Back to Journals » OncoTargets and Therapy » Volume 11
Reliability of using circulating tumor cells for detecting epidermal growth factor receptor mutation status in advanced non-small-cell lung cancer patients: a meta-analysis and systematic review
Authors Hu F, Mao XW , Zhang YJ, Zheng XX, Gu P, Wang HM, Zhang XY
Received 30 November 2017
Accepted for publication 9 January 2018
Published 14 March 2018 Volume 2018:11 Pages 1373—1384
DOI https://doi.org/10.2147/OTT.S158479
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Carlos E Vigil
Fang Hu,* Xiaowei Mao,* Yujun Zhang, Xiaoxuan Zheng, Ping Gu, Huimin Wang, Xueyan Zhang
Department of Pulmonary Medicine, Shanghai Chest Hospital, Shanghai Jiao Tong University, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Purpose: To evaluate the clinical value of circulating tumor cells as a surrogate to detect epidermal growth factor receptor mutation in advanced non-small-cell lung cancer (NSCLC) patients.
Methods: We searched the electronic databases, and all articles meeting predetermined selection criteria were included in this study. The pooled sensitivity, specificity, positive likelihood ratio, negative likelihood ratio, and diagnostic odds ratio were calculated. The evaluation indexes of the diagnostic performance were the summary receiver operating characteristic curve and area under the summary receiver operating characteristic curve.
Results: Eight eligible publications with 255 advanced NSCLC patients were included in this meta-analysis. Taking tumor tissues as reference, the pooled sensitivity, specificity, positive likelihood ratio, negative likelihood ratio, and diagnostic odds ratio of circulating tumor cells for detecting the epidermal growth factor receptor mutation status were found to be 0.82 (95% confidence interval [CI]: 0.50–0.95), 0.95 (95% CI: 0.24–1.00), 16.81 (95% CI: 0.33–848.62), 0.19 (95% CI: 0.06–0.64), and 86.81 (95% CI: 1.22–6,154.15), respectively. The area under the summary receiver operating characteristic curve was 0.92 (95% CI: 0.89–0.94). The subgroup analysis showed that the factors of blood volume, histological type, EGFR-tyrosine kinase inhibitor therapy, and circulating tumor cell and tissue test methods for EGFR accounted for the significant difference of the pooled specificity. No significant difference was found between the pooled sensitivity of the subgroup.
Conclusion: Our meta-analysis confirmed that circulating tumor cells are a good surrogate for detecting epidermal growth factor receptor mutation when tumor tissue is unavailable in advanced NSCLC patients, but more precise techniques are needed to improve their clinical efficiency.
Keywords: non-small-cell lung cancer, circulating tumor cell, epidermal growth factor receptor, meta-analysis
Introduction
Lung cancer has caused the most deaths related to cancer worldwide.1 Non-small-cell lung cancer (NSCLC) is the most common type of lung cancer.2 However, most NSCLC patients are diagnosed at an advanced stage and thus lose the chance to be eligible for surgery.3,4
A major progress in the last few years is the identification of epidermal growth factor receptor (EGFR) as a therapeutic target in a subgroup of NSCLC patients.5,6 Numerous clinical trials have confirmed that EGFR mutation is a reliable biomarker for EGFR-tyrosine kinase inhibitor (EGFR-TKI) therapy in NSCLC patients.7–9 Nowadays, EGFR-TKIs, such as gefitinib and erlotinib, have been suggested as first-line therapy in advanced NSCLC if patients harbor sensitive EGFR mutations.10 However, EGFR mutation status has to be known before administering EGFR-TKI therapy.5 Most NSCLC patients are diagnosed at advanced stage and cannot benefit from surgery. For advanced NSCLC patients the method of obtaining tumor tissue to detect EGFR mutation is limited, and it is sometimes also hard to get sufficient tumor tissue for further molecular testing.11 Besides, some patients cannot tolerate the invasive examination because of serious complications. In addition, most patients would develop acquired resistance during EGFR-TKI therapy,12 and so a repeat biopsy is required to understand the resistance mechanism, but patients always reject re-biopsy.13,14 Therefore, a viable and sensitive technique is required to monitor EGFR mutations in advanced NSCLC patients.
Liquid biopsy refers to the technology that makes full use of body fluids obtained noninvasively, such as peripheral blood, urine, etc, to study the tumor-related gene mutations.15 Circulating free DNA, circulating tumor cells (CTCs), and exosomes of peripheral blood are often selected to detect driver gene mutation.16–18 The CTC is the cell that the tumor mass sheds or spontaneously releases into the blood circulation;17 some research indicates that CTCs can reflect the mutation status in the tumor mass.17,19,20 Therefore, CTCs have been considered to be a potential surrogate for the detection of driver gene mutation.21 Some research groups have explored their clinical value and have shown that CTCs were reliable surrogates for detecting driver gene mutation status, such as EGFR mutations. Yet their sensitivity varies significantly in different studies, from 0.13 to 1.00.22,23
To address this issue, we performed this meta-analysis to evaluate the clinical value of CTCs as a surrogate for tumor tissue to detect EGFR in advanced NSCLC patients.
Materials and methods
Search strategy
We comprehensively searched the electronic databases PubMed, EMBASE, and Web of Science using the following search keywords: lung cancer, epidermal growth factor receptor or EGFR, circulating tumor cell or CTC, serum or plasma or blood, mutations. Alternative spellings and abbreviations were also considered. Reference lists of included studies and relevant reviews were also searched to identify additional studies. Besides, we also retrieved three meeting databases, including American Society of Clinical Oncology (ASCO), European Society for Medical Oncology (ESMO), and World Conference on Lung Cancer (WCLC). The literature search was conducted without any limitations, and the last search was performed on June 20, 2017.
Selection criteria
First, the records retrieved from the database and the reference list were filtered by title and summary, and then full-text articles of relevant studies were retrieved for further review. We selected eligible studies according to the following inclusion criteria: 1) all selected NSCLC patients should be diagnosed histopathologically or cytologically; 2) the study should compare the EGFR mutation between CTCs and tumor tissue; 3) the patients included in the study were either in the advanced stage or had relapsed and could not benefit from operation after multidisciplinary treatment; and 4) enough data was available to construct the diagnostic 2×2 table.
The exclusion criteria were as follows: 1) tumor tissues and CTCs were not paired; and 2) the reference EGFR status was not obtained from tumor tissue. The study selection process was performed independently by two authors, and any discrepancy was resolved by discussion with the third author.
Data extraction
Two operators extracted the following data independently from the included studies: author names, publication years, histological type, method of CTC isolation, the detection methods of EGFR in tissue and CTC specimens, blood volume, EGFR-TKI treatment between tissue and CTC specimens, true positive, false-positive (FP), false-negative (FN), and true negative. The methods with optimal sensitivity or specificity were extracted when a variety of methods were used to detect EGFR mutations in CTCs in one research article. The data was thoroughly examined and any discrepancies were resolved by a third person.
Quality assessment
Methodological quality of eligible studies was evaluated, using the quality assessment of diagnostic accuracy studies 2 (QUADAS-2), by two operators. QUADAS-2 is a useful tool which consists of four domains (patient selection, index test, reference standard, and flow and timing). Fourteen questions related to the quality of the article were judged as “yes,” “no,” or “unclear.”
Statistical analysis
True positive, FP, FN, and true negative from each eligible study were extracted. The pooled sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio, and corresponding 95% confidence intervals (95% CI) were calculated based on bivariate regression model. Summary receiver operative curve (SROC) and area under the SROC (AUSROC) were measured.
The Spearman correlation between the logit of sensitivity and logit of 1-specificity was calculated to determine the effect of threshold, and a P-value <0.05 indicated a significant threshold effect. The heterogeneity caused by non-threshold effect was measured by Q test and the inconsistency index I2, and a P-value ≤0.05 and I2 value ≥50% indicated that significant heterogeneity is not caused by the threshold effect. This was further verified by a Galbraith plot. If there is no heterogeneity between the studies, the points representing each study will fall within the range of the 95% boundaries. When there was significant heterogeneity, meta-regression was performed to detect the source.
All statistical analyses were performed using the STATA software (version 12.0, STATA Corp, College Station, TX, USA) with the MIDAS module.
Results
Study selection
Five hundred and eight studies were identified by the searches. After 125 duplicate reports were excluded, we excluded 348 inapposite publications by scanning titles and abstracts. After further investigation, 27 articles were removed (nine reviews, one non-English study, six articles that did not provide sufficient data to construct 2×2 tables, five in which CTC was not detected, and six that did not match CTCs and tissue). Besides, there was a study presented at a conference but not published in full, so this study was also not included. Finally, eight studies22–29 were eligible for analysis (Figure 1).
  | Figure 1 Flow diagram of study selection. |
Characteristics of the eligible studies
Baseline characteristics of eligible studies are shown in Table 1. All eligible studies were published between 2008 and 2017. A total of eight studies and 255 advanced NSCLC patients were included in the meta-analysis. Among them, three studies only enrolled adenocarcinoma patients, whereas the other five did not specify the type of cancer. The separation method for CTCs is classified as follows: three studies used Cellsearch, two studies used magnetic activated cell sorting, two studies used microfluidics, and one study used CTC-chip. Although the methods of CTC isolation were different in all the studies, all methods follow the same principle.30 The detection methods of EGFR in CTC specimens were classified as follows: five used Sanger sequencing, one used droplet digital polymerase chain reaction, one used next-generation sequencing (NGS), and one used magnetically sensed antibody sandwich assays. The detection methods of EGFR in tissue were classified as follows: four used Sanger sequencing, four used droplet digital polymerase chain reaction, and three did not specify the method. The blood volume used in six studies was 10 mL, one used 7.5 mL, and the other used 6 or 7.5 mL. Between the obtaining of tissue and CTC specimens, patients underwent EGFR-TKI treatment in five studies, while the patients in three studies were EGFR-TKI treatment-naïve (Table 1).
Six studies had QUADAS-2 scores ≥10. QUADAS-2 summary plot is presented in Figure S1. As shown, the methodological quality of eligible studies was not significantly affected by bias.
Accuracy of CTCs for detecting EGFR mutations
Taking tumor tissues as reference, the pooled sensitivity and specificity of CTCs for detecting the EGFR mutation status were 0.82 (95% CI: 0.50–0.95) and 0.95 (95% CI: 0.24–1.00), respectively (Figure 2). The PLR and NLR of CTCs were 16.81 (95% CI: 0.33–848.62) and 0.19 (95% CI: 0.06–0.64), respectively (Figure S2). The diagnostic odds ratio was 86.81 (95% CI: 1.22–6,154.15) (Figure S3). As shown in Figure 3, the AUSROC was 0.92 (95% CI: 0.89–0.94), indicating that CTCs had a high diagnostic accuracy. Fagan plot was generated for the visual presentation of diagnostic performance.
  | Figure 2 Forest plots of sensitivity and specificity. |
Threshold effect and heterogeneity
Threshold effect is the major source of heterogeneity. The Spearman correlation coefficient and P-value were calculated to evaluate the threshold effect, and were found to be −0.22 and 0.05, respectively, confirming that the threshold effect was not significant. As shown in the forest plots of accuracy data (Figure S2) and Galbraith plot (Figure 4), significant heterogeneity was detected. We used meta-regression to detect the source of heterogeneity. The data were divided into five subgroups, according to the volume of blood, histological type, sample size, whether CTC and tumor tissue test methods were consistent, and whether EGFR-TKI treatment was performed. Then, we analyzed the results of subgroup analysis. The subgroup analysis showed that the factors blood volume, histological type, EGFR-TKI, and CTC and tissue test methods for EGFR accounted for significant differences in the pooled specificities. No significant difference was found between the pooled sensitivity with regard to the subgroups (Table 2) (Figure 5).
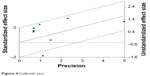  | Figure 4 Galbraith plot. |
  | Table 2 Subgroup analysis |
  | Figure 5 Forest plots of subgroup analysis. |
Publication bias
Deek’s funnel plot was used to test the publication bias. As shown in Figure S4, the funnel plot and P-value 0.793 (>0.05) suggested no evidence of publication bias.
Discussion
EGFR-TKIs are standard treatments for advanced NSCLC patients harboring activating EGFR mutations.31,32 EGFR mutation status should be known before adopting EGFR-TKI treatment.33 It is often considered that the tumor tissue is the gold standard sample for detecting EGFR mutations. But even in prospective, well-designed clinical trials, it is still impossible to obtain enough tumor tissue samples for molecular detection from one in three patients.34 This has become one of the major limitations of precision treatment for NSCLC.
CTCs are tumor cells that are shed from the tumor mass and circulate in the blood.17,35 Some studies have shown that CTCs can not only predict the prognosis in advanced NSCLC patients but also reflect the gene mutations in the tumor burden. A meta-analysis including 20 trials and 1,576 NSCLC patients confirmed that the presence of more CTCs indicates shorter progression-free survival and overall survival.36 The detection of driver gene mutation in CTC has been a hot topic in the last few years. Tissue biopsy is invasive and is limited by the intratumoral or intertumoral heterogeneity.15,37,38 However, CTCs overcome these shortcomings owing to the ease of obtaining the sample by noninvasive techniques, and they also provide an alternative for monitoring EGFR mutation in target therapy. Some studies have pointed out that CTCs are a good surrogate to monitor tumor gene mutations in advanced NSCLC owing to their noninvasive and reliable nature.35,39 Marchetti et al23 first reported that the CellSearch System coupled with NGS was a very sensitive diagnostic tool for EGFR mutation analysis in CTCs, and obtained a good result with sensitivity of 0.82. Furthermore, NGS showed the low-frequency mutations in tumor tissue which were missed by Sanger sequencing.23 In addition, a novel microfluidic device capable of specific selection and isolation of single rare cells within a mixed cell population was introduced by Yeo et al; it can not only detect genetic aberrations at the level of single cells, but also realize personalized therapies by tracking changes in anticancer therapies.28 Breitenbuecher et al26 have described a novel strategy based on CTC enrichment and highly sensitive detection of somatic mutations, and this strategy has shown that the problem of low CTC counts in stage IV NSCLC can be overcome. This may be another new step toward “liquid biopsy” for molecular diagnostics and disease monitoring in patients with advanced NSCLC.26 Another study conducted by Sundaresan et al27 found that CTCs as a liquid biopsy method can be combined with a tissue biopsy method and that this method provided a more complete assessment for patients with advanced lung cancer. Since the sensitivity of CTCs for detecting EGFR mutations varied significantly in different studies, we performed this meta-analysis to determine their diagnostic value. In our meta-analysis, good sensitivity and specificity results were seen. And, the high AUSROC (0.92) indicated a reliable diagnostic performance of CTCs. The PLR is high enough for clinical practice. However, the NLR is not low enough to exclude the FN cases. That is to say, if a negative result of EGFR is obtained, it should sometimes be confirmed by tumor tissue biopsy. In addition, since the NLR is 0.19, which is above 0.1, and the specificity ranges from 0.24 to 1.00, this means the specificity of CTCs in EGFR detection is not stable enough. In subgroup analysis, significant differences were found between the pooled specificities in terms of blood volume, histological type, EGFR-TKI-therapy, and CTC and tissue test methods for EGFR. No significant difference was found between the pooled sensitivity of the subgroups. Taken together, CTCs might be a suitable surrogate of tumor tissue for detection of EGFR mutation status in the real world, due to thier noninvasive features and high diagnostic value.
We observed that the sensitivity varied in different studies.22,23,26,28 The small sample size of these studies may have accounted for this. However, the meta-analysis overcomes the bias caused by insufficient sample size in individual studies, and no significant difference in the pooled sensitivity was shown in the subgroup analysis. Besides, a time interval was found in this low-sensitivity study between tumor tissue analysis and peripheral blood collection, and EGFR-TKI treatment was administered in this time interval. We inferred that the treatment was effective in killing tumor cells harboring active EGFR mutations, resulting in the decreased sensitivity of CTCs in EGFR detection. The three high-sensitivity studies were done in patients who had not undergone any targeted therapy. This may partly explain the differences in sensitivity that were obtained.40,41
Liu et al42 investigated the clinical value of detecting EGFR mutation status by CTCs in NSCLC patients and revealed that CTCs were a feasible and highly specific biomarker. In their study, they included cases regardless of tumor staging. However, only advanced NSCLC patients were enrolled in our study. For early-stage patients, the driver mutation status could be obtained using tumor tissue recovered during surgery. Also, liquid biopsy is applied in advanced NSCLC patients mostly owing to the difficulties in obtaining enough tumor biopsy samples, especially for target therapy patients. So, this study may be more meaningful for clinical practice. Some FN or FP cases were also observed in this study. Sundaresan et al27 pointed out that discordant detection results between tumor biopsy and blood-based analyses may result from technological differences, as well as sampling different tumor cell populations.
There are also some limitations in using CTCs to detect driver gene mutations. These include the fact that the heterogeneity of CTCs complicates identification, and the disparity of techniques used for CTC isolation will also limit the clinical use of CTCs.43,44 In addition, in some advanced NSCLC patients, CTCs in peripheral blood are not detected, due to the extreme limit of technical sensitivity.28 The limitations of this meta-analysis should also be emphasized. Only studies published in the English language were included in our meta-analysis, which could lead to selection bias. In addition, the sample size of 255 participants is relatively small, which will affect the statistical power. Therefore, a larger sample size and further research are required.
Conclusion
In summary, CTCs can be a good surrogate for detection of EGFR mutation when tumor tissue is unavailable, in advanced NSCLC patients. In addition, more precise techniques are needed to improve their clinical efficiency.
Acknowledgment
The study was supported by the National Natural Science Foundation of China (No. 81502450).
Disclosure
The authors report no conflicts of interest in this work.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. | ||
Ma K, Cohen V, Kasymjanova G, et al. An exploratory comparative analysis of tyrosine kinase inhibitors or docetaxel in second-line treatment of EGFR wild-type non-small-cell lung cancer: a retrospective real-world practice review at a single tertiary care centre. Curr Oncol. 2015;22:e157–e163. | ||
Chermiti Ben Abdallah F, Ben Ali G, et al. Treatment and prognosis of advanced stage non-small-cell lung cancer. Rev Mal Respir. 2014;31:214–220. | ||
Carnio S, Novello S, Mele T, Levra MG, Scagliotti GV. Extending survival of stage IV non-small cell lung cancer. Semin Oncol. 2014;4:69–92. | ||
Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. | ||
Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. | ||
Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010;11:121–128. | ||
Zhang L, Ma S, Song X, et al. Gefitinib versus placebo as maintenance therapy in patients with locally advanced or metastatic non-small-cell lung cancer (INFORM; C-TONG 0804): a multicentre, double-blind randomised phase 3 trial. Lancet Oncol. 2012;13:466–475. | ||
Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12:735–742. | ||
Ettinger DS, Wood DE, Akerley W, et al. NCCN guidelines insights: non-small cell lung cancer, version 4.2016. J Natl Compr Canc Netw. 2016;14:255–264. | ||
Fenizia F, De Luca A, Pasquale R, et al. EGFR mutations in lung cancer: from tissue testing to liquid biopsy. Future Oncol. 2015;11:1611–1623. | ||
Hammerman PS, Janne PA, Johnson BE. Resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Clin Cancer Res. 2009;15:7502–7509. | ||
Sun W, Yuan X, Tian Y, et al. Non-invasive approaches to monitor EGFR-TKI treatment in non-small-cell lung cancer. J Hematol Oncol. 2015;8:95. | ||
Majewski SA, Zuley ML, Pinnamaneni N, Ganott MA. Frequency of carcinoma at secondary imaging-guided percutaneous breast biopsy performed after a high-risk pathologic result at primary biopsy. AJR Am J Roentgenol. 2013;201:439–447. | ||
Ansari J, Yun JW, Kompelli AR, et al. The liquid biopsy in lung cancer. Genes Cancer. 2016;7:355–367. | ||
Sorber L, Zwaenepoel K, Deschoolmeester V, et al. Circulating cell-free nucleic acids and platelets as a liquid biopsy in the provision of personalized therapy for lung cancer patients. Lung Cancer. 2017;107:100–107. | ||
Zhang Z, Ramnath N, Nagrath S. Current status of CTCs as liquid biopsy in lung cancer and future directions. Front Oncol. 2015;5:209. | ||
Giallombardo M, Chacartegui Borras J, Castiglia M, et al. Exosomal miRNA analysis in Non-small Cell Lung Cancer (NSCLC) patients’ plasma through qPCR: a feasible liquid biopsy tool. J Vis Exp. 2016;111:e53900. | ||
Alix-Panabieres C, Pantel K. Circulating tumor cells: liquid biopsy of cancer. Clin Chem. 2013;59:110–118. | ||
Young R, Pailler E, Billiot F, et al. Circulating tumor cells in lung cancer. Acta Cytol. 2012;56:655–660. | ||
Yang B, Qin A, Zhang K, et al. Circulating tumor cells predict prognosis following tyrosine kinase inhibitor treatment in EGFR-mutant non-small-cell lung cancer patients. Oncol Res. 2017;25:1601–1606. | ||
Earhart CM, Hughes CE, Gaster RS, et al. Isolation and mutational analysis of circulating tumor cells from lung cancer patients with magnetic sifters and biochips. Lab Chip. 2014;14:78–88. | ||
Marchetti A, Del Grammastro M, Felicioni L, et al. Assessment of EGFR mutations in circulating tumor cell preparations from NSCLC patients by next generation sequencing: toward a real-time liquid biopsy for treatment. PLoS One. 2014;9:e103883. | ||
Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366–377. | ||
Punnoose EA, Atwal S, Liu W, et al. Evaluation of circulating tumor cells and circulating tumor DNA in non-small cell lung cancer: association with clinical endpoints in a phase II clinical trial of pertuzumab and erlotinib. Clin Cancer Res. 2012;18:2391–2401. | ||
Breitenbuecher F, Hoffarth S, Worm K, et al. Development of a highly sensitive and specific method for detection of circulating tumor cells harboring somatic mutations in non-small-cell lung cancer patients. PLoS One. 2014;9:e85350. | ||
Sundaresan T, Sequist L, Heymach J, et al. Detection of T790M, the acquired resistance EGFR mutation, by tumor biopsy versus noninvasive blood-based analyses. Clin Cancer Res. 2016;22:1103–1110. | ||
Yeo T, Tan SJ, Lim CL, et al. Microfluidic enrichment for the single cell analysis of circulating tumor cells. Sci Rep. 2016;6:22076. | ||
He J, Tan W, Ma J. Circulating tumor cells and DNA for real-time EGFR detection and monitoring of non-small-cell lung cancer. Future Oncol. 2017;13:787–797. | ||
Esmaeilsabzali H, Beischlag TV, Cox ME, Parameswaran AM, Park EJ. Detection and isolation of circulating tumor cells: principles and methods. Biotechnol Adv. 2013;31:1063–1084. | ||
D’Arcangelo M, Cappuzzo F. Erlotinib in the first-line treatment of non-small-cell lung cancer. Expert Rev Anticancer Ther. 2013;13:523–533. | ||
Wood DE, Kazerooni E, Baum SL, et al. Lung cancer screening, version 1.2015: featured updates to the NCCN guidelines. J Natl Compr Canc Netw. 2015;13:23–34. | ||
Sculier JP, Berghmans T, Meert AP. Advances in target therapy in lung cancer. Eur Respir Rev. 2015;24:23–29. | ||
Costa DB, Kobayashi S, Tenen DG, Huberman MS. Pooled analysis of the prospective trials of gefitinib monotherapy for EGFR-mutant non-small cell lung cancers. Lung Cancer. 2007;58:95–103. | ||
Tanaka F, Yoneda K, Hasegawa S. Circulating tumor cells (CTCs) in lung cancer: current status and future perspectives. Lung Cancer. 2010;1:77–84. | ||
Wang J, Wang K, Xu J, Huang J, Zhang T. Prognostic significance of circulating tumor cells in non-small-cell lung cancer patients: a meta-analysis. PLoS One. 2013;8:e78070. | ||
Xu M, Wang DC, Wang X, Zhang Y. Correlation between mucin biology and tumor heterogeneity in lung cancer. Semin Cell Dev Biol. 2017;64:73–78. | ||
Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. | ||
Tartarone A, Rossi E, Lerose R, et al. Possible applications of circulating tumor cells in patients with non small cell lung cancer. Lung Cancer. 2017;107:59–64. | ||
Shaw AT, Friboulet L, Leshchiner I, et al. Resensitization to crizotinib by the lorlatinib ALK resistance mutation L1198F. N Engl J Med. 2016;374:54–61. | ||
Yam I, Lam DC, Chan K, et al. EGFR array: uses in the detection of plasma EGFR mutations in non-small cell lung cancer patients. J Thorac Oncol. 2012;7:1131–1140. | ||
Liu Y, Xing Z, Zhan P, et al. Is it feasible to detect epidermal growth factor receptor mutations in circulating tumor cells in non-small cell lung cancer? A meta-analysis. Medicine. 2016;95:e5115. | ||
Calabuig-Farinas S, Jantus-Lewintre E, Herreros-Pomares A, Camps C. Circulating tumor cells versus circulating tumor DNA in lung cancer-which one will win? Transl Lung Cancer Res. 2016;5:466–482. | ||
Zhang C, Guan Y, Sun Y, Ai D, Guo Q. Tumor heterogeneity and circulating tumor cells. Cancer Lett. 2016;374:216–223. |
Supplementary materials
  | Figure S1 QUADAS-2 summary plot. |
  | Figure S2 Forest plots of PLR and NLR. |
  | Figure S3 Forest plot of diagnostic score and DOR. |
  | Figure S4 Deek’s funnel plot showed no significant publication bias. |
References
He J, Tan W, Ma J. Circulating tumor cells and DNA for real-time EGFR detection and monitoring of non-small-cell lung cancer. Future Oncol. 2017;13:787–797. | ||
Yeo T, Tan SJ, Lim CL, et al. Microfluidic enrichment for the single cell analysis of circulating tumor cells. Sci Rep. 2016;6:22076. | ||
Sundaresan T, Sequist L, Heymach J, et al. Detection of T790M, the acquired resistance EGFR mutation, by tumor biopsy versus noninvasive blood-based analyses. Clin Cancer Res. 2016;22:1103–1110. | ||
Breitenbuecher F, Hoffarth S, Worm K, et al. Development of a highly sensitive and specific method for detection of circulating tumor cells harboring somatic mutations in non-small-cell lung cancer patients. PLoS One. 2014;9:e85350. | ||
Earhart CM, Hughes CE, Gaster RS, et al. Isolation and mutational analysis of circulating tumor cells from lung cancer patients with magnetic sifters and biochips. Lab Chip. 2014;14:78–88. | ||
Marchetti A, Del Grammastro M, Felicioni L, et al. Assessment of EGFR mutations in circulating tumor cell preparations from NSCLC patients by next generation sequencing: toward a real-time liquid biopsy for treatment. PLoS One. 2014;9:e103883. | ||
Punnoose EA, Atwal S, Liu W, et al. Evaluation of circulating tumor cells and circulating tumor DNA in non-small cell lung cancer: association with clinical endpoints in a phase II clinical trial of pertuzumab and erlotinib. Clin Cancer Res. 2012;18:2391–2401. | ||
Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366–377. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.