Back to Journals » Diabetes, Metabolic Syndrome and Obesity » Volume 13
Reporting and Methods in Developing Prognostic Prediction Models for Metabolic Syndrome: A Systematic Review and Critical Appraisal
Authors Zhang H, Shao J, Chen D, Zou P , Cui N , Tang L, Wang D, Ye Z
Received 25 September 2020
Accepted for publication 19 November 2020
Published 15 December 2020 Volume 2020:13 Pages 4981—4992
DOI https://doi.org/10.2147/DMSO.S283949
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Juei-Tang Cheng
Hui Zhang,1 Jing Shao,1 Dandan Chen,1 Ping Zou,2 Nianqi Cui,3 Leiwen Tang,1 Dan Wang,1 Zhihong Ye1
1Department of Nursing, Zhejiang University School of Medicine Sir Run Run Shaw Hospital, Hangzhou, Zhejiang, People’s Republic of China; 2Department of Scholar Practitioner Program, School of Nursing, Nipissing University, Toronto, Ontario, Canada; 3Department of Nursing, The Second Affiliated Hospital Zhejiang University School of Medicine, Hangzhou, Zhejiang, People’s Republic of China
Correspondence: Zhihong Ye
Department of Nursing, Zhejiang University School of Medicine Sir Run Run Shaw Hospital, Hangzhou, Zhejiang 310016, People’s Republic of China
Tel +86-13606612119
Email [email protected]
Purpose: A prognostic prediction model for metabolic syndrome can calculate the probability of risk of experiencing metabolic syndrome within a specific period for individualized treatment decisions. We aimed to provide a systematic review and critical appraisal on prognostic models for metabolic syndrome.
Materials and Methods: Studies were identified through searching in English databases (PubMed, EMBASE, CINAHL, and Web of Science) and Chinese databases (Sinomed, WANFANG, CNKI, and CQVIP). A checklist for critical appraisal and data extraction for systematic reviews of prediction modeling studies (CHARMS) and the prediction model risk of bias assessment tool (PROBAST) were used for the data extraction process and critical appraisal.
Results: From the 29,668 retrieved articles, eleven studies meeting the selection criteria were included in this review. Forty-eight predictors were identified from prognostic prediction models. The c-statistic ranged from 0.67 to 0.95. Critical appraisal has shown that all modeling studies were subject to a high risk of bias in methodological quality mainly driven by outcome and statistical analysis, and six modeling studies were subject to a high risk of bias in applicability.
Conclusion: Future model development and validation studies should adhere to the transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD) statement to improve methodological quality and applicability, thus increasing the transparency of the reporting of a prediction model study. It is not appropriate to adopt any of the identified models in this study for clinical practice since all models are prone to optimism and overfitting.
Keywords: prediction model, risk, prognosis, metabolic syndrome, systematic review
Introduction
Metabolic syndrome (MetS) is defined as a cluster of cardiometabolic risk factors related to type 2 diabetes and cardiovascular disease.1 These risk factors are waist circumference, blood pressure, high-density lipoprotein level, triglycerides level, and hyperglycemia.2 MetS is a growing public health problem and concern. The prevalence of MetS has increased from 25.3% to 34.2% among US adults,3 and the prevalence of metabolic syndrome was 33.9% among Chinese adults as of 2010.4 People with MetS are likely to suffer from cardiovascular disease, insulin resistance, and hypertension leading to increased risk of cardiovascular morbidity and mortality.5 As diseases related to MetS can impose enormous health and economic burden, it is important to adopt effective and early measures to prevent the onset of morbidities for at-risk individuals. A healthy lifestyle is recommended as a suitable first-line intervention for MetS prevention and management.6 However, the specific target population that can benefit from a healthy lifestyle has yet to be determined using an evidence-informed decision-making approach.
A prognostic prediction model provides a multivariate predictor equation to help healthcare providers make decisions and plan for lifestyle changes or therapeutics.7 The prognostic prediction model can calculate the probability of risk of experiencing a particular health outcome within a specific time period for individualized treatment decisions.8 As we are in an era of personalized medicine with substantial and accumulating evidence from prognosis studies, there has been a rapid emergence of prognostic prediction models for MetS. However, none of them has been used in clinical practice or routine care. Herein, we aim to conduct a systematic review of prognostic prediction models for MetS to summarize and identify available evidence and knowledge gaps to facilitate their use. This could help clinicians and nurses determine which prognostic prediction model can be used in clinical practice.9
A systematic review and review protocol of prediction model for MetS were published previously.10,11 However, they did not include EMBASE database, which is one of the most important databases in the medical field.12 Moreover, unlike other systematic reviews for prediction model studies,13,14 the previous systematic review did not recommend any candidate predictors for future modeling studies based on their conclusions. A set of candidate predictors can help researchers select predictors to develop prediction models instead of using a purely data-driven approach. Lastly, events per variable (EPV) for sample size, the relationship between predictors and outcome definition, and some appropriate index (eg, Harrell’s c-index, and calibration plot) are important details for prediction models, but they were missing in the previous systematic review.
To fill the gap, in our systematic review, we aimed to expand and update prognostic prediction models for MetS in several important English and Chinese database, describe characteristics and the performance of the prognostic prediction models, and critically appraise methods and reporting of identified studies to indicate what is needed in further modeling studies.
Methods
This systematic review protocol was registered on the PROSPERO on 22 July 2020, and the registration number was CRD42020193282 (some updates were submitted to the PROSPERO). To improve rigor and reproducibility of this systematic review, we used the checklist for critical appraisal and data extraction for systematic reviews of prediction modeling studies (CHARMS) and the prediction model risk of bias assessment tool (PROBAST) to form the review question, study design, data extraction, and appraisal.15,16 This study adheres to the Preferred Reporting Items for Systematic reviews and Meta-Analysis (PRISMA) statement.
Search Strategy
The search strategy combined concepts related to prognostic prediction modeling studies and metabolic syndrome. A medical librarian helped us choose databases, iteratively developed strategy, and refined the search strategy. We developed an English search strategy combining subject indexing terms (ie MeSH) and free-text search terms in the title and abstract fields in Pubmed. This search strategy was translated appropriately for Embase, CINAHL, Web of Science (Core Collection). We also developed a Chinese search strategy combining subject indexing terms and free-text search terms in the title and abstract fields in Sinomed. This search strategy was translated appropriately for WANFANG, CNKI, CQVIP. The search strategy is presented in Supplementary Table S1. We systematically searched electronic databases from inception to July 27, 2020.
Study Selection
Inclusion criteria contained prognostic multivariable prediction studies (eg, model development studies and validation studies) for metabolic syndrome. Multivariable prediction studies focus on predicting an outcome by at least two predictors.17 Exclusion criteria contained diagnostic prediction model, predictor finding studies, model impact studies, and studies investigating a single predictor, test, or marker (such as single diagnostic test accuracy or single prognostic marker studies). This systematic review was limited to studies conducted in humans and published in English or Chinese. Study timing or setting was not limited. The titles and abstracts of all retrieved articles were independently screened by two reviewers (HZ, DDC) based on the selection criteria. When the information of the titles and abstracts suggested that the study was eligible, full text of this article was retrieved for further assessment. If there is any doubt regarding eligibility or disagreement, two reviewers (HZ, DDC) discussed with an advisor (JS) to reach a consensus.
Data Extraction and Critical Appraisal
A standardized electronic form based on the CHARMS checklist was constructed to facilitate the data extraction process.15 Information about objective, source of data, participants, outcome(s) to be predicted, candidate predictors, sample size, missing data, model development, model performance (eg, discrimination, calibration, and classifications measures), results, and interpretation of presented models was filled to complete this standardized form. If important information is missing, we sought clarification from the authors using email communication. One reviewer (HZ) extracted the data from the included studies, and another reviewer (DDC) checked the extracted data. For any disagreements, an advisor (JS) was consulted to resolve the disagreement and to reach a consensus.
We adopted PROBAST to assess the risk of bias, which can cause distorted estimation of a prediction model’s performance. PROBAST can also evaluate concerns regarding applicability of a prediction model.16 For the risk of bias, there are four key domains included in this tool: participants, predictors, outcome and analysis. Each domain can be rated as “high”, “low” or “unclear” risk of bias. For applicability, there are three key domains included in this tool: participants, predictors, and outcome. Each domain can be rated as “high”, “low” or “unclear” concerns. When risk of bias and applicability are evaluated as “low” in all domains, a prediction model can be judged as “low risk of bias” and “low concerns regarding applicability”, respectively. When risk of bias and applicability are assessed as “high” in one or more domains, a prediction model can be judged as “high risk of bias” or “high concerns regarding applicability”, respectively. When the evaluation about one or more domains is unclear and the remaining domains are judged as “low”, a prediction model can be judged as “unclear risk of bias” or “unclear concerns regarding applicability”, respectively. Two reviewers (HZ, DDC) independently assessed the methodological quality (risk of bias) and applicability of included studies. If there were any disagreements, this was resolved by discussion and consultation with an advisor (JS) to reach a consensus.
Results
From the 29,668 retrieved articles using the search strategy, 31 full articles were reviewed. Finally, eleven studies meeting the selection criteria were included in this review (Figure 1).
 |
Figure 1 PRISMA flow diagram. Notes: Adapted from Moher D, Liberati A, Tetzlaff J, Altman DG. The PRISMA Group (2009) Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Med. 2009;6(7): e1000097. Creative commons license and disclaimer available from: http://creativecommons.org/licenses/by/4.0/legalcode.18 |
Study Characteristics
The 11 included studies reported 22 prognostic prediction models identified as prediction model development with or without external validation. There were no prediction model external validation studies. The duration of follow-up ranged from 2 years to 7 years. Data source and study design can be found in Table 1. Eight prediction modeling studies used a retrospective cohort study design using health examination data19–26 and one used a retrospective cohort study design from Isfahan Cohort Study (Table 1).27 One prediction modeling study used a case–control study design from the French occupational GAZEL cohort.28 Notably, one study adopted a prospective cohort study design to develop risk model, but this model was validated using a cross-sectional study design.29 All models predicted a single endpoint that was MetS.
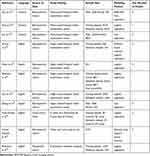 |
Table 1 Characteristics of Studies |
Characteristics of the Models
There were two studies including only males.20,28 Studies used different diagnostic criteria for metabolic syndrome (Table 2). One study aimed to develop a model that can both predict recovery from MetS and the risk of suffering MetS.22 An original score was developed from the samples without MetS, and then some of the predictors of this original score were reduced to form a final score based on the samples who recovered from MetS.22
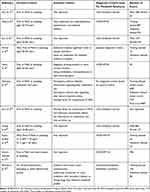 |
Table 2 Clinical Characteristics of the Study Population |
Ten studies aimed to develop prediction models for adults,19–28 and only one study aimed to predict risk probability for adolescent youths.29 The included studies used modeling methods encompassing logistic regression, cox regression, and machine learning techniques (eg, decision tree and support vector machine). A total of 48 predictors were selected in the final models (Figure 2), and 10 of these predictors are the criteria of the diagnostic criteria of MetS.
Analysis Methods
Two studies reported that participants without data about the diagnostic criteria of MetS and important examination results were excluded.22,26 One study reported that cases with missing values were excluded by applying listwise deletion.29 Others did not report the presence and methods for handling missing data. Continuous predictors were dichotomized or categorized in three studies.22,26,29 The number of participants with the outcome events and the number of candidate predictor parameters varied across the studies. Events per variable can be found in Figure 3, and statistical power was not sufficient in six studies due to EPV below 10.19–21,25,28,29 Univariate analysis was used to select candidate predictors in some studies22,24,26,28 and one study chose principal component analysis to select candidate predictors.25 One study did not report the number of outcome events,26 and we tried to contact the corresponding author via email to ask for more information about the number of events, but no response has been received.
 |
Figure 3 Events per variable for prediction modeling studies. Notes: This graphical presentation format was adapted from Ensor et al.14 A1 and A2 indicate the EPV for male and female, respectively in Gao et al.19; B, Yang et al.24; C1(male) and C2(female), sun et al.23; D, Hirose et al.21; E; Hsiao et al.20; F, Obokata et al.22; G1(male) and G2(female), Zhang et al.25; H, Pujos-Guillot et al.28; I, Karimi-Alavijeh et al.27; J, Efstathiou et al29; The EPV could not be calculated for Zou et al.26 |
Regarding model evaluation, two studies reported only apparent performance, which means the predictive performance has not been corrected (Table 3).19,21 Three studies reported only external validation without internal validation.24,28,29 Three performed internal validation using cross-validation,23,25,27 and the others used random split-sample for internal validation.20,22,26
 |
Table 3 The Presentation Format and Performance of Models |
Regarding model performance, there were three studies reporting calibration. However, only one used the calibration plot,22 while two studies used the Hosmer–Lemeshow test (Table 3). 21,29 Three studies report classification measures (eg, sensitivity and specificity) instead of c-statistic, and did not predefine a probability threshold.20,27,29 The c-statistic is also known as the area-under-the-curve of a receiver operating characteristic curve ranged from 0.67 to 0.95 in all studies. The full regression equations were presented in only four studies.20,21,23,24 One study presented a simplified scoring system,26 and two studies provided regression coefficients without baseline components.22,29 Others did not present their prediction models.19,25,27,28
Risk of Bias and Applicability of Model Studies
PROBAST was used to assess the risk of bias and applicability of model studies. As shown in Figure 4, most model studies had a high risk of bias driven by the analysis and outcome domains. Six studies were found to have high concerns of applicability due to inappropriate exclusion criteria, uncommon and unavailable predictors, and selected predictors being part of the outcome definition.20–22,24,28,29
 |
Figure 4 The risk of bias and the applicability of the model studies. |
Discussion
This systematic review identified and critically appraised 11 prediction modeling studies for MetS. Machine learning methods and regression methods were adopted to develop prognostic prediction models. Compared to a similar systematic review,10 we added six prognostic prediction models by searching English and Chinese databases.19,20,23,24,27,28 Three of them came from Japan, France, and Iran.20,27,28 The remaining studies came from China. Additionally, some of the important details were expanded and updated.
Model Development
One study adopted a nested case–control design from a pre-existing cohort, but it did not appropriately adjust for the original cohort outcome event frequency in the analysis. This can result in a high risk of bias for the prediction model.28 In such cases, it is recommended to reweight the control and case samples by the inverse sampling fraction from the original cohort as this can correct the estimation of baseline risk to obtain a corrected absolute predicted probability and model calibration measures.30 Some studies recruited only male participants or excluded participants who regularly drank alcohol or were current smokers.20,21,30 This means that the enrolled participants cannot be representative of this model’s targeted population resulting in a high risk of bias in participants’ domain and raising concern for applicability. One study adopted a prospective cohort study design to predict MetS in adolescence from Natal and Parental Profile, but this prognostic model was validated in a cross-sectional study that only recruited adolescence.29 The external validation may be inappropriate, due to study designs and populations issues. Researchers should choose a representative population, and a correct study design to improve the performance of the prediction model.13
Regarding sample sizes, EPV above 20 is recommended as the minimum sample size for model development.31 Moreover, for modeling studies using machine-learning techniques, a substantially higher EPV (often >200) is required to avoid overfitting.32 The sample size of five studies was not appropriate, because their EPVs were below 10 (Figure 3).19–21,25,28,29 Another study developed two prediction models based on a female cohort and a male cohort, and unfortunately, EPV of the female cohort is not appropriate.19 Inappropriate EPV can cause high risks of overfitting and biased predictions.33 This means that although researchers reported a C statistic which is close to 1, the performance of these models will probably be worse due to optimism when they are tested in another new dataset. Future studies are encouraged to determine an appropriate sample size, since different prediction modeling studies and different modeling techniques require different EPVs, for example, the EPV should be above 100 for model validation studies.34
Three studies used Cox proportional hazard models to develop a prognostic model.19,23,25 However, these modeling studies did not describe censored data, which are important for time-to-event analysis. Additionally, Harrell’s c-index or the D statistic is more appropriate than C statistic for evaluating survival model performance. Therefore, these models may suffer analysis bias resulting in a risk of bias when estimating predictive performance. Future studies should use appropriate statistical methods based on their study designs (eg, cross-sectional study or cohort study) and types of outcome data (binary or time-to-event outcomes), when performing statistical analysis.
Missing data are recognized as a common and increasingly vital problem in medical scientific research.35 However, simply excluding participants with missing data from the analysis called complete case (CC) analysis will cause biased predictor–outcome associations and biased model performance.16 Some studies adopted CC analysis,22,26,29 which discards valuable information from incomplete records. The remaining studies identified by this review did not report information about missing data. In such cases, participants with missing data are more likely to be deleted from statistical analysis because statistical packages automatically exclude individuals with any missing value.16 Multiple imputation can be a solution to missing data. The main advantage of multiple imputation is that it can obtain correct standard errors and P values, so it is regarded as the most appropriate method to handle missing data.36
One study dichotomized continuous predictors.22 Although dichotomization of continuous predictors can improve clinical interpretation and maintain simplicity, it is a suboptimal choice because of loss of information, lower predictive ability, and higher optimism.37 One study converted continuous predictors into categorized variables.26 However, it is recommended that predictors should be kept as continuous and the linear association between predictors and outcomes should be examined (eg, restricted cubic splines or fractional polynomials). If researchers consider categories in their studies, they should categorize continuous predictors into four or more groups based on widely accepted cut points.38
Researchers’ datasets usually have many features that could be selected as candidate predictors. The process of selecting predictors can be divided into two stages. First, researchers need to select candidate predictors for inclusion when the multivariable analysis is performed. To reduce the number of predictors, some studies adopted the univariate analysis to produce a simpler model.22,24,26,28 This strategy is not recommended because nuances in the data set or confounding by other predictors may exclude some important predictors causing predictor selection bias and increased overfitting.39 Future studies are encouraged to adopt some appropriate options, such as clinical reasons and a literature review.39 Secondly, predictors are selected during multivariable modeling. Common methods include stepwise selection techniques (eg, forward elimination), and these techniques were used in four studies.19,22,23,26,29 However, forward selection techniques are more likely to increase the risk of overfitting, especially in small sample size.40 Some popular penalized regression approaches should be adopted, such as ridge regression and lasso regression because these techniques can shrink each predictor effect differently and exclude some predictors entirely.41
This review identified 48 predictors from included prediction models (Figure 2). However, 10 of these predictors are included in the outcome definition. Ideally, outcomes should be determined without the need for predictor information; otherwise, the association between predictors and outcomes is prone to be overestimated and the model performance is more likely to be optimistic.17 After excluding the predictors mentioned above, a set of candidate predictors may be considered for future studies in adults if they were included at least two models:13 serum HMW-adiponectin, total adiponectin, HOMA-IR, serum insulin, free fatty acids, weight, glycated albumin, hip circumference, MCV, MCH, physical activity, AST, ALT, BMI, NGC, TC, serum uric acid, LDL-cholesterol, gender, smoking, WBC, LC, Hb, HCT, and age. These predictors are available in clinical practice. Because there was only one study about adolescence included in this review,29 we cannot recommend any predictor for the prognostic prediction model in adolescence.
Model Evaluation
After developing or validating prediction models, testing model performance is a vital step. Different measures to evaluate model performance may be used, such as calibration, discrimination, and (re)classification. It is recommended that both calibration and discrimination should be reported in all prediction model papers.42 However, only three studies reported both calibration and discrimination.21,22,29 For calibration, calibration plots are more appropriate than a statistical test of calibration (eg, Hosmer–Lemeshow test), because the direction or magnitude of miscalibration cannot be indicated by the Hosmer–Lemeshow test. For discrimination, Karimi-Alavijeh et al43 and Efstathiou et al29 reported sensitivity and specificity without a predefined probability. A predefined probability threshold is required when researchers adopt the (re)classification measures to avoid optimism and biases.
For modeling studies, external and internal validation are important. Two studies only evaluated apparent performance as model performance without internal validation.19,21 Regarding the reproducibility, internal validation is needed in model development studies. Three studies randomly split a dataset into a training group and a validation group.20,22,26 However, this approach is suboptimal especially in small samples, since this technique merely creates two smaller but similar datasets by chance, and does not use all available data to develop the prediction models.8 Bootstrapping and cross-validation techniques are recommended to conduct internal validation to correct the optimism of prediction models.44 To ensure transportability of a prediction model, external validation is needed. There were three studies that used external validation,24,28,29 but two of them only adopted external validation and omitted internal validation.24,29 Internal validation is vital to develop models, as models with only external validation may also be overfitting and optimism.17
Risk of Bias and the Applicability of Models
Eleven prognostic modeling studies in MetS were identified and these models are all at high risk of bias mainly driven by the analysis and outcome domains. The concerns regarding the applicability of several models are due to their inclusion/exclusion criteria (eg, male and BMI) and availability of predictors. Consequently, it is not appropriate to adopt any of them in clinical practice since each model performance is prone to be optimistic and overfitting.
Strengths and Limitations
This systematic review of prognostic prediction models in MetS adopted rigorous methods and a sensitive search strategy to search across several leading Chinese and English databases of biomedical literature. Compared to a previous systematic review,10 we expanded and updated many important details, such as EPV for sample size, the relationship between predictors and outcome definition, and some appropriate index (eg, Harrell’s c-index, and calibration plot). Additionally, this review added six prognostic prediction models by searching different databases.19,20,23,24,27,28 Another strength is that a set of candidate predictors are recommended for future modeling studies in this review. These candidate predictors are serum HMW-adiponectin, total adiponectin, HOMA-IR, serum insulin, free fatty acids, weight, glycated albumin, hip circumference, MCV, MCH, physical activity, AST, ALT, BMI, NGC, TC, serum uric acid, LDL-cholesterol, gender, smoking, WBC, LC, Hb, HCT, and age.
This systematic review did not search gray literature, so unpublished models were not included. Furthermore, we did not receive responses for information concerning the missing event number in a study from the corresponding author.26 Lastly, a quantitative analysis of the results was not performed because of the lack of homogeneity in the predictors, poor data, and model types.
Conclusion
This systematic review draws a map of the studies on multivariable prognostic models in MetS summarizing and appraising characteristics of studies, methodological characteristics, model performance, risk of bias, and the applicability of models. Future modeling development and validation studies are encouraged to adhere to the TRIPOD reporting guideline to improve the statistical methods of studies to increase the transparency of a prediction model study.
Acknowledgments
The first author wants to thank Mr Wei Tan, who gave her tremendous help.
Funding
This work was supported by the Zhejiang province medical technology project [grant numbers WKJ-ZJ-1925]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Zhejiang province medical technology project.
Disclosure
The authors have no conflicts of interest.
References
1. Wilson PWF, D’Agostino RB, Parise H, Sullivan L, Meigs JB. Metabolic syndrome as a precursor of cardiovascular disease and type 2 diabetes mellitus. Circulation. 2005;112(20):3066–3072. doi:10.1161/CIRCULATIONAHA.105.539528
2. Semnani-Azad Z, Khan TA, Blanco Mejia S, et al. Association of major food sources of fructose-containing sugars with incident metabolic syndrome: a systematic review and meta-analysis. JAMA Netw Open. 2020;3(7):e209993. doi:10.1001/jamanetworkopen.2020.9993
3. Moore JX, Chaudhary N, Akinyemiju TA. Metabolic syndrome prevalence by race/ethnicity and sex in the United States, National Health and Nutrition Examination Survey, 1988–2012. Prev Chronic Dis. 2017;14:160287. doi:10.5888/pcd14.160287
4. Lu JL, Wang LM, Li M, et al. Metabolic syndrome among adults in China: the 2010 China Noncommunicable Disease Surveillance. J Clin Endocr Metab. 2017;102(2):507–515.
5. Sergi G, Dianin M, Bertocco A, et al. Gender differences in the impact of metabolic syndrome components on mortality in older people: a systematic review and meta-analysis. Nutr Metab Cardiovasc Dis. 2020.
6. Zheng X, Yu H, Qiu X, Ying CS. The effects of a nurse-led lifestyle intervention program on cardiovascular risk, self-efficacy and health promoting behaviours among patients with metabolic syndrome: randomized controlled trial. Int J Nurs Stud. 2020;2020:103638.
7. Moons KG, Royston P, Vergouwe Y, Grobbee DE. Prognosis and prognostic research: what, why, and how? BMJ. 2009;338:b375.
8. Steyerberg EW, Moons KG, van der Windt DA, et al. Prognosis Research Strategy (PROGRESS) 3: prognostic model research. PLoS Med. 2013;10(2):e1001381.
9. Damen J, Hooft L. The increasing need for systematic reviews of prognosis studies: strategies to facilitate review production and improve quality of primary research. Diagn Progn Res. 2019;3:2.
10. Ibrahim MS, Pang D, Pappas Y, Randhawa G. Metabolic syndrome risk models and scores: a systematic review. Biomed J Sci Tech Res. 2020;26(1):19695–19707.
11. Ibrahim MS, Pang D, Randhawa G, Pappas Y. Risk models and scores for metabolic syndrome: systematic review protocol. BMJ Open. 2019;9(9):e027326. doi:10.1136/bmjopen-2018-027326
12. Cochrane handbook for systematic reviews of interventions: the Cochrane Collaboration; 2011. Available from: http://handbook-5-1.cochrane.org/.
13. Wynants L, Van Calster B, Collins GS, et al. Prediction models for diagnosis and prognosis of covid-19 infection: systematic review and critical appraisal. BMJ. 2020;369:m1328.
14. Ensor J, Riley RD, Moore D, Snell KI, Bayliss S, Fitzmaurice D. Systematic review of prognostic models for recurrent venous thromboembolism (VTE) post-treatment of first unprovoked VTE. BMJ Open. 2016;6(6):e011190. doi:10.1136/bmjopen-2016-011190
15. Moons KGM, de Groot JAH, Bouwmeester W, et al. Critical appraisal and data extraction for systematic reviews of prediction modelling studies: the CHARMS checklist. PLoS Med. 2014;11(10):e1001744. doi:10.1371/journal.pmed.1001744
16. Moons KGM, Wolff RF, Riley RD, et al. PROBAST: a tool to assess risk of bias and applicability of prediction model studies: explanation and elaboration. Ann Intern Med. 2019;170(1):W1–W33. doi:10.7326/M18-1377
17. Collins GS, Reitsma JB, Altman DG, Moons KGM. Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (TRIPOD): the TRIPOD Statement. Br J Surg. 2015;102(3):148–158. doi:10.1002/bjs.9736
18. Moher D, Liberati A, Tetzlaff J, Altman DG. The PRISMA Group (2009) Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Med. 2009;6(7):e1000097. doi:10.1371/journal.pmed.1000097
19. Gao YS, Bd L. Bayesian model averaging method for predicting 5 years of metabolic syndrome risk in men and women. Shandong Med J. 2016;56(39):91–94.
20. Hirose H, Takayama T, Hozawa S, Hibi T, Saito I. Prediction of metabolic syndrome using artificial neural network system based on clinical data including insulin resistance index and serum adiponectin. Comput Biol Med. 2011;41(11):1051–1056. doi:10.1016/j.compbiomed.2011.09.005
21. Hsiao F-C, Wu C-Z, Hsieh C-H, He C-T, Hung Y-J, Pei D. Chinese metabolic syndrome risk score. South Med J. 2009;102(2):159–164. doi:10.1097/SMJ.0b013e3181836b19
22. Obokata M, Negishi K, Ohyama Y, Okada H, Imai K, Kurabayashi M. A risk score with additional four independent factors to predict the incidence and recovery from metabolic syndrome: development and validation in large Japanese cohorts. PLoS One. 2015;10(7):7. doi:10.1371/journal.pone.0133884
23. Ohyama Y. Risk prediction model of metabolic syndrome in health management Population. J Shandong Univ. 2017;6(55):87–92.
24. Yang X, Tao Q, Sun F, Cao C, Zhan S. Setting up a risk prediction model on metabolic syndrome among 35–74 year-olds based on the Taiwan MJ Health-checkup Database. Zhonghua Liuxingbingxue Zazhi. 2013;34(9):874–878.
25. Zhang W, Chen Q, Yuan Z, et al. A routine biomarker-based risk prediction model for metabolic syndrome in urban Han Chinese population. BMC Public Health. 2015;15(1):64. doi:10.1186/s12889-015-1424-z
26. Zou -T-T, Zhou Y-J, Zhou X-D, et al. MetS risk score: a clear scoring model to predict a 3-year risk for metabolic syndrome. Hormone Metab Res. 2018;50(9):683–689. doi:10.1055/a-0677-2720
27. Karimi-Alavijeh F, Jalili S, Sadeghi M. Predicting metabolic syndrome using decision tree and support vector machine methods. ARYA Atheroscler. 2016;12(3):146–152.
28. Pujos-Guillot E, Brandolini M, Petera M, et al. Systems metabolomics for prediction of metabolic syndrome. J Proteome Res. 2017;16(6):2262–2272. doi:10.1021/acs.jproteome.7b00116
29. Efstathiou SP, Skeva II, Zorbala E, Georgiou E, Mountokalakis TD. Metabolic syndrome in adolescence can it be predicted from natal and parental profile? The Prediction of Metabolic Syndrome in Adolescence (PREMA) study. Circulation. 2012;125(7):902–910. doi:10.1161/CIRCULATIONAHA.111.034546
30. Ganna A, Reilly M, de Faire U, Pedersen N, Magnusson P, Ingelsson E. Risk prediction measures for case-cohort and nested case-control designs: an application to cardiovascular disease. Am J Epidemiol. 2012;175(7):715–724. doi:10.1093/aje/kwr374
31. Ogundimu EO, Altman DG, Collins GS. DG, GS C. Adequate sample size for developing prediction models is not simply related to events per variable. J Clin Epidemiol. 2016;76:175–182. doi:10.1016/j.jclinepi.2016.02.031
32. van der Ploeg T, Austin PC, Steyerberg EW. Modern modelling techniques are data hungry: a simulation study for predicting dichotomous endpoints. BMC Med Res Methodol. 2014;14(1):137. doi:10.1186/1471-2288-14-137
33. Courvoisier DS, Combescure C, Agoritsas T, Gayet-Ageron A, Perneger TV. Performance of logistic regression modeling: beyond the number of events per variable, the role of data structure. J Clin Epidemiol. 2011;64(9):993–1000. doi:10.1016/j.jclinepi.2010.11.012
34. Vergouwe Y, Steyerberg EW, Eijkemans MJC, Habbema JDF. Substantial effective sample sizes were required for external validation studies of predictive logistic regression models. J Clin Epidemiol. 2005;58(5):475–483. doi:10.1016/j.jclinepi.2004.06.017
35. Vergouwe Y, Royston P, Moons KGM, Altman DG. Development and validation of a prediction model with missing predictor data: a practical approach. J Clin Epidemiol. 2010;63(2):205–214. doi:10.1016/j.jclinepi.2009.03.017
36. Marshall A, Altman DG, Royston P, Holder RL. Comparison of techniques for handling missing covariate data within prognostic modelling studies: a simulation study. BMC Med Res Methodol. 2010;10.
37. Royston P, Altman DG, Sauerbrei W. Dichotomizing continuous predictors in multiple regression: a bad idea. Stat Med. 2006;25(1):127–141. doi:10.1002/sim.2331
38. Collins GS, Ogundimu EO, Cook JA, Manach YL, Altman DG. Quantifying the impact of different approaches for handling continuous predictors on the performance of a prognostic model. Stat Med. 2016;35(23):4124–4135. doi:10.1002/sim.6986
39. Steyerberg EW. Clinical Prediction Models: A Practical Approach to Development, Validation, and Updating. New York: Springer; 2019.
40. Royston P, Moons KG, Altman DG, Vergouwe Y. Prognosis and prognostic research: developing a prognostic model. BMJ. 2009;338(mar31 1):b604. doi:10.1136/bmj.b604
41. Pavlou M, Ambler G, Seaman SR, Guttmann O, Elliott PKM. How to develop a more accurate risk prediction model when there are few events. BMJ. 2015;351:h3868.
42. Steyerberg EW, Vickers AJ, Cook N, et al.Assessing the performance of prediction models: a framework for some traditional and novel measures. Epidemiology. 2010;21(1):128. doi:10.1097/EDE.0b013e3181c30fb2
43. Karimi-Alavijeh F, Jalili S, Sadeghi M. Predicting metabolic syndrome using decision tree and support vector machine methods. ARYA Atheroscler. 2016;12:3.
44. Moons KG, Kengne AP, Woodward M, et al. Risk prediction models: I. Development, internal validation, and assessing the incremental value of a new (bio)marker. Heart. 2012;98(9):683–690.
45. Zhang W, Chen Q, Yuan Z, et al. A routine biomarker-based risk prediction model for metabolic syndrome in urban Han Chinese population. BMC Public Health. 2015;15:64.
46. Pujos-Guillot E, Brandolini M, Pétéra M, et al. Systems metabolomics for prediction of metabolic syndrome. J Proteome Res. 2017;16(6):2262–2272.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

