Back to Journals » Infection and Drug Resistance » Volume 15
Relationship Between Antibiotic Resistance, Biofilm Formation, and Biofilm-Specific Resistance in Escherichia coli Isolates from Ningbo, China
Authors Qian W, Li X, Yang M, Liu C, Kong Y, Li Y, Wang T, Zhang Q
Received 3 March 2022
Accepted for publication 17 May 2022
Published 3 June 2022 Volume 2022:15 Pages 2865—2878
DOI https://doi.org/10.2147/IDR.S363652
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Weidong Qian,1 Xinchen Li,1 Min Yang,1 Chanchan Liu,2 Yi Kong,3 Yongdong Li,4 Ting Wang,1 Qian Zhang5
1School of Food and Biological Engineering, Shaanxi University of Science and Technology, Xi’an, 710021, People’s Republic of China; 2Xi’an Medical College, Xi’an, 710309, People’s Republic of China; 3Research Center for Tissue Repair and Regeneration Affiliated to the Medical Innovation Research Department, the General Hospital of the People’s Liberation Army, Beijing, 100048, People’s Republic of China; 4Ningbo Municipal Center for Disease Control and Prevention, Ningbo, 315010, People’s Republic of China; 5Department of Dermatology, Huazhong University of Science and Technology Union Shenzhen Hospital, Shenzhen, 518004, People’s Republic of China
Correspondence: Ting Wang; Qian Zhang, Tel +10 29-86168583, Email [email protected]; [email protected]
Purpose: Several Escherichia coli pathotypes still constitute an important public health concern owing to its pathogenicity and antimicrobial resistance. Moreover, biofilm formation of E. coli can allow the strains to interfere with host and antimicrobial eradication, thus conferring additional resistance. The association between the formation of biofilm and antimicrobial resistance determinants has been extensively exploited; nevertheless, there is still no definite conclusion. The purpose of this study was to provide additional data to augment the present knowledge about the subject.
Methods: Antibiotic resistance/susceptibility profiles of 81 isolates from pediatric individuals in China between 2011 and 2014 against 20 antibiotics were assessed using the VITEK 2 system. Biofilm-forming capacities were evaluated using the crystal violet staining method, confocal laser scanning microscopy (CLSM), and field emission scanning electron microscopy. Biofilm compositions inside the biofilm formed by representative strains were assessed using CLSM. The effects of antibiotics on biofilms generated by E. coli strains of different biofilm-forming ability were examined using CLSM in combination with gatifloxacin. The relationships between antibiotic resistance, biofilm formation, and biofilm-specific resistance in E. coli isolates were investigated.
Results: The results showed that 23 isolates were classified as multidrug-resistant, and 57 isolates were classified as extensively drug-resistant (XDR). Among the 69 isolates with the ability to form biofilms, 46 isolates were stronger biofilm formers. Correlation analysis demonstrated that strain populations exhibiting more robust biofilm formation likely contained larger proportions of XDR isolates.
Conclusion: Together, our study implies that there was an association between biofilm-formation and resistance to several antibiotics for XDR-E. coli isolates, and would provide novel insights regarding the prevention and treatment against E. coli-related infections.
Keywords: Escherichia coli, biofilm, antibiotic resistance, biofilm-specific resistance
Introduction
Antimicrobial agents play an important part in the treatment of bacterial infections to significantly reduce mortality and morbidity globally.1 However, the overuse of antibiotics in human and veterinary medicine has resulted in the selection of bacteria that are resistant to currently available antibiotics; in turn, this has allowed the propagation of drug-resistant strains in hospitals and communities, which poses a global challenge to public health.2–5 Among these infectious agents, one such bacterium is Escherichia coli (E. coli), a gram-negative, facultative anaerobic microbe that constitutes an intestinal commensal bacterium of humans and animals.6 However, infectious pathologies of E. coli also act as an opportunistic or true pathogen causing illnesses such as intestinal, extra-intestinal, urinary tract, and gastrointestinal infections, along with meningitis, peritonitis, and septicemia.7 For instance, uropathogenic E. coli (UPEC) is the most frequent cause of urinary tract infections (UTIs) and a common cause of severe, life-threatening sepsis.8,9 Especially, in the past two decades, E. coli has surged to the primary cause of early-onset bloodstream infection (BSI) in preterm infants, and the common and recognized cause of late-onset BSI.10–13 Comparatively, E. coli-caused BSIs are markedly associated with enhanced mortality compared with those triggered by gram-positive bacteria in hospitalized infants.14 In addition, E coli can lead to the potentially severe complication, including hemolytic uremic syndrome and acute diarrhea, which occurs most frequently in young children (age 5 and younger).15,16
Antibiotic resistance primarily derives from changes, or mutations, in the DNA of the bacteria in addition to acquisition of antibiotic resistance genes from other bacterial species through plasmid transfer. Particularly, E. coli can exhibit a powerful ability to obtain antibiotic resistance determinants, resulting in few remaining effective treatment options against its pathogenic variants.17 For instance, previous studies displayed that the E. coli ST131 clone is associated with resistance to multiple classes of antibiotics, including fluoroquinolones, third-generation cephalosporins and more recently last-line carbapenems and polymyxins.9,18–20 Fighting E. coli-related infections has been compromised by the increased emergence of resistance to various first-line antibiotics. Decreased susceptibility of E. coli to cephalosporins, including imipenem, meropenem, and ertapenem, among members of Enterobacteriaceae has emerged mainly due to the diffusion of extended-spectrum β-Lactamases (ESBLs) such as CTX-M, TEM, and SHV, and metallo-ß-Lactamases (MBLs).21,22 Recently, the emergence of ESBLs-producing E. coli has become a major concern and their emergence in infectious disease.23 MBLs are founded increasingly in gram-negative organisms and are recognized mostly in Pseudomonas and Klebsiella species.24,25 The emergence of Enterobacteriaceae harboring ESBL has resulted in reliance on colistin treatment in humans. Further, since a plasmid-borne colistin resistance gene (mcr-1) was detected in an E. coli isolate from China, several families of genes (mcr-1, mcr-2, mcr-3, mcr-4, and mcr-5) in E. coli isolates have been reported worldwide. Meanwhile, qnr and aac(6′)-Ib-cr genes are known to mediate the resistance to quinolones in Enterobacteriaceae, and are commonly found on the same resistance plasmids as ESBL, and thus, could represent a serious challenge for the public health.
Various kinds of bacteria can form biofilms as a survival strategy, distinct from other multidrug resistance mechanisms; this is especially relevant with regard to E. coli, which represents one of the most common causes of UTIs,26 and can cause biofilm-related contamination of medical devices, such as intravascular catheters, urinary catheters, and orthopedic implants.27,28 A biofilm is an assemblage of surface-associated microbial cells that are embedded in a self-produced matrix of extracellular polymeric substances. Importantly, bacterial cells within biofilms exhibit a set of emergent properties that differ substantially from their planktonic (freely suspended) counterparts.29 The biofilm formation ability is a feature common to most microorganisms in natural and medical systems, constituting a protected mode of growth that allows survival in the associated relatively harsh environmental conditions.30 Previous studies have demonstrated that extracellular matrix of biofilms aids in the adherence of E. coli to host cells, resists the shear forces associated with urine flow and contributes to the persistence of bacteria and chronicity of infection.31,32 Accordingly, biofilms were reported to be a major factor contributing to many chronic inflammatory diseases and involved in numerous serious consequences for public health.33,34 In China, some studies have reported the isolation of E. coli strains from humans; however, few studies have described E. coli strains isolated from infants and children younger than four years of age in conjunction with the associated biofilm formation ability.35,36
In addition, the relationship between antibiotic resistance and biofilm-production has attracted considerable interests from researchers. Previous studies have pointed out that biofilm formation can be induced by certain antibiotics under conditions of low doses, suggesting that regulation of biofilms may be involved in the global response to external stress, including antibiotics.37 Nevertheless, previous studies regarding quantitative correlation between biofilm formation and antibiotic resistance have yielded different and inconsistent results between different bacterial species, leaving researchers further explore the association.37,38 In this study, we investigated the relationship between antibiotic resistance, biofilm-formation, and biofilm-specific resistance in E. coli isolates derived from Chinese infants and children younger than four years of age in Ningbo. In addition, the possible relationship between the antibiotic resistance and the ability of biofilm-formation among these E. coli isolates was examined. The results could provide a foundation for further research regarding the potential mechanisms of biofilm-mediated enhanced resistance in E. coli.
Materials and Methods
Bacterial Strains and Growth Conditions
A total of 81 clinical strains were collected in the children’s hospital in Ningbo, Zhejiang province, China during 2011–2014 (Table S1). The range of age of the pediatric patients was from 11 minutes after birth to 4-year-old child; 34.6% were female and 65.4% were male. All isolates were grown at 37 °C and identified as E. coli using Matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry (MS) and Clinpro tools software (Bruker Biotyper; Bruker Daltonics, Bremen, Germany).39 The E. coli ATCC 19606 was used as the reference strain. One single bacterial colony was inoculated into a test tube containing 3 mL of tryptic soy broth (TSB) and grown at 37°C.
Antibacterial Susceptibility Testing with E. coli Isolates
The antibacterial susceptibility testing was performed usingthe disk diffusion and the VITEK 2 system (bioM’erieux, Marcy l’Etoile, France) and interpreted by the protocol specified in the guidelines of Clinical and Laboratory Standards Institute (CLSI, 2015). The antibacterial agents included cefoxitin, ceftriaxone, cefotaxime, cefepime, ceftazidime, cefazolin, moxifloxacin, ofloxacin, levofloxacin, gentamicin, amikacin, amoxycillin, minocycline, piperacillin, azithromycin, nitrofurantoin, polymyxin B, and meropenem (Thermo Fisher Scientific, USA). MDR isolates were defined as resistant to at least three different classes of antibiotics. Isolates were defined as extensively drug resistant (XDR) if they were not susceptible to at least one agent in all but two or fewer antibacterial categories (ie, bacterial isolates remained susceptible to only one or two antibacterial categories).
Biofilm Formation Assays
Biofilm production was assessed using a 96-well microtiter plate assay. After overnight growth in TSB, 150 μL of cell suspension were adjusted to the turbidity of a 0.5 McFarland standard and transferred into each microtiter plate well and incubated at 37 °C for 36 h. After three brief washes with 10 mM phosphate-buffered saline (PBS) solution and a 20-min fixation step with 180 μL methanol, the plate was stained with 180 μL 0.1% (v/v) crystal violet (CV) for 15 min and washed gently three times with 10 mM PBS. The formed biofilms were then dissolved with 180 μL 33% (v/v) acetic acid for 30 min. Sterile TSB was used as the negative control. The biofilm formation was measured at 550 nm optical density (OD550) using a microtiter plate reader (Thermo Fisher Scientific, Vantaa, Finland). The biofilm assays were performed in triplicate. According to the cut-off ODc value (the cut-off ODc value = the mean OD of the negative control + three standard deviations (SD) of the negative control), the biofilms were divided into the following four categories: OD ≤ ODc (non-biofilm producers), ODc < OD ≤ 2 ODc (low-biofilm producers), 2 ODc < OD ≤ 4 ODc (medium-biofilm producers), and OD > 4 ODc (strong-biofilm producers).40
Detection of Resistance Genes
Genomic DNA of E. coli isolates were extracted using the genomic DNA extraction kit (Magen Biotech, Guangzhou, China). All isolates were tested by PCR amplification of resistance genes that confer resistance to β-lactams (bla−TEM, bla−SHV, bla−OXA, bla−PSE, and bla−CTX-M), quinolones (qnrA, qnrB, qnrS, acc (6’)-Ib-cr, and qepA), aminoglycosides (acc (6’)-Ib, aac (3)-I, aac (3)-II, aac (3)-III, aac (3)-IV, and ant (2”)), tetracyclines (tetA and tetB), and polymyxins (mcr-1, mcr-2, mcr-3, mcr-4, and mcr-5). PCR primers were synthesized by Sangon Co., Ltd. (Shanghai, China), and shown in Table S2.
Confocal Laser Scanning Microscopy (CLSM) Analysis of Biofilm Structure
CLSM (LSM800, Carl Zeiss AG, Oberkochen, Germany) was applied to observe biofilm structure.41 After incubation overnight at 37°C, 1 mL of cell suspensions (approximately 1×106 CFU/mL) were added into each well of a 24-well microtiter plate containing a sterile coverslip and then cultured at 37 °C under 50 rpm for 36 h to form biofilms. Subsequently, the biofilms on the glass coverslips were washed three times with 10 mM PBS to remove unattached cells, and then stained with 5 μM SYTO 9 (Thermo Fisher Scientific, USA) for 15 min. Biofilms were visualized and imaged using CLSM, where the excitation/emission wavelength was 483/502 nm for SYTO 9.
Field Emission Scanning Electron Microscope (FESEM) Observations of Biofilms
For visualization of biofilms, biofilms of representative isolates were observed by FESEM (Nova Nano SEM-450, FEI, Hillsboro, OR, USA).41 One mL of cell suspension of E. coli isolates (approximately 1×106 CFU/mL) was transferred into each well of a 24-well microtiter plate containing a sterile glass coverslip, cultured at 37 °C under shaking conditions of 50 rpm for another 36 h, and fixed with 2.5% (v/v) glutaraldehyde solution for 4 h at 4 °C. The biofilms were then dehydrated in a series of washes with 30, 50, 70, and 90% ethanol for 10 min each, followed by 15 min rinses in 100% ethanol. Air-dried samples were immediately sputter-coated with platinum and subjected to FESEM.
CLSM Analysis of Biofilm Composition
CLSM analysis of the composition within biofilms was performed according to Qian et al with a few modifications.41 The biofilms of representative E. coli isolates were grown on a 24-well microtiter plate with glass coverslips for 36 h at 37 °C with the stirring speed of 50 rpm. Following incubation, the biofilms were rinsed with 10 mM PBS and stained with three types of fluorescent markers for 15 min in the dark: (i) 4′, 6-diamidino-2-phenylindole (DAPI, Sigma-Aldrich, USA) to stain extracellular DNA (eDNA); (ii) Film Tracer SYPRO Ruby biofilm matrix stain (SYPRO Ruby, Invitrogen, Paisley, UK), which labels most classes of proteins; and (iii) wheat germ agglutinin (WGA, Invitrogen, Paisley, UK), which stains N-acetyl-D-glucosamine residues. Finally, samples were washed with 10 mM PBS to remove residual dyes and observed under CLSM. The fluorescences were detected using the following combination of laser excitation and emission band-pass wavelengths: 358/461 nm for DAPI, 450/610 nm for SYPRO Ruby, and 495/519 for WGA.
Diffusion Assay of Gatifloxacin Within Biofilms
To evaluate the diffusion of the gatifloxacin within biofilms produced by the strong, medium and weak biofilm producer of E. coli, gatifloxacin with the intrinsic fluorescence was used to evaluate antibiotics diffusion within the biofilm by CLSM.41 The biofilms were formed on the glass coverslips inside each well of a 24-well microtiter plate and cultured at 37 °C, 50 rpm for 36 h. The coverslips were washed gently three times with 10 mM PBS, and gatifloxacin was added at a final concentration of 0.4 mg/mL, and the coverslips were further incubated at 37 °C for 4 h. Next, 5 μM SYTO 9 was added and co-incubated for 15 min, and the biofilms were then washed three times with 10 mM PBS and observed using CLSM. The emission peak for gatifloxacin was recorded at 495 nm upon excitation at 291 nm. Finally, three random fields were visualized for each biofilm, and representative images were captured.
Statistical Analysis
Spearman’s rank correlation tests were used for intergroup comparisons. The Wilcoxon rank sum test was used for comparison of biofilm formation between isolates susceptible/non-susceptible to each antimicrobial category. The chi-square test was used to analyze the correlation between biofilm formation ability, antibiotic resistance genes, and antibiotic resistance. Data analyses were performed using SPSS (version 20.0 SPSS Statistics, Inc., IBM, Armonk, NY, USA). p < 0.05 was considered statistically significant for all tests.
Results and Discussion
Antimicrobial Susceptibility of Planktonic E. coli Isolates
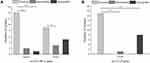
The resistance patterns of E. coli isolates to the tested antimicrobial agents are presented in Figure 1B and Table 1. The phenotypic resistance profiles of the E. coli isolates were as follows: amoxycillin, 96.30% (78/81); cefotaxime, 69.14% (56/81); ceftriaxone, 66.67% (54/81); cefazolin, 64.20% (52/81); cefepime, 61.73% (50/81); azithromycin, 61.73% (50/81); moxifloxacin, 50.62% (41/81); ceftazidime, 33.33% (27/81); ofloxacin, 30.86% (25/81); gentamicin, 29.63% (24/81); levofloxacin, 24.69% (20/81); minocycline, 22.22% (18/81); nitrofurantoin, 14.81% (12/81); cefoxitin, 6.17% (5/81); amikacin, 6.17% (5/81), and piperacillin, 3.70% (3/81). The relatively high resistance rates for cephalosporins and ofloxacin among 81 E. coli isolates are similar to several recent reports, indicating that the resistance levels of uropathogenic E. coli against fluoroquinolones and cephalosporins in Iran is ranging between 15–66% and 30–60%, respectively.32,42 Moreover, the highest and the relatively lower resistance rate among 81 E. coli isolates was related to amoxycillin, as well as to piperacillin and amikacin, respectively, which is inconsistent with the studies performed by others.43,44 These studies demonstrated that resistance to piperacillin and amikacin antibiotics was lower than other antibiotics. For instance, Allami et al reported that the uropathogenic E. coli isolates from southern Iraq were most resistant to piperacillin, ticarcillin, amoxicillin/clavulanic acid (92%, 91%, and 88%) and most sensitive to amikacin and imipenem, respectively. Accordingly, both piperacillin and amikacin may be recommended as the last-line option for clinical treatment of infections caused by E. coli when there is no effective treatment available in Ningbo. In addition, a worrisome finding from this study is a relatively high resistance rate for nitrofurantoin (14.81%) in isolates of E. coli. This is similar to a previous study demonstrating that E. coli isolates were resistant to nitrofurantoin (21%),45 but in contrary to a few previous studies with a low prevalence of nitrofurantoin resistance in E. coli isolates.46,47 The potential explanation of these resistance to antibiotics could be attributed to the inappropriate and excessive use of these antibiotics in the empirical treatment for E. coli-related infections in different parts of the world.48 Additionally, according to the limited data, antibiotic resistance rates of cefotaxime, cefepime, ceftazidime, and ofloxacin were higher in Zhejiang (69.14, 61.73, 33.33, and 30.86%) than in Shanghai city (24.5, 23.5, 12.4, and 3.4%), respectively.35 Thus, it is crucial to evaluate the local resistance rates of specific pathogens to antibiotics, and area-specific data on antimicrobial resistance could help physicians in prescribing choices for empirical antibiotic treatment, thereby reducing overuse of antibiotics for the treatment of infections. Meanwhile, among the 81 E. coli isolates tested, all isolates were resistant to at least one antibiotics; no isolates were resistant to all of the 18 antibiotics. Specifically, 23 isolates were classified as MDR, and 57 isolates were classified as XDR. As such, 98.77% (80/81) of the isolates were either MDR or XDR, indicating the prevalence rate of MDR or XDR isolates in Ningbo was higher in comparison to in North America and Europe. Such high resistance in E. coli isolates may be attributed to clinical pattern of overuse and inappropriate use of antibiotics in previous years.49
 |
Table 1 Correlation Between the Level of Biofilm Formation and Resistance to 16 Antibioticals in E. coli Clinical Isolates |
The Relationship Between Antibiotic Resistance and Resistance Genes
The frequency of resistance genes is shown in Table 2. Of the E. coli isolates analyzed, 61.7% (50/81) carried one or more resistance genes. Among the 20 resistance genes investigated, aac (3)-II, acc (6’)-Ib-cr, and ant (2”) were most frequently detected, demonstrating 32.10 (26/81), 20.99 (17/81), and 20.99% (17/81) frequency, respectively. The frequency of the other resistance genes was as follows: tetA (12.35%, 10/81), bla−TEM (8.64%, 7/81), bla−OXA (1.23%, 1/81), qepA (1.23%, 1/81), and aac (3)-IV (1.23%, 1/81). By contrast, bla−SHV, bla−PSE, bla−CTX-M, qnrA, qnrB, qnrS, aac (3)-I, aac (3)-III, acc (6’)-Ib, and tetB were not detected in E. coli isolates. Our results with low prevalence of ESBL-producing genes are inconsistent with several previous studies in different regions of the world, including Saudi Arabia, Vietnam, China, Iran, and Mexico, which highlighted the CTX-M, SHV, and TEM genes as the most critical mechanisms of ESBL production in E. coli strains.50 The prevalence of blaTEM and blaOXA genes was more common in our settings compared with blaCTX-M and blaSHV genes. This is similar to the report by Sid Ahmed et al, revealing that the recent emergence of CTX-M group was predominantly mediated through the mutations of blaTEM and blaSHV genes.51 The resistance gene tetA (12.35%) detection rate of clinical E. coli isolates was lower, but the aac (3)-IV (1.23%) was higher than that reported by Momtaz et al.52 In addition, the percentage of isolates carrying the bla−TEM gene was 8.64% in the present study, which is lower than that in a previous report by Nawaz et al.53
 |
Table 2 The Frequency of Antibiotic Resistance Genes Among 81 Isolates |
The Association Between Biofilm Formation and Antibiotic Resistance
The biofilm production ability of MDR or XDR strains enhances the overall resistance, potentially resulting in treatment failure. In this context, awareness of the definite relationship between antibiotic resistance and biofilm formation ability of bacteria could help clinicians in prescribing regimes that specifically address biofilm to the treatment arsenal against MDR or XDR infections. The correlation between biofilm formation and the pattern of antibiotic resistance has been the focus of significant research in recent years; however, to date, definite conclusions cannot be drawn regarding the relationship since there have been some studies that have reported conflicting results.32 In this study, the biofilm-forming capacity of E. coli isolates was summarized in Figure 1A. Sixty-nine isolates (85.19%, 69/81) exhibited the ability to produce biofilms, among which 46 isolates (56.79%, 46/81) were strong biofilm producers, 15 isolates (18.52%, 15/81) were medium biofilm producers, 8 isolates (9.88%, 8/81) represented weak biofilm producers, and 12 isolates (14.81%, 12/81) were non-biofilm producers. Among the 46 strong-biofilm producers, only 2.17% (1/46) were non-MDR isolates, 19.57% (9/46) constituted MDR isolates, and 78.26% (36/46) represented XDR isolates. The 15 medium-biofilm producers comprised 33.33% (5/15) MDR and 66.67% (10/15) XDR isolates. By contrast, the 8 weak-biofilm producers consisted of 50.00% MDR and 50.00% XDR isolates. The 12 strains that were negative for biofilm formation consisted of 58.33% (7/12) MDR and 41.67% (5/12) XDR isolates (Figure 1B). These compositional proportions indicated that XDR isolates tended to form stronger biofilms, revealing that the populations that exhibited more robust biofilm formation likely contained larger proportions of XDR isolates. The Spearman correlation coefficient (rs) for this comparison was 0.280 (p < 0.01). Similarly, Dumaru et al, found that 62.7% of isolates were biofilm-producers in gut bacteria, and that there was strong association between the MDR-status and biofilm-production.54 Avila-Novoa et al explored the correlation between biofilm-production and MDR in Acinetobacter baumannii (A. baumannii) using the phenotypic method, and found that 73.3% of isolates were biofilm-producers; however, in their study, there is no clear association between biofilm-formation and specific antibiotic resistance was revealed.55 Next, XDR E. coli isolates tended to form stronger biofilms than MDR and non-MDR strains; this conclusion was also confirmed by statistical analyses (rs = 0.243, p < 0.05; Table 2), indicating a positive correlation between biofilm formation capacity and antibiotic resistance phenotypes. Similarly, XDR E. coli isolates revealed a greater potential to be strong biofilm producers than MDR or non-MDR E. coli isolates.56 These results were in agreement with a previous study displaying that biofilm-forming E. coli isolates from clinical isolates in Uganda were more resistant than the non-biofilm formers with 64% being MDR as compared to 36% among the non-biofilm forming E. coli isolates. Similarly, among 130 E. coli isolates from patients having UTI symptoms in Iran, 80 (61.53%, 80/130) were able to form biofilms, and maximum resistance to ampicillin (87.5%), followed by tetracycline (75%), nalidixic acid (72.5%) and co-trimoxazole (71.25%).57 These observations indicate that the biofilm formation by E. coli isolates tested was potentially associated with MDR bacteria, as well as made their eradication difficult, suggesting that exploiting the factors associated with biofilm formation is key to the development of new therapies.58
Meanwhile, we also investigated the relationship between biofilm formation and specific antibiotic resistance. The results revealed that isolates resistant to cefoxitin, ceftriaxone, cefazolin, and gentamicin could form stronger biofilms than those susceptible or exhibiting intermediate resistance, being indicative of a positive correlation between biofilm quantity and resistance profile (rs = 0.233–0.253, p < 0.05; Table 3). A similar report demonstrated that gentamicin-resistance and ceftazidime-resistance were related to biofilm-formation in E. coli.59 Moreover, it was previously reported that, for E. coli strains with strong biofilm formation ability were more resistant to amoxicillin-clavulanic acid, norfloxacin, cotrimoxazole, gatifloxacin, and gentamicin, suggesting that the biofilms formed by these strains provide the ability to survive when exposed to these antibiotics.60 The correlation between biofilm formation and resistance to the eight antimicrobial categories was also evaluated. For six of the categories, including cephalosporins and quinolones, fluoroquinolones, aminoglycosides, tetracyclines, macrolides, and nitrofurans, non-susceptible isolates could form stronger biofilms than susceptible isolates (p < 0.05; Figure 3A–F). Conversely, for β-lactams and lipopeptides, no significant difference in biofilm formation between susceptible and non-susceptible isolates was observed (Figure 3G, and H). Perez et al exploited the correlation of meropenem-resistance and the ability to form biofilms,61 and found an inverse relationship between carbapenem-resistance and biofilm-production. In addition, Fábréga et al demonstrated an inverse relationship between biofilm-formation and quinolone-resistance in the context of Salmonella enterica.62 Together, there are discrepancies in the relationship between antibiotic resistance and biofilm formation among different strains,32 but further efforts are still needed to optimize the treatment regime for clinician.
 |
Table 3 Biofilm Forming Capacities of E. coli with Different Antibiotic Resistance Phenotypes |
CLSM and FESEM Analysis of Biofilm Morphology and Structure
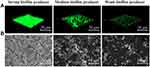
36-h biofilm structures of representative isolates observed using CLSM are presented in Figure 4A. Marked variability in three-dimensional biofilm architecture between the different isolates were displayed. Weak biofilm producers formed only a few, small scattered cell clusters, whereas medium biofilm producer formed rough biofilms containing several small aggregates and were of variable thickness. In contrast, strong biofilm producers displayed a high degree of variability in terms of biofilm structure. FESEM examination of biofilms cultivated for 36 h revealed that E. coli isolates exhibited disparate biofilm-forming abilities and adopted various structural conformations. Strong biofilm producers displayed thick-cell layers and medium biofilm producers shaped rough biofilms containing several small aggregates and were of variable thickness, while weak biofilm producers only a few, small scattered cell clusters (Figure 4B). Similarly, strong biofilm-producing bacteria such as Vibrio parahaemolyticus established thick 3-D structures, whereas poor-biofilm-forming strains produced thin and inconsistent biofilms.63
Biofilm Composition by CLSM
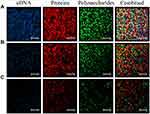
CLSM in conjugation with three different fluorescent dyes was used to investigate the distribution of eDNA, proteins, and polysaccharides within the biofilm. As demonstrated in Figure 5A, eDNA, proteins, and polysaccharides were at high levels and uniformly distributed in strong biofilm producers, whereas weak biofilm producers revealed lower biomass level and the proteins and polysaccharides were the major components within biofilms (Figure 5B, and C).
Diffusion of Gatifloxacin Within Biofilms
To evaluate the contribution of the resistance of biofilms to antibiotics, gatifloxacin diffusion within biofilms was visualized using CLSM. As presented in Figure 6, CLSM revealed that for the biofilms produced by the strong biofilm producers, gatifloxacin (blue fluorescence) was mainly confined to the outer periphery of the biofilm, with little or no gatifloxacin penetration into the biofilm interior. In contrast, more penetration of gatifloxacin into the biofilm by the weak-biofilm producers was observed. Moreover, CLSM clearly revealed extensive gatifloxacin diffusion within biofilms by the weak-biofilm producers, almost reaching the basal layers. The results indicated that dense biofilms limited the penetration antimicrobial agents through the biofilm matrix due to their dense and thick structure, and thus could contribute to the antimicrobial tolerance of biofilms.64
Conclusion
In this study, we evaluated the relationship between antibiotic resistance, biofilm formation, and biofilm-specific resistance of E. coli isolates from pediatric patients in Ningbo, China. The current study showed that the clinical isolates of E. coli from patients in Ningbo, China, demonstrated a positive association between the MDR or XDR phenotype and biofilm-production, and the correlation between biofilm-formation and cefoxitin, ceftriaxone, cefazolin, and gentamicin resistance was observed. Therefore, a local and regular surveillance of biofilm-formation in E. coli isolates and their antimicrobial resistance profiles should be performed. This might help to formulate therapeutic strategies against E. coli-related infections. Moreover, in this study, our sample size is a little small and only from a geographic location. Expanding the sample size and geographic area in order to cast light on how different geographical regions and patient populations contribute to the selection of pathogens with different profiles of biofilm and its resistance should await further study.
Ethics Approval and Consent to Participate
This study received approval from the Ethical Committee (IRB) of Ningbo Municipal Center for Disease Control and Prevention and was conducted in accordance with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. Since this study was observational and strains from redundant, abandoned individual excreta were employed in clinical diagnosis, the confidentiality of patient data is preserved. Considering this study did not affect patients’ health and privacy, the exemption criteria were met. After consulting with the IRB of the Ningbo Municipal Center for Disease Control and Prevention, written informed consent from the patient or parents was not required.
Acknowledgments
This work was supported in part by the national natural science foundation (11975177),the science and technology plan project of Xianyang science and technology bureau (2021ZDYF-NY-0007), the China postdoctoral science foundation (2019M653938), and Xi’an medical college (22Z01).
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval for the version to be published; have agreed on the journal to which the article has been submitted; and agreed to be accountable for all aspects of the work.
Disclosure
The authors claim no conflicts of interest in this study.
References
1. Tadesse DA, Zhao S, Tong E, et al. Antimicrobial drug resistance in Escherichia coli from humans and food animals, United States, 1950-2002. Emerg Infect Dis. 2012;18(5):
2. Kahn LH. Antimicrobial resistance: a one health perspective. Trans R Soc Trop Med Hyg. 2017;111(6):255–260. doi:10.1093/trstmh/trx050
3. Qiao M, Ying GG, Singer AC, et al. Review of antibiotic resistance in China and its environment. Environ Int. 2018;110:160–172. doi:10.1016/j.envint.2017.10.016
4. Pokharel S, Shrestha P, Adhikari B. Antimicrobial use in food animals and human health: time to implement ‘one health’ approach. Antimicrob Resist Infect Control. 2020;9(1):181. doi:10.1186/s13756-020-00847-x
5. Shin B, Park W. Zoonotic diseases and phytochemical medicines for microbial infections in veterinary science: current state and future perspective. Front Vet Sci. 2018;5:166. doi:10.3389/fvets.2018.00166
6. Jang J, Hur HG, Sadowsky MJ, et al. Environmental Escherichia coli: ecology and public health implications-a review. J Appl Microbiol. 2017;123(3):570–581. doi:10.1111/jam.13468
7. Denamur E, Clermont O, Bonacorsi S, et al. The population genetics of pathogenic Escherichia coli. Nat Rev Microbiol. 2021;19(1):37–54. doi:10.1038/s41579-020-0416-x
8. Flores-Mireles AL, Walker JN, Caparon M, et al. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol. 2015;13(5):269–284. doi:10.1038/nrmicro3432
9. Alvarez-Fraga L, Phan MD, Goh KGK, et al. Differential Afa/Dr fimbriae expression in the multidrug-resistant Escherichia coli ST131 clone. mBio. 2022;13:e0351921. doi:10.1128/mbio.03519-21
10. Stoll BJ, Hansen N, Fanaroff AA, et al. Changes in pathogens causing early-onset sepsis in very-low-birth-weight infants. N Engl J Med. 2002;347(4):240–247. doi:10.1056/NEJMoa012657
11. Hornik CP, Fort P, Clark RH, et al. Early and late onset sepsis in very-low-birth-weight infants from a large group of neonatal intensive care units. Early Hum Dev. 2012;88(Suppl):S69–S74. doi:10.1016/S0378-3782(12)70019-1
12. Ecker KL, Donohue PK, kim KS, et al. The impact of group B streptococcus prophylaxis on late-onset neonatal infections. J Perinatol. 2013;33(3):206–211. doi:10.1038/jp.2012.76
13. Bergin SP, Thaden JT, Ericson JE, et al. Neonatal Escherichia coli bloodstream infections: clinical outcomes and impact of initial antibiotic therapy. Pediatr Infect Dis J. 2015;34(9):933–936. doi:10.1097/INF.0000000000000769
14. Bauserman MS, Laughon MM, Hornik CP, et al. Group B Streptococcus and Escherichia coli infections in the intensive care nursery in the era of intrapartum antibiotic prophylaxis. Pediatr Infect Dis J. 2013;32(3):208–212. doi:10.1097/INF.0b013e318275058a
15. Eybpoosh S, Mostaan S, Gouya MM, et al. Frequency of five Escherichia Coli pathotypes in Iranian adults and children with acute diarrhea. PLoS One. 2021;16(2):e0245470. doi:10.1371/journal.pone.0245470
16. Ko H, Maymani H, Rojas-Hernandez C. Hemolytic uremic syndrome associated with Escherichia coli O157: h7 infection in older adults: a case report and review of the literature. J Med Case Rep. 2016;10(175). doi:10.1186/s13256-016-0970-z
17. Moradigaravand D, Palm M, Farewell A, et al. Prediction of antibiotic resistance in Escherichia coli from large-scale pan-genome data. PLoS Comput Biol. 2018;14(12):e1006258. doi:10.1371/journal.pcbi.1006258
18. Rogers BA, Sidjabat HE, Paterson DL. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J Antimicrob Chemother. 2011;66(1):1–14. doi:10.1093/jac/dkq415
19. Petty NK, Ben Zakour NL, Stanton-Cook M, et al. Global dissemination of a multidrug resistant Escherichia coli clone. Proc Natl Acad Sci USA. 2014;111(15):5694–5699. doi:10.1073/pnas.1322678111
20. Roer L, Hansen F, Stegger M, et al. Novel mcr-3 variant, encoding mobile colistin resistance, in an ST131 Escherichia coli isolate from bloodstream infection, Denmark, 2014. Euro Surveill. 2017;22(31). doi:10.2807/1560-7917.ES.2017.22.31.30584
21. Behzadi P, Garcia-Perdomo HA, Karpinski TM, et al. Metallo-β-lactamases: a review. Mol Biol Rep. 2020;47(8):6281–6294. doi:10.1007/s11033-020-05651-9
22. Issakhanian L, Behzadi P. Antimicrobial agents and urinary tract infections. Curr Pharm Des. 2019;25(12):1409–1423. doi:10.2174/1381612825999190619130216
23. Alizade H, Fallah F, Ghanbarpour R, et al. Phylogenetic groups, extended-spectrum β-lactamases and metallo-β-lactamase in Escherichia coli isolated from fecal samples of patients with diarrhea in Iran. Gastroenterol Hepatol Bed Bench. 2015;8(3):207–214.
24. Poirel L, Lagrutta E, Taylor P, et al. Emergence of Metallo-β-Lactamase NDM-1-producing multidrug-resistant Escherichia coli in Australia. Antimicrob Agents Chemother. 2010;54(11):4914–4916. doi:10.1128/AAC.00878-10
25. Nepal K, Pant ND, Neupane B, et al. Extended spectrum beta-lactamase and metallo beta-lactamase production among Escherichia coli and Klebsiella pneumoniae isolated from different clinical samples in a tertiary care hospital in Kathmandu, Nepal. Ann Clin Microbiol Antimicrob. 2017;16(1):62. doi:10.1186/s12941-017-0236-7
26. Wiedemann B, Heisig A, Heisig P. Uncomplicated urinary tract infections and antibiotic resistance-epidemiological and mechanistic aspects. Antibiotics. 2014;3(3):341–352. doi:10.3390/antibiotics3030341
27. Francolini I, Donelli G. Prevention and control of biofilm-based medical-device-related infections. FEMS Immunol Med Microbiol. 2010;59(3):227–238. doi:10.1111/j.1574-695X.2010.00665.x
28. Singhai M, Malik A, Shahid M, et al. A study on device-related infections with special reference to biofilm production and antibiotic resistance. J Glob Infect Dis. 2012;4(4):193–198. doi:10.4103/0974-777X.103896
29. Flemming HC, Wingender J, Szewzyk U, et al. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol. 2016;14(9):563–575. doi:10.1038/nrmicro.2016.94
30. McDougald D, Rice SA, Barraud N, et al. Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat Rev Microbiol. 2011;10(1):39–50. doi:10.1038/nrmicro2695
31. Delcaru C, Alexandru I, Podgoreanu P, et al. Microbial biofilms in urinary tract infections and prostatitis: etiology, pathogenicity, and combating strategies. Pathogens. 2016;5(4):65. doi:10.3390/pathogens5040065
32. Behzadi P, Urban E, Gajdacs M. Association between biofilm-production and antibiotic resistance in uropathogenic Escherichia coli (UPEC): an in vitro study. Diseases. 2020;8(2):17. doi:10.3390/diseases8020017
33. Omar A, Wright JB, Schultz G, et al. Microbial biofilms and chronic wounds. Microorganisms. 2017;5(1):9. doi:10.3390/microorganisms5010009
34. Vestby LK, Gronseth T, Simm R, et al. Bacterial biofilm and its role in the pathogenesis of disease. Antibiotics. 2020;9(2):59. doi:10.3390/antibiotics9020059
35. Huang Z, Pan H, Zhang P, et al. Prevalence and antimicrobial resistance patterns of diarrheagenic Escherichia coli in shanghai, China. Pediatr Infect Dis J. 2016;35(8):835–839. doi:10.1097/INF.0000000000001190
36. Tapiainen T, Hanni AM, Salo J, et al. Escherichia coli biofilm formation and recurrences of urinary tract infections in children. Eur J Clin Microbiol Infect Dis. 2014;33(1):111–115. doi:10.1007/s10096-013-1935-4
37. Qi L, Li H, Zhang C, et al. Relationship between antibiotic resistance, biofilm formation, and biofilm-specific resistance in Acinetobacter baumannii. Front Microbiol. 2016;7(483). doi:10.3389/fmicb.2016.00483
38. Abidi SH, Sherwani SK, Siddiqui TR, et al. Drug resistance profile and biofilm forming potential of Pseudomonas aeruginosa isolated from contact lenses in Karachi-Pakistan. BMC Ophthalmol. 2013;13:57. doi:10.1186/1471-2415-13-57
39. Khot PD, Fisher MA. Novel approach for differentiating shigella species and Escherichia coli by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2013;51(11):3711–3716. doi:10.1128/JCM.01526-13
40. Stepanovic S, Vukovic D, Hola V, et al. Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS. 2007;115(8):891–899. doi:10.1111/j.1600-0463.2007.apm_630.x
41. Qian W, Yang M, Li X, et al. Anti-microbial and anti-biofilm activities of combined chelerythrine-sanguinarine and mode of action against Candida albicans and Cryptococcus neoformans in vitro. Colloids Surf B Biointerfaces. 2020;191:111003. doi:10.1016/j.colsurfb.2020.111003
42. Alizade H. Escherichia coli in Iran: an overview of antibiotic resistance: a review article. Iran J Public Health. 2018;47(1):1–12.
43. Ahmed N. Antibiotic resistance profile in relation to virulence genes fimH, hlyA and usp of uropathogenic E. coli isolates in Lahore, Pakistan. Trop Biomed. 2020;36(2):559–568.
44. Salehzadeh A, Zamani H. Characterization of (Uropathogenic) E. coli isolated from urinary tract infections: phylogenetic typing and distribution of virulence-associated traits. Br J Biomed Sci. 2018;75(1):40–42. doi:10.1080/09674845.2017.1336834
45. Allami M, BahreiniM, Sharifmoghadam MR. Antibiotic resistance, phylogenetic typing, and virulence genes profile analysis of uropathogenic Escherichia coli isolated from patients in southern Iraq. J Appl Genet. 2022; 63(2):401–412. doi:10.1007/s13353-022-00683-2
46. Ho PL, Chu YPS, Lo WU, et al. High prevalence of Escherichia coli sequence type 131 among antimicrobial-resistant E. coli isolates from geriatric patients. J Med Microbiol. 2015;64:243–247. doi:10.1111/j.1469-0691.2009.02941.x
47. Zhanel GG, Hisanaga TL, Laing NM, et al. Antibiotic resistance in Escherichia coli outpatient urinary isolates: final results from the North American urinary tract infection collaborative alliance (nautica). Int J Antimicrob Agents. 2006;27(6):468–475. doi:10.1016/j.ijantimicag.2006.02.009
48. Huttner A, Verhaegh EM, Harbarth S, et al. Nitrofurantoin revisited: a systematic review and meta-analysis of controlled trials. J Antimicrob Chemother. 2015;70(9):2456–2464. doi:10.1093/jac/dkv147
49. He P, Sun Q, Shi L, et al. Rational use of antibiotics in the context of China’s health system reform. BMJ. 2019;365:l4016. doi:10.1136/bmj.l4016
50. Sadeghi M, Ebrahim-Saraie HS, Mojtahedi A. Prevalence of ESBL and AmpC genes in E. coli isolates from urinary tract infections in the north of Iran. New Microbes and New Infections. 2021;45:100947. doi:10.1016/j.nmni.2021.100947
51. Sid Ahmed MA, Bansal D, Acharya A, et al. Antimicrobial susceptibility and molecular epidemiology of extended-spectrum beta-lactamase-producing Enterobacteriaceae from intensive care units at Hamad Medical Corporation, Qatar. Antimicrob Resist Infect Control. 2016;5(4). doi:10.1186/s13756-016-0103-x
52. Momtaz H, Rahimi E, Moshkelani S. Molecular detection of antimicrobial resistance genes in E. coli isolated from slaughtered commercial chickens in Iran. Vet Med-Czech. 2012;57(4):193–197. doi:10.17221/5916-Vetmed
53. Nawaz Z, Zahoor MK, Siddique AB, et al. Molecular identification of blactx-m and blatem genes among multi-drug resistant enteropathogenic Escherichia coli isolated from children. Pak J Pharm Sci. 2019;32(3):1215–1218.
54. Dumaru R, Baral R, Shrestha LB. Study of biofilm formation and antibiotic resistance pattern of gram-negative Bacilli among the clinical isolates at BPKIHS, Dharan. BMC Res Notes. 2019;12(1):38. doi:10.1186/s13104-019-4084-8
55. Avila-Novoa MG, Solis-Velazquez OA, Rangel-Lopez DE, et al. Biofilm formation and detection of fluoroquinolone- and carbapenem-resistant genes in multidrug-resistant Acinetobacter baumannii. Can J Infect Dis Med Microbiol. 2019;2019:3454907. doi:10.1155/2019/3454907
56. Katongole P, Nalubega F, Florence NC, et al. Biofilm formation, antimicrobial susceptibility and virulence genes of uropathogenic Escherichia coli isolated from clinical isolates in Uganda. BMC Infect Dis. 2020;20(1):453. doi:10.1186/s12879-020-05186-1
57. Tajbakhsh E, Ahmadi P, Abedpour-Dehkordi E, et al. Biofilm formation, antimicrobial susceptibility, serogroups and virulence genes of uropathogenic E. coli isolated from clinical samples in Iran. Antimicrob Resist Infect Control. 2016;5(11). doi:10.1186/s13756-016-0109-4
58. Sharma G, Sharma S, Sharma P, et al. Escherichia coli biofilm: development and therapeutic strategies. J Appl Microbiol. 2016;121(2):309–319. doi:10.1111/jam.13078
59. Cepas V, Lopez Y, Munoz E, et al. Relationship between biofilm formation and antimicrobial resistance in gram-negative bacteria. Microb Drug Resist. 2019;25(1):72–79. doi:10.1089/mdr.2018.0027
60. Mittal S, Sharma M, Chaudhary U. Biofilm and multidrug resistance in uropathogenic Escherichia coli. Pathog Glob Health. 2015;109(1):26–29. doi:10.1179/2047773215Y.0000000001
61. Perez LR. Acinetobacter baumannii displays inverse relationship between meropenem resistance and biofilm production. J Chemother. 2015;27(1):13–16. doi:10.1179/1973947813Y.0000000159
62. Fabrega A, Soto SM, Balleste-Delpierre C, et al. Impact of quinolone-resistance acquisition on biofilm production and fitness in Salmonella enterica. J Antimicrob Chemother. 2014;69(7):1815–1824. doi:10.1093/jac/dku078
63. Mizan MFR, Jahid IK, Kim M, et al. Variability in biofilm formation correlates with hydrophobicity and quorum sensing among Vibrio parahaemolyticus isolates from food contact surfaces and the distribution of the genes involved in biofilm formation. Biofouling. 2016;32(4):497–509. doi:10.1080/08927014.2016.1149571
64. Ciofu O, Rojo-Molinero E, Macia MD, et al. Antibiotic treatment of biofilm infections. APMIS. 2017;125(4):304–319. doi:10.1111/apm.12673
65. Borowiak M, Fischer J, Hammerl JA, et al. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting salmonella enterica subsp. Enterica serovar Paratyphi B. J Antimicrob Chemother. 2017;72(12):3317–3324. doi:10.1093/jac/dkx327
66. Literak I, Manga I, Wojczulanis-Jakubas K, et al. Enterobacter cloacae with a novel variant of act ampc beta-lactamase originating from glaucous gull (Larus hyperboreus) in Svalbard. Vet Microbiol. 2014;171(3–4):432–435. doi:10.1016/j.vetmic.2014.02.015
67. Zhao X, Yang J, Ju Z, et al. Molecular characterization of antimicrobial resistance in Escherichia coli from rabbit farms in Tai’an, China. Biomed Res Int. 2018;2018:8607647. doi:10.1155/2018/8607647
68. Mlynarcik P, Kolar M. Molecular mechanisms of polymyxin resistance and detection of mcr genes. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2019;163(1):28–38. doi:10.5507/bp.2018.070
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.