Back to Journals » Infection and Drug Resistance » Volume 11
Resistance phenotype and clinical molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae among pediatric patients in Shanghai
Authors Tian D, Pan F, Wang C , Sun Y, Zhang H
Received 28 May 2018
Accepted for publication 2 August 2018
Published 24 October 2018 Volume 2018:11 Pages 1935—1943
DOI https://doi.org/10.2147/IDR.S175584
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Joachim Wink
Dongxing Tian, Fen Pan, Chun Wang, Yan Sun, Hong Zhang
Department of Clinical Laboratory, Shanghai Children’s Hospital, Shanghai Jiaotong University, Shanghai, China
Background: Carbapenem-resistant Klebsiella pneumoniae (CRKP) has caused wide global disseminations and serious clinical outcomes in pediatric patients, and the purpose of this study was to analyze drug resistance, molecular epidemiology, and clinical characteristics of CRKP from children in Shanghai, China.
Methods: A retrospective study was conducted from January 2016 to December 2017, and a total of 170 CRKP isolates were collected. Antimicrobial susceptibility was determined by the broth microdilution method. MAST D73C and polymerase chain reaction were used for the analysis of carbapenemase types. Multilocus sequence typing of K. pneumoniae was performed for genetic relationship. Clinical data were also reviewed.
Results: Of the 170 CRKP isolates, blaOXA-232 was mainly detected with a proportion of 42.35%, followed by blaNDM-1 (20.59%), blaKPC-2 (17.65%), blaNDM-5 (16.47%), and blaIMP-4 (1.18%). The predominant gene was blaOXA-232 in 2016 (54.46%; 55/101) and blaNDM-1 in 2017 (31.88%; 22/69). All these 170 CRKP isolates showed high resistance to cephalosporins and carbapenems (>95%), except for tigecycline and colistin. Sixteen distinct sequence types were observed with ST15 being mostly identified (41.76%). Most CRKP harboring OXA-232 type carbapenemase belonged to ST15, while NDM-1 type belonged to ST37 and KPC-2 type belonged to ST11. Furthermore, other β-lactamase genes including blaTEM, blaCTX-M, and DHA-1 were also found in this study. Clinical data reviewed that more than half of the patients produced clinical infections (112/170), mainly lower respiratory tract (58/112) and bloodstream (21/112) infections. A majority of these children had received therapy of antibiotics before CRKP isolation, especially for carbapenems (76/170) and β-lactam/β-lactamase inhibitor combinations (91/170).
Conclusions: Our data revealed the increasing incidence of OXA-232-producing K. pneumoniae from pediatric patients in Shanghai, and infection control measures should be conducted to limit the spread of CRKP strains.
Keywords: Klebsiella pneumoniae, carbapenemases, drug resistance, OXA-232, NDM-5, children
Erratum for this paper has been published
Introduction
Infections caused by carbapenem-resistant Klebsiella pneumoniae (CRKP) isolates are becoming an evolving crisis of global dimensions, especially for pediatric patients, due to high morbidity and mortality.1 Carbapenemase production is the main cause of carbapenem resistance; K. pneumoniae carbapenemases (KPCs), oxacillinase type 48 (OXA-48), and New Delhi metallo-β-lactamase (MβL) (NDM) carbapenemase have been reported worldwide. The prevalence of each carbapenemase varies geographically, and its resistance profiles also differ. The frequencies of KPC- and NDM-producing K. pneumoniae were significantly higher in the USA, Canada, Greece, Taiwan, Colombia, and China, whereas OXA-48-like-producing strains have significantly spread in Turkey and North Africa. In China, despite the extensive spread of KPC-producing strains, NDM-producing K. pneumoniae are the type mainly reported in children, though they are rarely found in adults.2–4 The detection rate of CRKP isolates in pediatrics increased from 5.3% to 15.9% according to data from the CHINET antimicrobial resistance surveillance program for 2005–2014.5 Recently, K. pneumoniae isolates producing OXA-232 carbapenemase, a new variant of OXA-48 that was first described in a patient who had traveled from India to France,6 were reported in a children’s hospital in China.7 To date, several countries, such as the UK, Switzerland, Brunei, and Italy,8–11 have reported sporadic OXA-232-harboring cases, often emerging together with NDM-1.
Because children are a naturally vulnerable population, carbapenem antibiotics are the first choice for treating infections caused by multidrug-resistant bacteria due to the side effects of other antibiotics, such as fluoroquinolones and aminoglycosides,2 and this has led to a burgeoning carbapenem resistance among pathogenic bacteria. The co-harboring of other resistance genes in CRKP has further increased the difficulty of delivering effective antibiotic therapy. Unfortunately, the dearth of effective treatment and extensive use of invasive procedures have increased the incidence of CRKP colonizations or infections and have caused high mortality in patients.12,13
Although CRKP has caused wide global disseminations and serious clinical outcomes in pediatric patients, there remained limited data available on the susceptibility and molecular epidemiology of these pathogens in China. Carbapenemase identification greatly aids in targeted drug use and helps to prevent further dissemination. Therefore, we conducted this study to investigate the resistance profiles, molecular epidemiology, and clinical characteristics of CRKP isolates obtained from children in the Shanghai region.
Materials and methods
Study population and bacterial isolates
This retrospective study was conducted in Shanghai Children’s Hospital, a 700-bed general university-affiliated hospital, with an estimated population of 2,088,000 patient visits per year. From January 2016 to December 2017, 523 non-duplicated strains of K. pneumoniae were isolated from inpatients in our hospital. All isolates were first identified using the matrix-assisted laser desorption ionization-time of flight-mass spectrometry (MALDI-TOF MS; Bruker Daltonik GmbH, Bremen, Germany). Among these strains, 170 (32.50%) were identified as resistant (zone diameter: ≤19 mm) to imipenem or meropenem by the disk-diffusion method. Escherichia coli ATCC 25922 was used as the quality control for identification and antimicrobial susceptibility testing.
This study was approved by the Shanghai Children’s Hospital Ethics Committee (Shanghai Jiao Tong University School of Medicine). The Review Board exempted this retrospective study from requiring informed consent because it only focused on the bacteria and did not have an impact on the patients.
Antimicrobial susceptibility testing and phenotypic analysis
Antimicrobial susceptibility was determined using the broth microdilution method according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI).14 Bacterial colonies selected from an 18- to 24-hour agar plate were adjusted to a turbidity equivalent 0.5 McFarland standard and the bacteria suspension was further diluted to a final inoculum density of approximately 105 CFU/mL in each well. Each well of a plate was inoculated with 100 µL inoculum, and the plate was incubated for 24 hours at 35°C. After incubation, the minimum inhibitory concentration (MIC) was read as the lowest concentration of antibiotic at which there was no visible growth. The results were determined and interpreted as follows: colistin according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST),15 tigecycline according to the US Food and Drug Administration, and all the others according to the CLSI M100-S27 criteria.14 MASTDISCS combi Carba plus disc system D73C (MASTCarba plus; Mast Group Ltd., Liverpool, UK) was used for the identification of carbapenemase types. Mast D73C is a five-disc detection set comprised of a penem antibiotic-only disc as well as MβL inhibitor, KPC inhibitor, and AmpC inhibitor discs. The fifth disc is a temocillin disc with a MβL inhibitor. D73C can detect MβL-positive strains, KPC-positive strains, and OXA-48-like-positive strains, and it can also differentiate KPC-positive isolates from isolates expressing AmpC coupled with porin loss.
Detection of antimicrobial resistance genes and multilocus sequence typing (MLST)
Polymerase chain reaction (PCR) assays were performed to detect drug resistance genes, such as carbapenemases (blaKPC , blaAIM , blaGIM , blaSIM , blaNDM , blaIMP , blaVIM, and blaOXA-48), common extended-spectrum β-lactamase (ESBL) genes (blaCTX groups, blaTEM-1, and blaSHV), AmpC genes (MOX, FOX, DHA, CIT, AAC, and EBC), and plasmid-mediated colistin resistance gene (MCR-1), according to previously described conditions and primers.16–19 The positive amplicons screened by electrophoresis on a 1.5% agarose gel were sequenced, and the DNA sequences obtained were analyzed and compared with those available in the NCBI GenBank database (https://www.ncbi.nlm.nih.gov/genbank/) using BLAST searches.
MLST was performed according to the protocols available at the MLST Pasteur website. Alleles and sequence types (STs) were assigned using the database (http://bigsdb.pasteur.fr/perl/bigsdb/bigsdb.pl?db=pubmlst_klebsiella_seqdef_public). Novel alleles and STs were submitted to the administrator of the database and assigned new designations. The eBURST version 3.0 software was used to analyze the clustering of related STs. In this study, isolates were grouped together if six of the seven alleles were homologues.
Clinical and epidemiological data
Both clinical and epidemiological information were obtained from the medical records of each patient. This information included patient demographics, neonatal birth information, brief hospital course, infection or colonization by CRKP, and underlying and epidemiologic characteristics. CRKP infections and colonizations were defined in accordance with the CDC/NHSN definitions.20
Results
Isolate information
During the study period, 523 non-duplicated strains of K. pneumoniae were isolated from inpatients in our hospital, of which 170 (32.50%) were identified as CRKP. The percentages of CRKP isolates were 34.47% (101/293) in 2016, and 30.00% (69/230) in 2017. The distribution of carbapenemase-producing K. pneumoniae isolates between January 2016 and December 2017 is shown in Figure 1. More carbapenemase-producing K. pneumoniae isolates were detected between July and December 2016, after which a significant decrease in detection occurred. Later, a substantial increase in detection occurred in December 2017 due to NDM-5-producing K. pneumoniae isolates.
Phenotypic characteristics and antimicrobial susceptibility
Phenotypic analysis revealed that of the 170 CRKP isolates, 169 were carbapenemase producers. The remaining isolate expressed AmpC coupled with porin loss. Among the 169 carbapenemase producers, KPC, MβL, and OXA-48-like producers accounted for 17.75% (30/169), 39.65% (67/169), and 42.61% (72/169), respectively. Overall, these 170 CRKP isolates showed high resistance to all tested drugs, especially cephalosporins and carbapenems, except for tigecycline and colistin. The resistance rates of ertapenem, imipenem, and meropenem were 99.41%, 95.88%, and 95.29%, respectively. One isolate was resistant to tigecycline and five isolates were resistant to colistin (Table 1). Comparative analyses revealed different hydrolysis profiles among the three types of carbapenemase producers. The MβL-producing isolates exhibited a lower resistant rate for amikacin (4.48%), gentamicin (13.43%), nitrofurantoin (35.82%), sulfamethoxazole/trimethoprim (61.69%), aztreonam (88.06%), ciprofloxacin (11.94%), and levofloxacin (7.46%) than the other two types of carbapenemase producers. In addition, OXA-48-like-producing K. pneumoniae isolates showed a lower resistant rate of cefmetazole as well as a relatively lower carbapenem minimum inhibitory concentration (Table 2).
Genotypes and MLST
PCR results confirmed that the KPC producers all harbored the blaKPC-2 gene (100%; 30/30), and the OXA-48-like producers all carried the blaOXA-232 gene (100%; 72/72). The blaNDM-1 (53.73%; 36/67), blaNDM-5 (41.79%; 28/67), and blaIMP-4 (4.48%; 3/67) genes were detected in MβL-producing K. pneumoniae isolates.
Among these CRKP strains, blaOXA-232, blaKPC-2, blaNDM-1, blaNDM-5, and blaIMP-4 were detected in 42.35% (72/170), 17.65% (30/170), 20.59% (35/170), 16.47% (28/170), and 1.18% (2/170) of the strains, respectively; additionally, one isolate co-harboring blaKPC-2 and blaNDM-1 and one isolate coharboring blaKPC-2 and blaIMP-4 were identified; no carbapenemase gene was identified in the remaining isolate. The predominant gene was blaOXA-232 in 2016 (54.46%; 55/101) and blaNDM-1 in 2017 (31.88%; 22/69) (Table 3). The most prevalent ESBL genes were blaTEM-1 (90.59%; 154/170), followed by blaCTX-M-15 (68.24%; 116/170), blaSHV-1 (58.26%; 99/170), blaSHV-11 (30.59%; 52/170), blaCTX-M-14 (14.12%; 24/170), blaSHV-12 (7.65%; 13/170), and blaSHV-5 (0.59%; 1/170). Most isolates harbored more than one ESBL gene (95.88%; 163/170), while seven isolates (4.12%; 7/170) possessed only one ESBL gene. Plasmid-borne AmpC β-lactamases genes, such as DHA-1 (4.12%; 7/170) and CMY-6 (1.18%; 2/170), were also found in this study. Fortunately, the recently reported MCR-1 gene was not detected in any of the isolates from this study.
  | Table 3 Distribution of different CRKP genotypes from 2016 to 2017 Abbreviation: CRKP, carbapenem-resistant Klebsiella pneumoniae. |
Sixteen distinct STs were observed among the 170 CRKP isolates. The predominant ST in CRKP isolates was ST15 (41.76%; 71/170), followed by ST48 (17.65%; 30/170) and ST11 (17.65%; 30/170) (Table 4). Three novel alleles (infB138, tonB447, and tonB448) and three novel STs (ST3245, ST3246, and ST3247) were detected in this study. Most isolates carrying blaOXA-232 belonged to ST15; only one belonged to ST3245. Likewise, most CRKP isolates carrying blaKPC-2 belonged to ST11, and only one isolate belonged to ST18. The isolates carrying NDM genes (including blaNDM-1 and blaNDM-5) belonged to ST14, ST17, ST37, ST43, ST45, ST48, ST105, ST2673, ST3246, and ST3247. The blaIMP-4- positive isolates were observed in two distinct STs (ST101 and ST198). The eBURST analysis indicated that these STs could be clustered into two groups (ST14, ST15, ST3245, and ST3246 in group one; ST17, ST18, and ST3247 in group two) and 10 singletons.
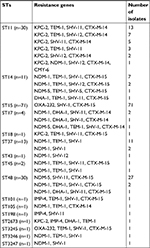  | Table 4 Genotypes and MLST of 170 CRKP isolates Abbreviations: CRKP, carbapenem-resistant Klebsiella pneumoniae; MLST, multilocus sequence typing; STs, sequence types. |
Clinical and epidemiological characteristics
The clinical and epidemiological characteristics of the patients from whom the isolates were obtained are summarized in Table 5. During the study period, a total of 170 patients developed infections or colonizations caused by CRKP; most of them were male (54.71%; 93/170) and aged less than 3 months (69.41%; 118/170). Most isolates were from respiratory specimens (68.23%; 116/170). The median length of hospital stay was 27 days (12–62 days), and the patients developed CRKP colonization or infection an average of 8 days (4–21 days) after admission.
Most CRKP isolates were detected in the neonatal intensive care unit (NICU) (65.88%; 112/170), followed by the pediatric intensive care unit (PICU) (16.47%; 28/170). Among the NICU patients whose samples were used for this study, the median birth weight was 2090 g (1330–2999 g), and 63.39% (71/112) were classified as premature. A high percentage of these 112 neonates were born via a cesarean delivery (69.64%; 78/112). Additionally, premature membrane rupture, low Apgar score, and history of drug use during pregnancy were reported for a small portion of these neonates, with percentages of 16.96% (19/112), 16.07% (28/112), and 24.11% (27/112), respectively.
Underlying conditions or diseases in the sampled patients are noted in Table 5. The most common disease was pneumonia (49.41%; 84/170); 54 patients (31.76%) had congenital heart disease, and 18 patients (10.58%) had gastroenteritis. Most patients had a history of invasive procedures, including central venous catheter (78.65%; 132/170), intubation/mechanical ventilation (75.71%; 127/170), surgery (26.47%; 45/170), urinary catheterization (24.12%; 41/170), and thoracic tube drainage (3.53%; 6/170). Over half of the patients developed CRKP infections (65.88%; 112/170), mainly lower respiratory tract (51.72%; 58/112), and bloodstream (18.75%; 28/112) infections. The remaining patients were colonized by CRKP (34.12%; 58/170), mainly respiratory tract colonization. Carbapenems (44.71%; 76/170) and β-lactam/β-lactamase inhibitor combinations (53.53%; 91/170) were the most frequently used antibiotics before CRKP isolation, whereas third-generation cephalosporin (29.41%; 50/170) was relatively less used.
Discussion
CRKP is a critical threat to pediatric patients, especially neonates, due to limited therapeutic options.1 Dissemination of NDM-1-producing K. pneumoniae ST76 and ST37 was observed in our hospital between March and June in 2014.2 Although NDM-1 was previously reported as the most common type of carbapenemase in children,21 our current study revealed that OXA-232 (42.35%; 72/170) was the most prevalent type in our patient population. In China, there are limited epidemiological characteristic data available on pediatric CRKP infections, especially those caused by OXA-232-producing K. pneumoniae. In our study, we aimed to provide comprehensive microbial resistance profiles, genotypes, and epidemiological clinical characteristics of CRKP infections mainly caused by OXA-232-producing K. pneumoniae isolates, because this information may help to prevent the expanded spread of OXA-232.
Overall, our study showed very high antimicrobial-resistant rates for common antibiotics, except for the low resistant rates for colistin and tigecycline. Notably, the antimicrobial resistance profiles significantly varied among different carbapenemase-producing K. pneumoniae isolates. NDM-producing K. pneumoniae showed the lowest drug resistance to aminoglycosides and fluroquinolones, a finding which is not consistent with previous reports. In earlier studies, NDM producers were usually also resistant to aminoglycosides as they frequently harbor 16S rRNA methylases.22–24 However, aminoglycosides and fluroquinolones are not recommended for pediatrics in China due to their side effects. The rare clinical usage of these drugs in children may be the reason for the low resistance to aminoglycosides and fluroquinolones observed in our study. In contrast, the newly prevalent OXA-232 producers in our hospital were all resistant to aminoglycosides and fluroquinolones. Compared with KPC-2 and NDM producers, OXA-232 producers had lower MIC values for carbapenems, a finding which is in accordance with the reported characteristics of OXA-48.25 Indeed, OXA-232 has a weaker ability to hydrolyze imipenem and temocillin than do OXA-48 or OXA-181 in the previous study.6 OXA-232 producers are now receiving increasingly more attention because of their higher resistance to most antibiotics than KPC-2 and NDM producers. It is of great urgency to strengthen surveillance and undertake strict infection control measures to prevent a more threatening spread of OXA-232-producing K. pneumoniae isolates in China.
Several countries, such as the UK (mainly ST14, ST147, ST231, and ST15),8 Switzerland (ST231),9 Italy (ST16),11 Brunei (ST231),10 and Tunisia (ST147),26 have reported infections caused by OXA-232-producing K. pneumoniae since the initial report of this carbapenemase type in France (ST14).6 In China, Yin et al7 first reported a small clonal dissemination of OXA-232-producing K. pneumoniae among five neonates between April and June 2016. Here, we revealed an outbreak of OXA-232-producing ST15 K. pneumoniae involving 72 patients in the NICU. The first OXA-232-producing K. pneumoniae in our study was obtained from the sputum of a 2-month-old patient from NICU in March 2016. We speculated that this strain caused the outbreak of OXA-232-producing K. pneumoniae in our hospital and even in Shanghai. Most of NDM-5 producers were identified as ST48 (90%; 27/30). NDM-5 has mostly been found in E. coli,27 and this is the first reported outbreak of NDM-5-producing ST48 K. pneumoniae in China. In addition, KPC-2, the predominant carbapenemase among adults in China, was detected in isolates from patients in the PICU and charity ward, and most of these isolates belonged to ST11, which is different from the pandemic sequence type ST258. A single KPC-2-producing K. pneumoniae isolate was identified as belonging to ST18. To the best of our knowledge, ST18 K. pneumoniae has not been reported before in China.
This study described the monthly distribution of CRKP isolates between January 2016 and December 2017 based on the molecular typing results, and the data suggest that three clonal disseminations of CRKP isolates have occurred in our hospital. KPC-2-producing ST11, OXA-232 producing ST15, and NDM-producing ST48 and ST14 K. pneumoniae isolates were responsible to these clonal disseminations at different times. Afterward, a substantial decrease was observed in the number of CRKP infections or colonizations in our hospital. Early detection and strict infection controls may have succeeded in preventing the further dissemination of CRKP during this time. It was worth noting that OXA-232-producing K. pneumoniae ST15 gradually disappeared in 2017 while NDM-5-producers appeared. The rapid emergence and spread of NDM-5-producing ST48 K. pneumoniae and NDM-1-producing ST37 K. pneumoniae in the NICU were observed between November and December 2017. Our study findings support the proposition that high-risk clones should be taken as the major consideration when developing a strategy to prevent the further dissemination of pathogenic isolates.
Most of the patients whose isolates were used in this study had carbapenem exposure and underwent invasive procedures. Previous work has found that antibiotic exposure and invasive procedures were both independent risk factors for developing CRKP infections and colonizations.28,29 Additionally, a 5-year retrospective case–control study in Turkey showed that 39.0% of CRKP-colonized patients in the PICU and 18.1% of CRKP-colonized patients in the NICU developed systemic CRKP infection after a mean of 10.6±1.9 days following detection of colonization.30 During our study period, 34.12% of all cases were identified as CRKP colonizations, and 58.93% (66/112) of infected patients had CRKP colonizations previously. The patients colonized with CRKP should be monitored for the possible further development of CRKP infection. A study performed in China reported that very low birth weight, preterm birth, and total parenteral nutrition were associated with nosocomial infections with carbapenem-resistant Enterobacteriaceae.28 In our study, 10.71%, 63.39%, and 67.86% of the neonates had very low birth weight, a preterm birth, and total parenteral nutrition, respectively. When implementing procedures to prevent CRKP infections, patients at high risk should be of the greatest concern.
Limitations
There are several limitations in our study. First, it was performed in a single hospital in Shanghai, and the prevalence and molecular characteristics of CRKP isolates in our pediatric patients may not be generalizable to pediatrics throughout our country. Additionally, our retrospective analysis briefly summarized the clinical characteristics of the patients from whom the CRKP isolates were obtained, and we did not determine independent risk factors for CRKP infections and colonizations, because there has been a lot of relative reports, and our study focused on the resistance phenotype and clinical molecular epidemiology of CRKP.
Conclusions
Our study described the antimicrobial resistance profiles and STs of CRKP isolates from pediatric patients as well as the clinical characteristics of these patients. OXA-232 was identified as being the predominant carbapenemase type in our isolates. Our results highlight the high antimicrobial-resistant rates of OXA-232-producing K. pneumoniae compared with CRKP that produces other types of carbapenemase. Furthermore, our data reveal the occurrence of several clonal disseminations in our hospital, which highlights the need to pay more attention to the newly emerging carbapenemase OXA-232 and NDM-5 and to determine which patients are at high risk of infection so that we can promptly take strict precautions.
Acknowledgments
We thank all members of the Clinical Laboratory of Shanghai Children’s Hospital for their cooperation and technical help. This work was supported by the Youth Foundation of Shanghai Municipal Commission of Health and Family Planning (20154Y0159) and the Shanghai Municipal Commission of Health and Family Planning (2015ZB0203). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
Logan LK. Carbapenem-resistant Enterobacteriaceae: an emerging problem in children. Clin Infect Dis. 2012;55(6):852–859. | ||
Zhu J, Sun L, Ding B, et al. Outbreak of NDM-1-producing Klebsiella pneumoniae ST76 and ST37 isolates in neonates. Eur J Clin Microbiol Infect Dis. 2016;35(4):611–618. | ||
Zhou J, Li G, Ma X, Yang Q, Yi J. Outbreak of colonization by carbapenemase-producing Klebsiella pneumoniae in a neonatal intensive care unit: Investigation, control measures and assessment. Am J Infect Control. 2015;43(10):1122–1124. | ||
Zhang X, Li X, Wang M, et al. Outbreak of NDM-1-producing Klebsiella pneumoniae causing neonatal infection in a teaching hospital in mainland China. Antimicrob Agents Chemother. 2015;59(7):4349–4351. | ||
Hu FP, Guo Y, Zhu DM, et al. Resistance trends among clinical isolates in China reported from CHINET surveillance of bacterial resistance, 2005–2014. Clin Microbiol Infect. 2016;22(Suppl 1):S9–S14. | ||
Potron A, Rondinaud E, Poirel L, et al. Genetic and biochemical characterisation of OXA-232, a carbapenem-hydrolysing class D β-lactamase from Enterobacteriaceae. Int J Antimicrob Agents. 2013;41(4):325–329. | ||
Yin D, Dong D, Li K, et al. Clonal dissemination of OXA-232 carbapenemase-producing Klebsiella pneumoniae in neonates. Antimicrob Agents Chemother. 2017;61(8):e00385-17. | ||
Findlay J, Hopkins KL, Loy R, et al. OXA-48-like carbapenemases in the UK: an analysis of isolates and cases from 2007 to 2014. J Antimicrob Chemother. 2017;72(5):1340–1349. | ||
Mancini S, Poirel L, Tritten ML, Lienhard R, Bassi C, Nordmann P. Emergence of an MDR Klebsiella pneumoniae ST231 producing OXA-232 and RmtF in Switzerland. J Antimicrob Chemother. 2018;72(3):821–823. | ||
Abdul Momin MHF, Liakopoulos A, Phee LM, Wareham DW. Emergence and nosocomial spread of carbapenem-resistant OXA-232-producing Klebsiella pneumoniae in Brunei Darussalam. J Glob Antimicrob Resist. 2017;9:96–99. | ||
Avolio M, Vignaroli C, Crapis M, Camporese A. Co-production of NDM-1 and OXA-232 by ST16 Klebsiella pneumoniae, Italy, 2016. Future Microbiol. 2017;12:1119–1122. | ||
Ulu-Kilic A, Alp E, Percin D, et al. Risk factors for carbapenem resistant Klebsiella pneumoniae rectal colonization in pediatric units. J Infect Dev Ctries. 2014;8(10):1361–1364. | ||
Kontopidou F, Giamarellou H; Group for the Study of KPC-producing Klebsiella pneumoniae infections in intensive care units. Infections caused by carbapenem-resistant Klebsiella pneumoniae among patients in intensive care units in Greece: a multi-centre study on clinical outcome and therapeutic options. Clin Microbiol Infect. 2014;20(2):O117–O123. | ||
Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing twentieth informational supplement. Wayne, PA: CLSI; 2017. | ||
The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. European Committee on Antimicrobial Susceptibility Testing (EUCAST); 2017. | ||
Pérez-Pérez FJ, Hanson ND. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002;40(6):2153–2162. | ||
Doosti A, Pourabbas M, Arshi A, Chehelgerdi M, Kabiri H. TEM and SHV genes in Klebsiella pneumoniae isolated from cockroaches and their antimicrobial resistance pattern. Osong Public Health Res Perspect. 2015;6(1):3–8. | ||
Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70(1):119–123. | ||
Woodford N, Fagan EJ, Ellington MJ. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum (beta)-lactamases. J Antimicrob Chemother. 2006;57(1):154–155. | ||
Horan TC, Andrus M, Dudeck MA. CDC/NHSN surveillance definition of health care-associated infection and criteria for specific types of infections in the acute care setting. Am J Infect Control. 2008;36(5):309–332. | ||
Dong F, Lu J, Wang Y, et al. A five-year surveillance of carbapenemase-producing Klebsiella pneumoniae in a pediatric hospital in China reveals increased predominance of NDM-1. Biomed Environ Sci. 2017;30(8):562–569. | ||
Al-Marzooq F, Ngeow YF, Tay ST. Emergence of Klebsiella pneumoniae-producing dual carbapenemases (NDM-1 and OXA-232) and 16S rRNA methylase (armA) isolated from a Malaysian patient returning from India. Int J Antimicrob Agents. 2015;45(4):445–446. | ||
Tängdén T, Giske CG. Global dissemination of extensively drug-resistant carbapenemase-producing Enterobacteriaceae: clinical perspectives on detection, treatment and infection control. J Intern Med. 2015;277(5):501–512. | ||
Berçot B, Poirel L, Nordmann P. Updated multiplex polymerase chain reaction for detection of 16S rRNA methylases: high prevalence among NDM-1 producers. Diagn Microbiol Infect Dis. 2011;71(4):442–445. | ||
Poirel L, Castanheira M, Carrër A, et al. OXA-163, an OXA-48-related class D β-lactamase with extended activity toward expanded-spectrum cephalosporins. Antimicrob Agents Chemother. 2011;55(6):2546–2551. | ||
Lahlaoui H, Bonnin RA, Moussa MB, Khelifa ABH, Naas T. First report of OXA-232-producing Klebsiella pneumoniae strains in Tunisia. Diagn Microbiol Infect Dis. 2017;88(2):195–197. | ||
Ahmad N, Khalid S, Ali SM, Khan AU. Occurrence of blaNDM Variants among enterobacteriaceae from a neonatal intensive care unit in a northern India hospital. Front Microbiol. 2018;9:407. | ||
Nour I, Eldegla HE, Nasef N, Shouman B, Abdel-Hady H, Shabaan AE. Risk factors and clinical outcomes for carbapenem-resistant Gram-negative late-onset sepsis in a neonatal intensive care unit. J Hosp Infect. 2017;97(1):52–58. | ||
Vanegas JM, Parra OL, Jiménez JN. Molecular epidemiology of carbapenem resistant gram-negative bacilli from infected pediatric population in tertiary-care hospitals in Medellín, Colombia: an increasing problem. BMC Infect Dis. 2016;16:463. | ||
Akturk H, Sutcu M, Somer A, et al. Carbapenem-resistant Klebsiella pneumoniae colonization in pediatric and neonatal intensive care units: risk factors for progression to infection. Braz J Infect Dis. 2016;20(2):134–140. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.




