess an RNF domain. Recently, RING finger protein family members have been proved implicated in tumorigenesis and tumor progression.8 Overexpression of RNF38 enhances HCC malignant features by facilitating TGF-β signaling pathway and is associated with poor prognosis.9 TRAIP, another RING-type E3 ubiquitin ligase, promotes HCC progression by regulating the expression of cell cycle related protein to affect G1/S transition.10 Therefore, further studies into the role of RNF family members in tumor invasion and metastasis are advocated.
RNF128, also known as gene related to anergy in lymphocytes (GRAIL), is a RING finger protein family E3 ubiquitin ligase that participates in T cell anergy induction. Deficiency of RNF128 promotes anti-tumor immune response of CD8+ T cells.11 Besides that, RNF128 interacts with p53 and activates EGFR/MAPK/MMP-2 pathway, promoting the invasiveness and migratory ability of esophageal squamous cell carcinoma.12 In melanoma, however, RNF128 regulates Wnt/β-catenin signaling to suppress EMT and stemness through a mechanism involving CD44 ubiquitination.13 RNF128 downregulation is associated with poor prognosis in upper urinary tract and bladder cancer,14 whereas there is a lack of research about the role of RNF128 in HCC.
Herein, we aimed to detect the expression of RNF128 in HCC tissues and evaluate its relationship with prognosis. We found that RNF128 expression was markedly higher in tumor in contrast to peritumor tissues, which was correlated with a poor clinical outcome. Furthermore, RNF128 promoted the invasiveness and metastatic potential of HCC in vivo and in vitro by activating EGFR/MEK/ERK signaling cascade.
Methods and Materials
Patients and Samples
Twenty-eight pairs of frozen HCC tissues and corresponding normal adjacent tissues were collected from Clinical Medical College, Yangzhou University for quantitative realtime polymerase chain reaction (qRT-PCR), and 16 tissue pairs were selected for Western blotting. One hundred and seventy-one pairs of paraffin-embedded HCC tissues and matched normal adjacent tissues were collected from HCC patients underwent complete surgical excision from January 2006 to December 2008 at Zhongshan Hospital Fudan University (Shanghai, China). HCC diagnosis was independently confirmed by 2 clinical pathologists. Corresponding clinicopathological information was gathered for follow-up statistical analysis. All subjects provided a written agreement to participate in the study and were fully informed of the details of the study. Approval for the study was provided by the ethics committee of Clinical Medical College, Yangzhou University and Zhongshan Hospital Research Ethics Committee, Fudan University. The research conformed to the guidelines of the Declaration of Helsinki.
Tissue Microarray
Tissue microarrays (TMA) were established as follows: Histological examination of HCC samples was performed by H&E staining to select sites that were not necrotic or hemorrhagic selected on paraffin blocks. In each case, 1-mm-diameter repeated punches were taken from two distinct sites, the central part of the tumor and the closest non-cancerous margin (referred to as intratumor and peritumor, respectively, 4 holes in total) to ensure homogenous staining and reproducibility (Shanghai Biochip Co., Ltd., Shanghai, China). In this way, we designed 4 tissue microarray blocks, with 171 cylinders in each block. These blocks (4µm thick) were mounted on glass slides coated with 3-aminopropyltriethoxysilane.
Immunohistochemistry (IHC)
Overnight dewaxing of the slides was performed by heating them at 60°C and then rinsed twice with xylene, 10 minutes each. Tissues were rehydrated by rinsing for 5 minutes in 95%, 80%, and 75% ethanol in distilled water. The tissues were treated with 3% hydrogen peroxide for 20 minutes to inactivate endogenous peroxidase. Samples were heated in EDTA (pH 8.0) (Absin Biotechnology Co., Ltd. Shanghai, China) at 95°C for 15 minutes for antigen recovery. They were then blocked with universal blocking serum for 1 hour and incubated with anti-RNF128 (Abcam, Cat. No. ab72,533) at 1:200, at 4°C overnight. Then, the samples were incubated with biotin-labeled secondary antibody and streptavidin-peroxidase (Goldenbridge Biotechnology, Zhongshan, China) for 30 minutes. Signal was developed using diaminobenzidine and tissues counterstained with hematoxylin. Samples were then dehydrated according to standard procedures and sealed with coverslips. They were then examined on a Leica QWin Plus v3 software (Leica Microsystems Imaging Solutions). Image J pro plus was used to analyze immunohistochemical staining intensity. Expression was considered high if staining intensity was greater than the median, and low if the staining intensity was less than or equal to the median.
Cell Lines
The human HCC cell lines MHCC97-H, MHCC97-L, HepG2, PLC/PRF/5, and HCCLM3 were obtained from the cell bank of the Chinese Academy of Sciences (Shanghai, China). All cells were cultured in DMEM supplemented with 10% FBS (Gibco, Thermo Fisher Scientific, USA) and 1% pen/strep (Beyotime Institute of Biotechnology, Shanghai, China). Cells were cultured at 37°C, 5% CO2, in a humified incubator.
Lentivirus Infection
All lentiviral vectors were bought from Shanghai Genomeditech Company (Shanghai, China). MHCC97-L and MHCC97-H cells were transfected with RNF128 shRNA lentiviral vectors and its control vector, and PLC/PRF/5 and HepG2 cells with RNF128 cDNA lentiviral vector and its control vector. RNF128 shRNA target sequences were: shRNA1, 5ʹ-TCTTAACGTGCAACCATATTT-3ʹ; shRNA2, 5ʹ-GCAGTGGATGTTATTCCTCAT-3ʹ; shRNA3, 5ʹ-GCAGTAGACATTGTTGCAATC-3ʹ. Antibiotic-resistant transfected cells were selected by treating with puromycin for 3 days.
Western Blot
Protein samples were obtained from the tissues using cell lysis buffer (Beyotime Institute of Biotechnology, Cat. No. P0013). Proteins were separated on 10% SDS PAGE (Beyotime Institute of Biotechnology, Shanghai, China). Protein was transferred onto 0.2µM polyvinylidene fluoride membranes (Merck-millipore, Darmstadt, Germany) and blocked with 5% skim milk in 0.1% TBST at room temperature for 1 hour. The membranes were then incubated with the following primary antibodies at 4°C for 12 hours: anti-RNF128 (Abcam, Cat. No. 72533) at 1:1000, anti-EGFR (Cell signaling, Cat. No. #2085) at 1:1000, anti-p-EGFR (Cell signaling, Cat. No. #3777) at 1:1000, anti-MEK (Cell signaling, Cat. No. # 9126) at 1:1000, anti-p-MEK (Cell signaling, Cat. No. #9154) at 1:1000, anti-ERK (Cell signaling, Cat. No. #4695) at 1:1000, anti-p-ERK (Cell signaling, Cat. No. #4370) at 1:1000, and anti-GAPDH (Cell signaling, Cat. No. #5174) at 1:1000. Membranes were then rinsed and incubated with secondary antibody (Cell signaling, Cat. No. #7074) at 1:2000, at room temperature for 2 hours. Signal was developed by enhanced Chemiluminescent Substrate (NCM Biotech, Suzhou, China), examined on a Biorad Chemidoc XRS Gel Imaging System and analyzed using ImageJ.
qRT-PCR
RNA was extracted using TRIzol reagent (Invitrogen, USA). cDNA was synthesized using prime Script RT kit (TaKaRa, Japan). qRT-PCR was performed with SYBR Green Realtime PCR Master Mix (Yeasen, Shanghai, China) using the following PCR primers: RNF128, fwd: 5ʹ-CTGCTCGAAGGCTACGGAATG-3′ and rvs: 5ʹ-GTGTGCGTAGTTGAAGCCTTCC-3ʹ. GAPDH, fwd: 5ʹ-GTCTCCTCTGACTTCAACAGCG-3ʹ and rvs: 5ʹ-ACCACCCTGTTGCTGTAGCCAA-3ʹ.
Cell Counting Kit-8 (CCK8) and Colony Formation Assay
In the CCK8 test, 2 × 103 cells per well were cultured in 96-well plate and incubated with CCK8 reagent for 2 hours. Absorbance was read at 450nm using an automatic plate reader.
In the colony formation assay, 1 × 103 cells per well were cultured in 6-well plates for 2 weeks. This was followed by fixation with 4% PFA and staining with 0.1% crystal violet. Colonies were counted under a microscope.
Wound-Healing Assays and Invasion Assays
In the wound-healing test, a sterile 200μL pipette tip was used to create a linear wound by scratching the surface of HCC cells. Wound healing was then examined under a microscope and imaged at appropriate times. Image J was used to calculate average area of cell migration.
In the cell transwell test, cells were seeded into 8-μm pore size, 24-well transwell plates, pre-coated with matrix gel for invasion assays and left uncoated for migration assays. 5×104 cells suspending in 200μL of DMEM were seeded into the upper chamber and 600μL DMEM+10% FBS added into the lower chamber. The cells were cultured for 48h, fixed with 4% PFA and stained with crystal violet. Invading and migrating cells were counted and imaged under a microscope. Results of 3 independent experiments were presented a mean ± SD.
Flow Cytometry
Flow cytometry was used to assess apoptosis. Cells were collected and stained with annexin V-FITC/PI. The upper right quadrant indicates annexin V/PI positive, hence late apoptotic cells whereas the lower right quadrant indicates annexin V positive, hence early apoptotic cells. Both quadrants are used to assess level of apoptosis among groups.
Animal Model
Four weeks old nude mice were obtained from the Translational Medical Center of Yangzhou University and housed in a pathogen-free room. 5×106 HCC cells were subcutaneously xenografted into the mice. The growth of tumors was measured at intervals of 3 days post-injection until 4 weeks. The animal experiments were approved by the Animal Experimentation Ethics Committee of Yangzhou University. All experiments were conducted followed the NIH Guide for the Care and Use of Laboratory Animals and Department of Science and Technology of Jiangsu Province Guidelines on the Treatment of Laboratory Animals.
Statistical Analysis
SPSS software 22.0 was used for statistical analysis. Differences of quantitative data between 2 groups were compared with Student’s t-test. The relationship between 2 categorical variables was assessed by the Fisher’s exact test and Pearson’s correlation coefficient. The Kaplan–Meier method and the Log rank test were utilized to analyze overall survival and cumulative recurrence rate. Cox proportional hazard regression was used to investigate independent prognostic factors. P values < 0.05 were considered statistically significant.
Results
RNF128 is Highly Expressed in HCC and Signifies Poor HCC Prognosis
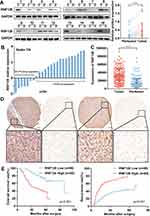
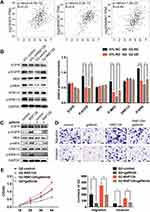
First, we assessed RNF128 protein and mRNA levels in the 24 pairs of HCC and paired adjacent normal tissues by Western blot and qRT-PCR. Results showed that RNF128 protein levels were elevated in HCC tissues relative to adjacent normal tissues (Figure 1A, p<0.001). Consistently, RNF128 mRNA levels were elevated in 17 samples of HCC tissues, accounting for 70.83% of all, compared with corresponding peritumor normal tissues (Figure 1B). IHC analysis on a tissue microarray slide of samples from 171 HCC patients was performed to further identify that RNF128 was elevated in tumor tissues relative to matched peritumor normal tissues (Figure 1C and D, p<0.001).
Additionally, we evaluated association between RNF128 expression and clinicopathological features of 171 HCC patients. We found RNF128 expression levels were significantly correlated with tumor size (p=0.011), Edmondson–Steiner Grade (p=0.002), and advanced TNM stage (p=0.003, Table 1). Kaplan-Meier analysis revealed that HCC patients with high RNF128 expression (n=85) tended to herald a shorter survival time and a higher recurrence rate compared with low RNF128 expression (n=86, Figure 1E, p<0.001). Univariate and multivariate analyses showed that RNF128 expression level is an independent risk factor for HCC prognosis (Table 2).
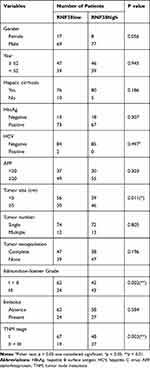 |
Table 1 Correlation Between RNF128 and Clinicopathological Characteristics in 171 HCC Patients |
 |
Table 2 Univariate and Multivariate Analyses of Factors Associated with Overall Survival and Cumulative Recurrence |
High Levels of RNF128 Promote HCC Cells Proliferation
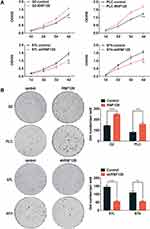
To further investigate the impact of RNF128 on the development of HCC, we detected RNF128 expression in different HCC cell lines (Supplementary Figure 1A). Next, we used shRNA to knock down RNF128 in MHCC97-L and MHCC97-H cells, which express high level of RNF128. Reciprocally, RNF128 was overexpressed in HepG2 and PLC/PRF/5 cell as they express low level of RNF128. The transfection efficiency of lentiviral infection was successfully verified by Western blot (Supplementary Figure 1B).
Then, we preformed CCK8 tests and colony formation assays to examine the impact of RNF128 on proliferative potential of HCC cells. CCK8 test revealed that overexpression of RNF128 promoted HepG2 and PLC/PRF/5 cells proliferation (Figure 2A, p=0.0015, p=0.0007), while its knockdown significantly suppressed MHCC97-L and MHCC97-H cell proliferation (p=0.0033, p=0.0031). Same trends were observed in the colony formation assay, the colony formation of MHCC97-H and MHCC97-L cells in the shRNF128 group were significantly lower than control group (Figure 2B, p=0.0003, p=0.0029), while the RNF128 overexpression significantly increased colony formation of HepG2 and PLC/PRF/5 cells (p<0.0001, p=0.0006). These results indicated that RNF128 might be critical for promoting cell proliferation and tumor growth.
High Levels of RNF128 Promote HCC Cells Migration and Invasion
To explore the impact of RNF128 on invasion and motility of HCC, transwell and wound-healing assays were constructed. In the transwell assay, PLC/PRF/5 and HepG2 cells in the overexpressed RNF128 group had more cells passing through the wells than control group, while MHCC97-H and MHCC97-L cells in the RNF128 knockdown group had less cells passing through both before and after matrix gel addition (Figure 3A). Results about the HCC cells mobility in wound-healing assay showed an alike consequence (Figure 3B). These data indicated that RNF128 enhanced migration and invasion of HCC cells.
High Levels of RNF128 Inhibit Apoptosis of HCC Cells
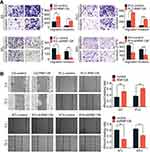
Next, flow cytometry was employed to evaluate whether RNF128 has an impact on anti-apoptotic capacity of HCC cells. In HepG2 cells, the percentage of annexin V positive cells upon RNF128 group vs controls was 4.13±0.31% and 11.38±0.51%, respectively (Figure 4A, p=0.0021). In PLC/PRF/5 cells, the percentage of annexin V positive cells upon RNF128 group vs controls was 14.45±0.59% and 17.88±0.34%, respectively (Figure 4A, p<0.001). For the shRNF128 group vs corresponding control group, the percentage of annexin V positive cells was 11.67±0.57% vs 7.50±0.45% and 7.06±0.86% vs 3.75±0.63% in MHCC97-L cells and MHCC97-H cells (Figure 4B, p=0.0013, p=0.0118), respectively, indicating that RNF128 remarkably inhibited the apoptosis of HCC cells.
High Levels of RNF128 Promote HCC Progression in Nude Mice
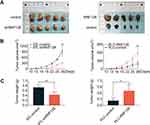
A subcutaneous xenograft model was constructed to confirm whether the oncogenic effects of RNF128 are consistent with the malignant behaviors of HCC in vivo. To this end, MHCC97-L-shRNF128 cells, PLC/PRF/5-RNF128 cells and corresponding control transfected cells were injected via subcutaneous route into BALB/c nude mice. Tumor volumes were then measured at intervals of three days for two weeks. Consistent with the observations in vitro, RNF128 was found to be involved in modulating transplanted tumor growth in nude mice (Figure 5A). Compared with MHCC97-L-control cells (943.15±252.96mm3), MHCC97-L-shRNF128 cells (307.57±75.92mm3) exhibited lower proliferation ability with 67% reduction in volume and 42% in weight (Figure 5B, p=0.0013, and Figure 5C, p=0.0066). In contrast, compared with PLC/PRF/5-control cells (189.75±107.73mm3), PLC/PRF/5-RNF128 cells (598.76± 171.36mm3) showed higher proliferation ability with 68% growth in volume and 69% in weight (Figure 5B, p=0.0037, Figure 5C, p=0.0172).
EGFR/MEK/ERK Pathway Activation is Involved in RNF128 Enhanced HCC Progression
In order to explore the potential mechanism of RNF128 in HCC progression, meanwhile refer to previous studies,12 we analyzed the correlation between RNF128 and the marker genes from EGFR/MEK/ERK signaling pathway by Gene Expression Profiling Interactive Analysis (GEPIA). The results showed that RNF128 was significantly correlated with the expression of EGFR, MAP2K1 and MAPK1 in HCC (Figure 6A, all p<0.001).
To further investigate whether RNF128 is involved in EGFR signaling pathway, we examined activation of EGFR/MEK/ERK in MHCC97-L and HepG2 cells by Western blot. We observed that p-EGFR, p-MEK and p-ERK1/2 were significantly upregulated in RNF128 overexpressed cells (Figure 6B, p=0.0012, p<0.001, p=0.0029), but decreased in RNF128 knockdown cells (all p<0.001).
Then gefitinib (1μM), the EGFR inhibitor, was used to confirm whether EGFR/MEK/ERK signaling is involved in RNF128 promoting effect on HCC malignant behavior. Expression of p-EGFR, p-MEK and p-ERK1/2 was decreased after gefitinib treatment, and EGFR/MEK/ERK pathway could be reactivated by RNF128 (Figure 6C). Cell function experiments revealed that the inhibitory effect of gefitinib on EGFR/MEK/ERK signaling counteracts the enhancement on migration, invasion and proliferation caused by RNF128 overexpression in HepG2 (Figure 6D, and E), suggesting that RNF128 might promote HCC progression via the EGFR/MEK/ERK pathway.
Discussion
The high metastasis and recurrence rate are responsible for the poor prognosis of HCC patients.5 In this study, we demonstrated that the expression of RNF128 was significantly upregulated in HCC tissues and associated with the HCC malignancy via EGFR/MEK/ERK signal pathway.
RNF128, a member of E3 ubiquitin ligases, is a type I transmembrane protein located in endosomal compartment.15–17 It has been confirmed to have an important role in both adaptive and innate immune responses. RNF128 induces the formation of immune tolerance in naïve T cells and the maintenance of Treg cells immunoregulatory function by mediating the ubiquitination and degradation of TCR-CD3. In mice, RNF128 deficiency is associated with higher risk of autoimmune disease.18 RNF128 also promotes TBK1 activation by mediating k63-linked polyubiquitination of TBK1, which promotes IRF3 activation and INF-β production. Thus, RNF128 is considered an enhancer of the innate immunity to RNF and DNA viruses.19 In addition, RNF128 also regulates energy metabolism20 and cytoskeletal reorganization.21
RNF128 has recently been documented to be involved in the progression of diverse cancers, including esophageal cancer,12 melanoma,13 urothelial carcinoma14 and lymphoma.11 Nonetheless, the role of RNF128 in HCC remains unexplored. Here, we clarified the function and mechanism of RNF128 in HCC for the first time. We showed that RNF128 expression was significantly elevated in HCC tissues, and higher expression of RNF128 in more malignant HCC cell lines, suggesting elevated RNF128 level may positively correlate with tumor malignancy. Analysis of clinical data from 171 HCC patients revealed RNF128 as an independent risk factor in HCC recurrence and survival. Simultaneously, high RNF128 levels were significantly correlated to malignant phenotypes, including tumor size, Edmondson–Steiner Grade and TNM stage.
Next, we explored the impact of RNF128 on HCC tumorigenesis and progression by overexpressing and knocking down RNF128 in HCC cell lines to conduct a series of cell function experiments. To ensure the reliability of the results, we selected two cell lines for knockdown and two cell lines for overexpression. Compared with the control group, RNF128 overexpression significantly enhanced invasion, metastasis, proliferation, and apoptosis resistance of HCC cells, while RNF128 knockdown had the opposite effect. In nude mouse xenograft model, influence of RNF128 on tumor growth was consistent with the cell experiments in vitro.
RNF128 physically interacts with the N-terminus of p53, ubiquitinating it and thus, suppresses its transactivation activity.22 It is known that P53 interacts with the EGFR promoter and regulates EGFR phosphorylation at multiple sites.23–25 Therefore, we further investigated the relationship between RNF128 and EGFR signaling in HCC and detected that phosphorylation of EGFR, ERK, and MEK were enhanced in HCC cells stably overexpressing RNF128, and vice versa. GEPIA analysis also showed that RNF128 expression in HCC was significantly correlated with MAP2K1, EGFR, and MAPK1 genes. To further clarify the mechanism by which RNF128 promotes HCC invasion, metastasis, proliferation, and anti-apoptosis, we treated HCC cells stably overexpressing RNF128 with gefitinib, an EGFR inhibitor, and observed a reversal of the malignant phenotypes enhanced by RNF128. These results suggest that RNF128 promotes HCC progression via EGFR/MEK/ERK pathway, which is consistent with the study in esophageal squamous cell carcinoma that showed RNF128 activated the EGFR/MEK/ERK pathway, promoting invasion and metastasis by direct interaction with p53. However, whether such EGFR/MEK/ERK pathway activated by p53-RNF128 interaction is present in HCC was not clarified in-depth in this article. The exact molecular interaction between RNF128 and EGFR signaling is our main contents of research of next stage. EGFR signaling activation can regulate various adapter molecules, triggering multiple downstream signal pathways, including phospholipase C cascades, JAK-STAT, and PI3K-AKT.26 Besides that, RNF128, as an E3 ubiquitin ligase, can affect various cellular biological functions by recognizing and binding multiple target proteins. Previous studies showed that lack of RNF128 enhanced the antitumor activity of CD8+ T cells in lymphomas.11 All these suggest that RNF128 may modulate additional signal transduction pathways or participate in other biological processes in HCC, which needs to be explored further.
In conclusion, we relatively comprehensively explored the effects and potential mechanism of RNF128 in HCC tumorigenesis and progression. Our findings show that RNF128 promotes HCC migration, invasion, proliferation, and anti-apoptotic ability via EGFR/MEK/ERK signaling. Our study provides valuable insight into the role of RNF128 in HCC and highlights RNF128 as a potential novel target for HCC treatment.
Abbreviations
RNF, RING finger protein; HCC, hepatocellular carcinoma; EGFR, epithelial growth factor; MEK, mitogen-activated protein kinase; ERK, extracellular regulated protein kinases; TMA, tissue microarray; IHC: immunohistochemistry; CCK8: Cell Counting Kit-8; qRT-PCR, quantitative real-time polymerase chain reaction.
Data Sharing Statement
All data and materials used in this study are included in the published article.
Ethics Approval
This study was approved by the ethics committee of Clinical Medical College, Yangzhou University and Zhongshan Hospital Research Ethics Committee, Fudan University (Shanghai, China). Xenograft experiments in nude nice were approved by the Animal Experimentation Ethics Committee of Yangzhou University.
Consent for Publication
All of HCC patients in the study have given their consent to publish their data.
Acknowledgments
We thank professor Guo-Ming Shi (Liver Cancer Institute, Zhongshan Hospital, Fudan University) for assistance with the sample and phenotype information collection.
Funding
This study was supported by The National Natural Science Foundation of China (grant number 81871909and 81702861); “13th five-year Plan” Science and Education strong Health Project leading personnel of Yangzhou (LJRC20181); Provincial-level discipline leader of the NJPH (DTRC201809); Chinese foundation for hepatitis prevention and control-TianQing liver disease research fund subject (No. TQGB20200180); Key talents in science and education of Yangzhou (RCC201818).
Disclosure
The authors report no conflicts of interest for this work.
References
1. Guo H, Li P, Su L, et al. Low expression of IL-37 protein is correlated with high Oct4 protein expression in hepatocellular carcinoma. Gene 2020;737:144445. doi:10.1016/j.gene.2020.144445
2. Liu LL, Zhu JM, Yu XN, et al. UBE2T promotes proliferation via G2/M checkpoint in hepatocellular carcinoma. Cancer management and research. 2019; 11:8359-8370. Cancer J Clin. 2018;68(6):394–424.
3. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi:10.3322/caac.21590
4. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA.
5. Sia D, Villanueva A, Friedman SL, Llovet JM. Liver cancer cell of origin, molecular class, and effects on patient prognosis. Gastroenterology. 2017;152(4):745–761. doi:10.1053/j.gastro.2016.11.048
6. Buetow L, Huang DT. Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat Rev Mol Cell Biol. 2016;17(10):626–642. doi:10.1038/nrm.2016.91
7. Vyacheslav A, Inigo BH, Hansen SVF, et al. UbiSite approach for comprehensive mapping of lysine and N-terminal ubiquitination sites. Nat Struct Mol Biol. 2018;25(7):631–640. doi:10.1038/s41594-018-0084-y
8. Senft D, Qi J, Ronai ZA. Ubiquitin ligases in oncogenic transformation and cancer therapy. Nat Rev Cancer. 2018;18(2):69–88.
9. Peng R, Zhang PF, Yang X, et al. Overexpression of RNF38 facilitates TGF-β signaling by Ubiquitinating and degrading AHNAK in hepatocellular carcinoma. CR. 2019;38(1):113. doi:10.1186/s13046-019-1113-3
10. Gou Z, Zeng Y, Chen Y, et al. TRAIP promotes malignant behaviors and correlates with poor prognosis in liver cancer. Biomed Pharmacother. 2020;124:109857. doi:10.1016/j.biopha.2020.109857
11. Haymaker C, Yang Y, Wang J, et al. Absence of Grail promotes CD8 T cell anti-tumour activity. Nat Commun. 2017;8(1):239. doi:10.1038/s41467-017-00252-w
12. Gao J, Wang Y, Yang J, et al. RNF128 promotes invasion and metastasis via the EGFR/MAPK/MMP-2 pathway in esophageal squamous cell carcinoma. Cancers. 2019;11:6. doi:10.3390/cancers11060840
13. Wei CY, Zhu MX, Yang YW, et al. Downregulation of RNF128 activates Wnt/β-catenin signaling to induce cellular EMT and stemness via CD44 and CTTN ubiquitination in melanoma. J Hematol Oncol. 2019;12(1):21. doi:10.1186/s13045-019-0711-z
14. Lee YY, Wang CT, Huang SK, et al. Downregulation of RNF128 predicts progression and poor prognosis in patients with urothelial carcinoma of the upper tract and urinary bladder. J Cancer. 2016;7(15):2187–2196. doi:10.7150/jca.16798
15. Anandasabapathy N, Ford GS, Bloom D, et al. GRAIL: an E3 ubiquitin ligase that inhibits cytokine gene transcription is expressed in anergic CD4+ T cells. Immunity. 2003;18(4):535–547. doi:10.1016/S1074-7613(03)00084-0
16. Heissmeyer V, Macián F, Im SH, et al. Calcineurin imposes T cell unresponsiveness through targeted proteolysis of signaling proteins. Nat Immunol. 2004;5(3):255–265. doi:10.1038/ni1047
17. Schartner JM, Simonson WT, Wernimont SA, Nettenstrom LM, Huttenlocher A, Seroogy CM. The gene related to anergy in lymphocytes, an E3 ubiquitin ligase, is necessary for anergy induction in CD4 T cells. J Immunol. 2004;173(1):79–85. doi:10.4049/jimmunol.173.1.79
18. Nurieva RI, Zheng S, Jin W, et al. The E3 ubiquitin ligase GRAIL regulates T cell tolerance and regulatory T cell function by mediating T cell receptor-CD3 degradation. Immunity. 2010;32(5):670–680. doi:10.1016/j.immuni.2010.05.002
19. Song G, Liu B, Li Z, et al. E3 ubiquitin ligase RNF128 promotes innate antiviral immunity through K63-linked ubiquitination of TBK1. Nat Immunol. 2016;17(12):1342–1351. doi:10.1038/ni.3588
20. Nakamichi S, Senga Y, Inoue H, et al. Role of the E3 ubiquitin ligase gene related to anergy in lymphocytes in glucose and lipid metabolism in the liver. J Mol Endocrinol. 2009;42(2):161–169. doi:10.1677/JME-08-0145
21. Ichikawa D, Mizuno M, Yamamura T, Miyake S. GRAIL (gene related to anergy in lymphocytes) regulates cytoskeletal reorganization through ubiquitination and degradation of Arp2/3 subunit 5 and coronin 1A. J Biol Chem. 2011;286(50):43465–43474. doi:10.1074/jbc.M111.222711
22. Chen YC, Chan JY, Chiu YL, et al. Grail as a molecular determinant for the functions of the tumor suppressor p53 in tumorigenesis. Cell Death Differ. 2013;20(5):732–743. doi:10.1038/cdd.2013.1
23. Sheikh MS, Carrier F, Johnson AC, Ogdon SE, Jr AJF. Identification of an additional p53-responsive site in the human epidermal growth factor receptor gene promotor. Oncogene. 1997;15(9):1095–1101. doi:10.1038/sj.onc.1201264
24. Nishi H, Senoo M, Nishi KH, et al. p53 Homologue p63 represses epidermal growth factor receptor expression. J Biol Chem. 2001;276(45):41717–41724. doi:10.1074/jbc.M101241200
25. Ludes-Meyers JH, Subler MA, Shivakumar CV, et al. Transcriptional activation of the human epidermal growth factor receptor promoter by human p53. Mol Cell Biol. 1996;16(11):6009–6019. doi:10.1128/MCB.16.11.6009
26. Wang DZ, Xia DR, Dubois RN. The crosstalk of PTGS2 and EGF signaling pathways in colorectal cancer. Cancers. 2011;3:3894–3908. doi:10.3390/cancers3043894
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.