Back to Journals » Infection and Drug Resistance » Volume 15
Report of a Fatal Purulent Pericarditis Case Caused by ST11-K64 Carbapenem-Resistant Hypervirulent Klebsiella pneumoniae
Authors Liang S, Cao H, Ying F , Zhang C
Received 22 June 2022
Accepted for publication 12 August 2022
Published 22 August 2022 Volume 2022:15 Pages 4749—4757
DOI https://doi.org/10.2147/IDR.S379654
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Shiwei Liang,1,2,* Huijun Cao,1,* Fei Ying,1 Chenchen Zhang2
1Centre for Clinical Laboratories, the Affiliated Hospital of Guizhou Medical University, Guiyang, People’s Republic of China; 2School of Clinical Laboratory Science, Guizhou Medical University, Guiyang, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Fei Ying, Tel +86 13985509788, Email [email protected]
Abstract: The report describes a 44-year-old female patient who died of the rare acute purulent pericarditis caused by Klebsiella pneumoniae (KP). The genomic analysis revealed an extensively drug-resistant ST11-K64 KP strain from five isolates (blood cultures, urine, ascites, pericardial effusion, and sputum). Several high virulence (hv) and carbapenem-resistant (CR) genes were identified in the pericardial effuse isolate. The isolates showed low resistance to healthy human serum. This study highlights the potential lethality of CR-hvKP infections in patients suffering from underlying comorbidities such as diabetes mellitus and chronic ailments.
Keywords: purulent pericarditis, Klebsiella pneumoniae, carbapenem-resistance, hypervirulence, ST11, K64
Introduction
Purulent pericarditis is a bacterial infection in the pericardium with gross or microscopic purulence, usually resulting from thoracic trauma, contagious spread of pathogens or hematogenous dissemination.1 Purulent pericarditis more frequently occurs in populations with compromised immune function or other predisposing factors such as diabetes and drug or alcohol abuse.2 It is a rare disease but has a very poor prognosis, requiring life-saving early diagnosis and effective treatment.2 Acute purulent pericarditis can rapidly progress into cardiac tamponade, systemic toxicity and cardiac diastolic dysfunction that lead to almost 100% mortality if left untreated and 40% in treated patients.3 The sources of the infection are often non-cardiac but via systemic dissemination from a primary pulmonary or abdominal infection. In recent years, the ascendancy of pathogens causing purulent pericarditis tends to drift from the Gram-positive bacteria such as Streptococcus pneumonia and Staphylococcus aureus,1 to Gram-negative bacteria such as Proteus, Escherichia coli, Pseudomonas, and in rare cases Haemophilus influenzae Acinetobacter baumannii and Klebsiella pneumoniae (KP),2,4–6 a phenomenon that can be attributed to the extensive use of broad-spectrum antimicrobials.7
Klebsiella pneumoniae (KP) is an Enterobacteriaceae that causes opportunistic infection with community or nosocomial acquirement. According to the types and amount of the mucoid polysaccharide capsule produced, they are characterised into classic KP (cKP) and hypermucousviscous KP (hmKP), the latter often referred to as hypervirulent KP (hvKP) due to its metastatic pathogenicity.8 The sequence type (ST) and the capsular antigen (K), as well as other virulence factors, including the extracapsular polysaccharide synthesis regulator genes, the ferric iron uptake system genes and the fimbriae expression genes, are used to classify a range of virulent KP lineages.9,10 hvKP is mostly implicated in pyogenic liver abscesses, septicaemia, pneumonia, cystitis, surgical wound infection, endophthalmitis, endocarditis and urinary tract infections.11 The acquisition, exchange, accumulation and convergence of plasmids or transposons coding for advanced virulence and multi-drug resistance of hvKP pose an urgent challenge for clinicians, as there are no evaluated treatment solutions. Such that the carbapenem-resistant hvKP (CR-hvKP) infection is considered a serious public health threat in Southeast Asia and beyond.10,12,13
Purulent pericarditis caused by KP has been scarcely documented.14 One report described a positive outcome of a 67-year-old man treated with surgical pericardiectomy and intradiaphragmatic abscess draining, but the virulence and genetic details of the KP strain were not determined.15 In another report, a capsule genotype K1 hvKP was detected in the isolates of a 43-year-old man with diabetes mellitus and alcoholism. Despite the presence of the virulence-attributing genes, the patient responded well to cefazolin, ceftriaxone and ciprofloxacin and was discharged after the treatment.6
We present here the clinical and microbiological findings of a 44-year-old female diabetic patient who developed purulent pericarditis caused by ST11-K64 CR-hvKP infection and died soon after the drainage surgery.
Case Presentation
A 44-year-old woman was admitted to the emergency department of the affiliated hospital of Guizhou Medical University, with complaints of coughing, vomiting, tightness in the chest, shortness of breath, abdominal pain, chills and fevers in the morning and at night that had persisted for half a month and worsened over the past three days. She was subsequently hospitalized into nephrology department with diagnosis of chronic renal failure (CRF). The patient had a medical history of type 2 diabetes mellitus and had been treated for CRF during the last two years. She was taking Euthyrox for treatment of hypothyroidism converted from hyperthyroidism that was diagnosed 20 years ago. Her surgical traumas included caesarean section and inferior vena cava filter placement for venous thrombosis. The physical examination the next day revealed moist rales in both lungs and serum measurement found neutrophilic leukocytosis (WBC count 11.14×109/L with 96.40% neutrophils), high interleukin-6 (IL-6, 251.20 pg/mL) and procalcitonin (PCT, 100 ng/L). Ceftizoxime was administrated for anti-infection treatment, which was replaced by meropenem after microbial detection of Klebsiella pneumoniae from the blood and urine isolates. The patient was discharged after 23 days of treatment when the acute symptoms disappeared and serum parameters returned to normal.
Twelve days later, the patient was re-admitted into the emergency department with a complaint of >10 hours of diarrhoea and >3 hours of impaired consciousness. Soon the patient was sent to the intensive care unit (ICU) due to her condition deteriorating into severe consciousness disorders. The vital signs and physical parameters on admission were the following: body temperature 37.2°C, hypotensive blood pressure 84/47 mmHg, heavy and moist rales in both lungs audible through auscultation, no abnormal bulge/depression or pulsation in the precordial area, no obvious pathological murmur heard by auscultation in all the cardiac valve areas and no oedema in the lower extremities. Laboratory analysis indicated neutrophilic leukocytosis (WBC count 35.22×109/L with 96.80% neutrophils) and suspicion of sepsis (interleukin-6 5000 pg/mL and PCT higher than 100 ng/L in serum). Computed tomography (CT) of the chest showed a large pleural effusion and abdominal effusion.
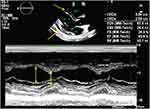
The emergency treatments included phlegm removal, sedation, analgesia, vasoactive drugs, anti-inflammation, and anti-infection drug imipenem. A closed drainage of the right thoracic cavity was performed to improve the right pneumothorax. The patient had to be subsequently submitted to orotracheal intubation and an invasive ventilator due to severe hypoxia. Despite all these treatments, her general condition deteriorated. A cardiac ultrasound exam revealed a significantly enlarging pericardial effusion (Figure 1).
Pericardiocentesis drainage was immediately employed, and 150 mL of purulent liquid was withdrawn. Bacterial identification and drug sensitivity test were conducted using MicroScan WalkAway 96 plus automatic analysis system. The drug susceptibility results were verified later by broth microdilution method. A Klebsiella pneumoniae strain was confirmed and cultured from the pericardial effusion, which was resistant to multiple antibiotics, especially carbapenems (Table 1). Therefore, the patient was diagnosed with purulent pericarditis caused by CR-KP. CR-KP were also detected in the patient’s blood, ascites, and urine.
 |
Table 1 Results of Antimicrobial Susceptibility Testing (Broth Microdilution) |
Within one to two hours after the pericardiocentesis drainage, the patient’s heart rate gradually dropped to 40 bpm. The patient failed to regain consciousness despite active resuscitation (ACDCRP) and eventually died, likely from multiorgan failure associated with sustained septic shock.
Microbiological and Molecular Characterization
Five isolates (blood cultures, urine, ascites, pericardial effusion, and sputum) collected from this patient were positive for ST11 Klebsiella pneumoniae, confirmed by PCR screening and DNA sequencing analysis of the seven house-keeping genes of KB (rpoB, phoE, gapA, infB, mdh, tonB, and pgi) according to the Institut Pasteur Multilocus Sequence Typing (MLST) databases (https://bigsdb.pasteur.fr/klebsiella/). Using the primers described previously for PCR,16 the capsular serotype of all the isolates was confirmed to be K64, which was validated by whole-genome sequencing. All isolates demonstrated hypermucoviscosity as shown by string length >5 mm in the string test (Figure 2). Common carbapenem resistance genes and virulence genes were identified by PCR using primers reported in the literature.17–19 All isolates presented β-lactam resistance gene blaKPC but were negative for other carbapenemases including OXA-48, IMP, NDM and VIM. The KP strain from all five isolates possessed virulence factor plasmids rmpA, mrkD, entB and ybtS, which are related to mucoid phenotype, type-3 fimbrial adhesin, enterobactin and yersiniabactin, respectively.20,21
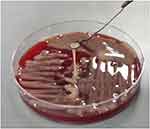 |
Figure 2 Representative image string test. Strings >5mm indicates the hypermucoviscous phenotype of the KP strain. |
The bacterial genome sequencing data obtained from pericardial effusion culture was compared against the Comprehensive Antibiotic Resistance Database (CARD) and Virulence Factor Database (VFDB). A total of 27 putative resistance genes were identified (Table 2), associated with three main resistance mechanisms: 1. The antibiotic efflux pump systems, prominently genes belonging to the ATP-binding cassette (ABC) and the major facilitator superfamily (MFS), 2. antibiotic inactivation or reduced permeability to target, which is induced by β-lactamase gene amplification and porin disruption, such as blakpc-1, blaTEM-1 and blaCTX-M-6522 and 3. alteration in the target sites of antibiotics by rmtB gene encoding the aminoglycoside-modifying 16S rRNA methylase23 and sul2 gene encoding altered forms of dihydropteroate synthase that are not inhibited by the sulfonamides.24 Furthermore, 31 virulence-associated genes were predicted, which suggested the employment of several iron uptake mechanisms in the patient isolate, including production of aerobactin (iuc), enterobactin (entB, entA, fepC, etc.) and secretion of yersiniabactin (ybtS, ybtE, irp1, 1rp2, etc.)25,26 (Table 3).
 |
Table 2 Potential Resistance Genes from Pericardial Effusion Isolate, Predicted by CARD Annotation |
 |
Table 3 Potential Virulence-Associated Genes from Pericardial Effusion Isolate, Predicted by VFDB Annotation |
Based on the evidence of carbapenem resistance, the hypervirulence determinants, and the mucoid phenotype of the bacteria, we conclude that the infective agent identified in the isolates is sequence type 11 (ST11) capsular serotype K64 carbapenem-resistant hypervirulent Klebsiella pneumoniae (CR-hvKP).
The serum survival of the CR-hvKP strain from the pericardial effusion, blood and ascites isolates was tested according to the previous description.27 As shown in Figure 3, the colony-forming unit (CFU) recovery of all isolates was inhibited by the human serum time-dependently. After three hours of incubation, barely any CFU colony was visible in two samples and no colony growth was observed by the next day (Figure 3A and B), indicating a susceptibility of the KP strain to normal human serum.
Discussion
There are few reported cases of CR-hvKP infection related to purulent pericarditis. The patient reported here, with a history of type 2 diabetes mellitus and chronic renal failure, as well as recent thoracic surgery, died within a few hours of acute purulent pericarditis onset. The strain detected in the multi-site infection was confirmed to be the sequence type 11 (ST11) capsular serotype K64 carbapenem-resistant hypervirulent Klebsiella pneumoniae (CR-hvKP).
Cardiac tamponade secondary to massive pericardial effusion is the most severe complication of bacterial pericarditis. Without efficient treatment, the patient develops toxic shock, dyspnea and a sharp drop in heart rate, and death.2 Great clinical acumen necessitates prompt diagnosis followed by emergent aggressive interventions of intravenous antibiotics to constrict infection deterioration or dissemination and adequate drainage to relieve the patient’s cardiac compression.5,25,28,29 Both empirical use of broad-spectrum antibiotics and pathogen-specific antibiotic regimen should be considered.
The extensive use of antibiotics has rendered purulent pericarditis rare but also enabled the survival and multiplying of multi-drug resistant (MDR) bacteria. Another example of selective pressure favouring the survival of antibiotic-resistant bacteria is the irrational use of antibiotics in both clinical and agricultural settings.30 The elderly population and individuals with inferior fitness or underlying health conditions such as diabetes mellitus, immunosuppression, chronic renal failure and previous surgical trauma are under the greatest threat.5 The shifting landscape of KP infection has drawn concerns over the emergence of KP “superbugs” armed with hypervirulence (hv) phenotypes, comprehensive resistance against common antibiotics, and the high transmissibility of the aggressive genetic elements.8,31–33 As carbapenem is currently reserved as physicians’ last weapon against difficult-to-treat infections, the lurking danger of CR-hvKP could lead the infection from morbid to lethal. In a retrospective study performed in 2016, more than half (57%) of the hvKP infections were carbapenemase-producing.34
According to the recent literature, serotypes K1/K2 and MLST-11 are the most referred to as hypervirulent KP strains (hvKP) causing severe nosocomial infections.35,36 While K1 and K2 KP cause mostly liver abscesses or further disseminated infection37 and ST11 (predominantly K64 serotype) imposes the highest hvKP prevalence in isolates from various sources including blood sputum, urine, stool, wound and drainage.38 ST11 CR-hvKP and its descendent sequence types such as ST258 have become the dominant clonal lineage in Asia, responsible for the recent outbreaks in some Chinese hospitals.12,18,39
The ST11 hvKP strain detected in all five isolates of the patient in the present report carries the plasmid-borne rmpA gene accounting for the overproduction of extracapsular polysaccharide, which has been considered an independent factor conferring the hypermucoviscosity phenotype, thereby predisposing abscess formation.40 Notably, the use of the terms hypermucoviscous KP (hmKP) and hvKP interchangeably to distinguish them from classic KP (cKP) is debatable. Compared to cKP, hmKP/hvKP is considered to insult mostly otherwise healthy populations and display hypermucoid appearance on agar plates.10 However, in vitro/in vivo and clinical studies have found that the morphological phenotype is not necessarily associated with high virulence. On the spectrum of many critical virulent factors that might constitute the minimum requirement for hypervirulence of hvKP, aerobactin is the most sensitive defining trait of hvKP.41,42 Genes encoding aerobactin (iuc) belong to the siderophore system, which mediates the elevated iron acquisition demanded by the survival of hvKP in host.43 Indeed, the strain from the pericardial effuse isolate of the patient reported here has a positive VFDB- annotated iuc identity. Although the bacteria did not display resistance to healthy human serum, the patient’s immune defence failed to fight against the dissemination because of her multimorbidity of diabetes mellitus, chronic kidney disease and complex surgery history.
The CR-hvKP strain was firstly detected from the blood and urine samples of the patient one month before the fatal incidence, which displayed the same virulence and carbapenem resistance genes. Although a full comparison of genetic mapping was not applied, we suspect that the occult bacteria at pulmonary focus multiplied and spread contiguously to pericardial space and other sites haematogenously, while the infection symptoms were controlled upon her discharge from the hospital. A lesson we have learned from this tragic case is that, in cases of CR-hvKP infections, vigilance may need to be extended to a follow-up tracing of the bacterial survival even after the patient appears symptom-free.
Conclusion
The Klebsiella pneumoniae strain identified in the fatal case of purulent pericardial was ST11-K64 CR-hvKP, consistent with the findings that ST11 dominates the carbapenem-resistant KP infections in China. Sporadic infection cases by similar strains have been identified in our hospital and seem to be increasing. As the advent and dissemination of the multidrug-resistant and highly virulent KP raises an alert, against the backdrop of population ageing and the rising prevalence of chronic conditions such as diabetes,44 surveillance at epidemiological and molecular levels is paramount to avoid an endemic outbreak.
Abbreviations
KP, Klebsiella pneumoniae; PP, purulent pericarditis; MLST, multilocus sequence typing; ST, sequence type; cKP, classic Klebsiella pneumoniae; CRKP, carbapenem-resistant Klebsiella pneumoniae; hvKP, hypervirulent Klebsiella pneumoniae; EICU, emergency intensive care unit; PCR, polymerase chain reaction; CARD, comprehensive antibiotic resistance database; VFDB, virulence factor database; CR-hvKP, carbapenem-resistant hypervirulent Klebsiella pneumoniae.
Ethnic and Patient Consent
This case report was approved by the Ethics Committee of The Affiliated Hospital of Guizhou Medical University. Since the patient is deceased, written informed consent for publication of her clinical details and clinical images was obtained from the next of kin.
Author Contributions
SL and HC have contributed equally to this work and they are co-first authors. SL, HC and CZ acquired clinical data and performed the laboratory analyses. SL wrote the manuscript. HC reviewed the patient notes and revised the manuscript. YF and HC conceived the study, carried out the literature search, and processed the patient record. All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Funding
This work was supported by the National Natural Science Foundation of China NO. 81460334, Foundation for the project of Natural Science Research Innovation Group of General Undergraduate College in Guizhou Province (Qianjiaohe KY character [2021]016) and the project of Guizhou Provincial Innovative Talents Team (Grant No.2019-5610). The funders had no role in the study design, conduct, analysis, interpretation of study results, decision to publish, or preparation of the manuscript. No additional external funding was received for this study.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Boyadzhieva G, Stickel M, Christ M, Minervini F, Rare A. Case of Pyopericardium After Blunt Thoracic Trauma. Ann Thorac Surg. 2021;111(4):e259–e261.
2. Latif A, Patel AD, Mahfood Haddad T, Lokhande C, Del Core M, Esterbrooks D. Massive purulent pericarditis presenting as cardiac tamponade. Proc. 2020;33(4):662–663.
3. Pankuweit S, Ristic AD, Seferovic PM, Maisch B. Bacterial pericarditis: diagnosis and management. Am J Cardiovasc Drugs. 2005;5(2):103–112.
4. Liu J, Xiao X, Cen C, Yuan H, Yang M. Rare purulent pericarditis caused by carbapenem-resistant Acinetobacter baumannii: a case report. Medicine. 2019;98(38):e17034.
5. Shah S, Shah P, Green J. Haemophilus influenzae purulent pericarditis in an immunocompetent individual. J Community Hosp Intern Med Perspect. 2021;11(1):96–98.
6. Yu WL, Cheng CC, Chuang YC. First report of acute purulent pericarditis by capsule genotype K1 Klebsiella pneumoniae in an alcoholic patient. Diagn Microbiol Infect Dis. 2009;63(3):346–347.
7. Varela MF, Stephen J, Lekshmi M, et al. Bacterial Resistance to Antimicrobial Agents. Antibiotics. 2021;10(5):548.
8. de Campos TA, Goncalves LF, Magalhaes KG, et al. A Fatal Bacteremia Caused by Hypermucousviscous KPC-2 Producing Extensively Drug-Resistant K64-ST11 Klebsiella pneumoniae in Brazil. Front Med. 2018;5:265.
9. Rivero A, Gomez E, Alland D, Huang DB, Chiang T. K2 serotype Klebsiella pneumoniae causing a liver abscess associated with infective endocarditis. J Clin Microbiol. 2010;48(2):639–641.
10. Choby JE, Howard-Anderson J, Weiss DS. Hypervirulent Klebsiella pneumoniae - clinical and molecular perspectives. J Intern Med. 2020;287(3):283–300.
11. Effah CY, Sun T, Liu S, Wu Y. Klebsiella pneumoniae: an increasing threat to public health. Ann Clin Microbiol Antimicrob. 2020;19(1):1.
12. Gu D, Dong N, Zheng Z, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. 2018;18(1):37–46.
13. Yang ZQ, Huang YL, Zhou HW, Zhang R, Zhu K. Persistent carbapenem-resistant Klebsiella pneumoniae: a Trojan horse. Lancet Infect Dis. 2018;18(1):22–23.
14. Brook I, Frazier EH. Microbiology of acute purulent pericarditis. A 12-year experience in a military hospital. Arch Intern Med. 1996;156(16):1857–1860.
15. Caixal Vila G, Flores E, Caldentey G. Purulent pericarditis due to Klebsiella pneumoniae pulmonary infection. Revista Colombiana de Cardiología. 2017;24(4):417–418.
16. Yu F, Lv J, Niu S, et al. Multiplex PCR Analysis for Rapid Detection of Klebsiella pneumoniae Carbapenem-Resistant (Sequence Type 258 [ST258] and ST11) and Hypervirulent (ST23, ST65, ST86, and ST375) Strains. J Clin Microbiol. 2018;56(9):658.
17. Indrajith S, Mukhopadhyay AK, Chowdhury G, et al. Molecular insights of Carbapenem resistance Klebsiella pneumoniae isolates with focus on multidrug resistance from clinical samples. J Infect Public Health. 2021;14(1):131–138.
18. Zhan L, Wang S, Guo Y, et al. Outbreak by Hypermucoviscous Klebsiella pneumoniae ST11 Isolates with Carbapenem Resistance in a Tertiary Hospital in China. Front Cell Infect Microbiol. 2017;7:182.
19. Fu L, Huang M, Zhang X, et al. Frequency of virulence factors in high biofilm formation blaKPC-2 producing Klebsiella pneumoniae strains from hospitals. Microb Pathog. 2018;116:168–172.
20. Yan Q, Zhou M, Zou M, Liu WE. Hypervirulent Klebsiella pneumoniae induced ventilator-associated pneumonia in mechanically ventilated patients in China. Eur J Clin Microbiol Infect Dis. 2016;35(3):387–396.
21. Wang G, Zhao G, Chao X, Xie L, Wang H. The Characteristic of Virulence, Biofilm and Antibiotic Resistance of Klebsiella pneumoniae. Int J Environ Res Public Health. 2020;17:17.
22. Zhang Y, Jiang X, Wang Y, et al. Contribution of beta-lactamases and porin proteins OmpK35 and OmpK36 to carbapenem resistance in clinical isolates of KPC-2-producing Klebsiella pneumoniae. Antimicrob Agents Chemother. 2014;58(2):1214–1217.
23. Habeeb MA, Haque A, Nematzadeh S, Iversen A, Giske CG. High prevalence of 16S rRNA methylase RmtB among CTX-M extended-spectrum beta-lactamase-producing Klebsiella pneumoniae from Islamabad, Pakistan. Int J Antimicrob Agents. 2013;41(6):524–526.
24. Antunes P, Machado J, Sousa JC, Peixe L. Dissemination of sulfonamide resistance genes (sul1, sul2, and sul3) in Portuguese Salmonella enterica strains and relation with integrons. Antimicrob Agents Chemother. 2005;49(2):836–839.
25. Latyshev Y, Mathew A, Jacobson JM, Sturm E. Purulent pericarditis caused by Haemophilus parainfluenzae. Tex Heart Inst J. 2013;40(5):608–611.
26. Huang SH, Wang CK, Peng HL, et al. Role of the small RNA RyhB in the Fur regulon in mediating the capsular polysaccharide biosynthesis and iron acquisition systems in Klebsiella pneumoniae. BMC Microbiol. 2012;12:148.
27. Liu Y, Liu PP, Wang LH, Wei DD, Wan LG, Zhang W. Capsular Polysaccharide Types and Virulence-Related Traits of Epidemic KPC-Producing Klebsiella pneumoniae Isolates in a Chinese University Hospital. Microb Drug Resist. 2017;23(7):901–907.
28. Cilloniz C, Rangel E, Barlascini C, Piroddi IM, Torres A, Nicolini A. Streptococcus pneumoniae-associated pneumonia complicated by purulent pericarditis: case series. J Bras Pneumol. 2015;41(4):389–394.
29. Chang SA. Tuberculous and Infectious Pericarditis. Cardiol Clin. 2017;35(4):615–622.
30. Manyi-Loh C, Mamphweli S, Meyer E, Okoh A. Antibiotic Use in Agriculture and Its Consequential Resistance in Environmental Sources: potential Public Health Implications. Molecules. 2018;23:4.
31. Du P, Zhang Y, Chen C. Emergence of carbapenem-resistant hypervirulent Klebsiella pneumoniae. Lancet Infect Dis. 2018;18(1):23–24.
32. Yang X, Sun Q, Li J, et al. Molecular epidemiology of carbapenem-resistant hypervirulent Klebsiella pneumoniae in China. Emerg Microbes Infect. 2022;11(1):841–849.
33. Gao H, Liu Y, Wang R, Wang Q, Jin L, Wang H. The transferability and evolution of NDM-1 and KPC-2 co-producing Klebsiella pneumoniae from clinical settings. EBioMedicine. 2020;51:102599.
34. Li J, Ren J, Wang W, et al. Risk factors and clinical outcomes of hypervirulent Klebsiella pneumoniae induced bloodstream infections. Eur J Clin Microbiol Infect Dis. 2018;37(4):679–689.
35. Wang TC, Lin JC, Chang JC, et al. Virulence among different types of hypervirulent Klebsiella pneumoniae with multi-locus sequence type (MLST)-11, Serotype K1 or K2 strains. Gut Pathog. 2021;13(1):40.
36. Dong N, Zhang R, Liu L, et al. Genome analysis of clinical multilocus sequence Type 11 Klebsiella pneumoniae from China. Microb Genom. 2018;4(2):3264.
37. Paczosa MK, Mecsas J. Klebsiella pneumoniae: going on the Offense with a Strong Defense. Microbiol Mol Biol Rev. 2016;80(3):629–661.
38. Sanikhani R, Moeinirad M, Shahcheraghi F, et al. Molecular epidemiology of hypervirulent Klebsiella pneumoniae: a systematic review and meta-analysis. Iran J Microbiol. 2021;13(3):257–265.
39. Zhao Y, Zhang X, Torres VVL, et al. An Outbreak of Carbapenem-Resistant and Hypervirulent Klebsiella pneumoniae in an Intensive Care Unit of a Major Teaching Hospital in Wenzhou, China. Front Public Health. 2019;7:229.
40. Yu WL, Ko WC, Cheng KC, et al. Association between rmpA and magA genes and clinical syndromes caused by Klebsiella pneumoniae in Taiwan. Clin Infect Dis. 2006;42(10):1351–1358.
41. Liu C, Guo J. Hypervirulent Klebsiella pneumoniae (hypermucoviscous and aerobactin positive) infection over 6 years in the elderly in China: antimicrobial resistance patterns, molecular epidemiology and risk factor. Ann Clin Microbiol Antimicrob. 2019;18(1):4.
42. Zhu J, Wang T, Chen L, Du H. Virulence Factors in Hypervirulent Klebsiella pneumoniae. Front Microbiol. 2021;12:642484.
43. Russo TA, MacDonald U, Hassan S, et al. An Assessment of Siderophore Production, Mucoviscosity, and Mouse Infection Models for Defining the Virulence Spectrum of Hypervirulent Klebsiella pneumoniae. mSphere. 2021;6(2):658.
44. Pulingam T, Parumasivam T, Gazzali AM, et al. Antimicrobial resistance: prevalence, economic burden, mechanisms of resistance and strategies to overcome. Eur J Pharm Sci. 2022;170:106103.
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.