Back to Journals » Cancer Management and Research » Volume 13
Regulatory Mechanisms of LncRNAs in Cancer Glycolysis: Facts and Perspectives
Authors Huang P , Zhu S, Liang X, Zhang Q, Luo X, Liu C, Song L
Received 6 April 2021
Accepted for publication 19 June 2021
Published 5 July 2021 Volume 2021:13 Pages 5317—5336
DOI https://doi.org/10.2147/CMAR.S314502
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Kenan Onel
Peng Huang, Shaomi Zhu, Xin Liang, Qinxiu Zhang, Xiaohong Luo, Chi Liu, Linjiang Song
Reproductive & Women-Children Hospital, School of Medical and Life Sciences, Chengdu University of Traditional Chinese Medicine, Chengdu, Sichuan, 611137, People’s Republic of China
Correspondence: Linjiang Song; Chi Liu
Reproductive & Women-Children Hospital, School of Medical and Life Sciences, Chengdu University of Traditional Chinese Medicine, Chengdu, Sichuan, 611137, People’s Republic of China
Email [email protected]; [email protected]
Abstract: Cancer cells exhibit distinct metabolic characteristics that employ glycolysis to provide energy and intermediary metabolites. This aberrant metabolic phenotype favors cancer progression. LncRNAs are transcripts longer than 200 nucleotides that do not encode proteins. LncRNAs contribute to cancer progression and therapeutic resistance and affect aerobic glycolysis via multiple mechanisms, including modulating glycolytic transporters and enzymes. Further, dysregulated signaling pathways are vital for glycolysis. In this review, we highlight regulatory mechanisms for lncRNAs in aerobic glycolysis that provide novel insights into cancer development. Moreover, a comprehensive understanding of the regulatory mechanisms of lncRNAs in aerobic glycolysis can provide new strategies for clinical cancer management.
Keywords: lncRNA, cancer progression, glycolysis, signaling pathway
Introduction
Metabolism consists of multiple interconnected cellular chemical transformations that provide energy to sustain life.1 Cancer is a malignant disease characterized by infinite proliferation.2 To satisfy the increasing energy demands and biomass, cancer cells show metabolic plasticity to reprogram the metabolic pathways that include the metabolism of glucose, glutamine and fatty acids.3 Among these, aberrant glucose metabolism is one well-known metabolic reprogramming in cancer. While most normal cells depend primarily on mitochondrial oxidative metabolism to generate energy, cancer cells employ glycolysis to provide energy and intermediary metabolites, even under aerobic conditions. This phenomenon was discovered by Warburg in the 1920s and is termed the Warburg effect or aerobic glycolysis.4 The process often leads to enhanced glucose uptake, faster adenosine triphosphate (ATP) production and the accumulation of lactate in tumor cells, all of which are essential for tumorigenesis.5 Enhanced glycolysis provides energy for accelerated proliferation,6 and establishes an acidic microenvironment that promotes tumor cell invasion and metastasis.7 Further, enhanced glycolysis helps tumor cells resist adjuvant therapy.8 Therefore, glycolysis favors cancer cell survival under cellular stress conditions. However, mechanisms of the phenomenon await further investigation.
Non-coding RNAs (ncRNAs) are functional RNA transcripts that have no ability to encode peptides.9 NcRNAs can be divided into two categories based on their length: short ncRNAs and long ncRNAs (lncRNAs). Short ncRNAs contain less than 200 nucleotides including microRNAs (miRNAs), small interfering RNAs (siRNAs), and small nuclear RNAs.9 Conversely, lncRNAs are a novel group of regulatory RNA molecules that are more than 200 nucleotides in length.10 Moreover, circular RNAs (circRNAs) are a novel type of ncRNAs. Unlike other RNA molecules, circRNAs have covalently closed circular structures.11
As a special type of ncRNAs, lncRNAs play a pivotal role in regulating gene expression through multiple mechanisms, such as remodeling chromatin structure, regulating DNA methylation, and transcriptional activation or interference.12 They are also implicated in post-transcriptional modulation of gene expression via regulating mRNA processing. Further, they are closely correlated with post-translational modification of proteins, including phosphorylation and ubiquitination.12 LncRNAs also act as molecular sponges of miRNAs to modulate target gene expression and may function as precursors for miRNAs or siRNAs.12 Finally, lncRNA also regulate alternative splicing processes that contribute to the spatial and temporal expression of genes.12
LncRNAs regulate cancer cell proliferation, invasion, metastasis, and therapeutic resistance. Such activities are partially dependent on their ability to regulate aerobic glycolysis.13–16 These molecules regulate glycolytic transporters and metabolic enzymes, thus interfering with metabolic signaling pathways. Underlying mechanisms of lncRNAs function in aerobic glycolysis are complicated and await systematic and comprehensive research. In this review, we highlight molecular mechanisms by which lncRNAs modulate aerobic glycolysis. Also, the potential therapeutic role of lncRNAs in aerobic glycolysis is discussed. Understanding these processes may support a comprehensive theoretical basis for clinical cancer management.
Overview of Glycolysis in Cancer
Glycolysis in Cancer Progression
Tumor cells exist in an unfavorable microenvironment that is hypoxic, acidic, and nutrient deficient. As an adaptive mechanism, tumor masses exhibit remarkable plasticity in converting to aerobic glycolysis.17,18 This specific metabolic reprogramming allows infinite proliferation of cancer cells.
Glycolysis is an inefficient way to provide energy but maintains rapid ATP production under energy stress. The rapid ATP production is required for cell proliferation.19 Additionally, glycolytic metabolism supplies large quantities of building blocks for biosynthesis in proliferating cancer cells. Activated glycolytic flux accompanied by 3-phosphoglycerate and dihydroxyacetone phosphate production sustains the production of amino acids, triglycerides, and phospholipids needed to accelerate proliferation.6 Under energy stress, enhanced glycolysis also favors tumor survival. Glycolysis produces fewer reactive oxygen species (ROS) than mitochondrial oxidative metabolism, reducing senescence and apoptosis.6
Cancer cells are apt to metastasize to remote sites to escape energy restrictions in primary tumor tissues. The glycolytic phenotype is advantageous for invasion and metastasis.7 Excessive lactic acid production by aerobic glycolysis generates higher hydrogen ion concentration (lower pH), thereby establishing an acidic extracellular environment. This environment favors degradation of extracellular matrices and enhances metastasis by inducing metalloproteinases.20 The acidic environment also sustains metastasis by helping cancer cells evade immune surveillance.21 Moreover, aerobic glycolysis reduces ROS production and increases antioxidant activity in cancer cells. These changes confer resistance to anoikis and enable distant metastasis.22
Glycolysis and Cancer Therapeutic Resistance
Cancer cell survival under altered glucose metabolism is a pivotal phase in oncogenesis and the development of treatment resistance.23 Adjuvant chemotherapy is the primary treatment for cancers; resistance of cancer cells to chemical agents significantly reduces survival of cancer patients.24 Theoretically, a tumor mass, composed of distinct chemo-resistant cells, is vital for chemoresistance.25 Actually, suppression of glycolysis inhibits cell proliferation and reduces resistance to anticancer drugs.5 In hepatocellular carcinoma (HCC) cells, enhanced aerobic glycolysis promotes sorafenib-resistance, and suppression of glycolysis reverses this resistance.26
Radiotherapy is also used to treat cancers. Radiation can kill cancer cells through breaking DNA double-strands and can thus prolong the survival of cancer patients and improve their quality of life.27 Glycolysis generates a chemically reduced milieu that favors development of radio-resistance in multiple types of cancer cells, including colorectal and breast cancer and melanoma.28–30 For example, activation of AKT enhances aerobic glycolysis and may induce acquired radio-resistance in human cancer cells.31
Adoptive T cell therapy (ACT) is a personalized and innovative immunotherapy strategy for cancer treatment. ACT uses transfusion of ex vivo-expanded autologous tumor-infiltrating lymphocytes into patients to regress tumors.32 Unfortunately, many patients show regrowth of tumors because of acquired ACT resistance.33 Cascone et al found that enhanced glycolysis attenuates the effectiveness of ACT, while repression of glycolysis promotes antitumor activity both in vivo and in vitro. Further, tumor cells from ACT-refractory melanoma patients display enhanced glycolytic metabolism compared with ACT responding patients.34 Thus, glycolysis may be a crucial regulator of ACT resistance. Cytotoxic T-lymphocyte antigen 4 (CTLA-4) blockade may inhibit immunosuppression of activation of naive T cells by CTLA-4, thus obstructing tumor development.35,36 Zappasodi et al showed that CTLA-4 blockade increases the number of infiltrating immune cells in glycolysis-deficient tumors. Additionally, CTLA-4 blockade enhances therapeutic activity against glycolysis-defective tumors in mice, suggesting that inhibition of tumor glycolysis may be effective for improving the therapeutic effects of CTLA-4 blockade.37
Overall, aerobic glycolysis is an optimal strategy for tumor cell survival in stressful microenvironments. It supports continued cell proliferation, growth, and metastasis and enhances resistance to antitumor treatment. However, the current understanding of aerobic glycolysis remains limited, which impedes the development of more effective cancer therapy. Understanding regulatory mechanisms of cancer glycolysis is thus significant and essential.
Regulatory Mechanisms of LncRNAs in Cancer Glycolysis
Cancer cells are known for accelerated energy metabolism and enhanced glycolysis.6 Altered glycolysis supplies energy for proliferation and influences cancer progression.38 In this section, we summarize the regulatory mechanisms of lncRNAs regulation of glycolysis via altering the expression of glycolytic enzymes and transporters. Such transporters include glucose transporters (GLUTs) and monocarboxylate transporters (MCTs). The actions of lncRNAs in control of carcinogenesis and cancer progression via regulation of transporters and glycolytic enzymes are summarized in Table 1.
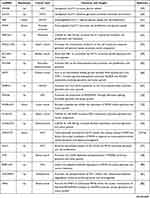 | 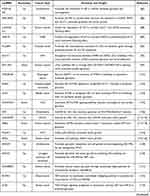 |  |
Table 1 The Regulatory Mechanisms of lncRNAs in Aerobic Glycolysis |
LncRNAs-Mediated Regulatory Mechanisms of Glycolytic Transporters
LncRNAs and GLUTs
GLUTs are important membrane proteins responsible for transporting glucose. These proteins play a critical role in regulating glucose homeostasis and metabolism.39 Three GLUTs family members are involved in cancer glucose metabolism—GLUT1, GLUT3, and GLUT4.40,41 GLUT1 is widely expressed in multiple tissues and participates in the regulation of basal glucose uptake.42 GLUT3 is mainly expressed in nerve tissues, while GLUT4 is an important glucose metabolism regulator in skeletal muscle and adipose tissues.43 However, GLUT4 is regulated by insulin, whereas the activity of GLUT1 and GLUT3 is not related to insulin.44 Under physiological conditions, GLUTs transport glucose into cells to meet energy requirements. However, during tumorigenesis, GLUTs expression are often deregulated.40 GLUT1 and GLUT3 are upregulated in multiple types of cancer.40 Upregulated GLUT1 and GLUT3 promote glucose uptake, cancer metastasis and growth.45,46 Moreover, GLUT4 is also upregulated in various cancers, leading to elevated glucose uptake and enhanced cancer progression.47,48 Several lncRNAs affect glucose uptake by regulating GLUT expression or distribution.
LncRNA colorectal neoplasia differentially expressed (CRNDE) shows increased glucose uptake by enhancing the expression of GLUT4.49 In oral squamous cell carcinoma (OSCC), a novel lncRNA-p23154, enhances glucose uptake and glycolysis through upregulating GLUT1. This upregulation promotes cell invasion and metastasis.50 Moreover, lncRNA Neighboring Enhancer of FOXA2 (NEF) downregulates GLUT1 expression, resulting in less glucose uptake and reduced cell proliferation in non-small-cell lung cancer (NSCLC).51 Further, overexpression of lncRNA mortal obligate RNA transcript (MORT) inhibits cell proliferation and glucose uptake in prostate carcinoma. GLUT1 overexpression reverses this anticancer function of MORT, lncRNA MORT/GLUT1 may play a crucial role in prostate carcinoma.52
MiRNAs are small RNA molecules of approximately 22 nucleotides that are important in physiological or pathological processes via negative regulation of mRNA expression.53 LncRNA may function as competitive endogenous RNA (ceRNA), bind with miRNA, and alter target gene expression.54 This ceRNA mechanism is correlated with GLUT-mediated glycolysis. For example, dopamine β hydroxylase antisense RNA 1 (DBH-AS1) enhances the glycolytic activity of melanoma cells by GLUT1 and facilitates cell proliferation and metastasis. Further, lncRNA DBH-AS1 suppresses the transcription of miR-223-3p through interacting with its promoter. MiR-223-3p binds directly to the 3’ untranslated region (UTR) of GLUT1, leading to decreased GLUT1 expression.55
Moreover, the localization of GLUT1 can affect its function. Zhang et al demonstrated that inhibition of translocation of GLUT1 to the cell membrane reduces both glucose uptake and glycolysis in lung cancer cells.56 LncRNA MACC1 antisense RNA 1 (MACC1-AS1) is overexpressed in gastric cancer tissues and correlated with the malignant behavior of cancer cells.57 Mechanistically, MACC1-AS1 increases the abundance of GLUT1 on cell membranes by facilitating translocation of GLUT1.57 MACC1-AS1 thus stimulates glycolysis, in part, through promoting the distribution of GLUT1 to the cell membrane. However, whether lncRNA modulates the distribution of other GLUTs, such as GLUT3 and GLUT4, to affect glucose uptake needs in-depth study.
LncRNAs and MCTs
MCTs, proton-coupled integral membrane proteins, control lactic acid transport across the plasma membrane. MCTs, particularly MCT1, MCT3 and MCT4, are upregulated in human cancers.58 Increased MCT expression promotes metastasis, angiogenesis, and metabolic reprogramming by facilitating lactic acid exchange.58
Logotheti et al identified a new lncRNA, SLC16A1 antisense RNA 1 (SLC16A1-AS1), transcribed by the antisense strand of the SLC16A1 gene. SLC16A1 encodes MCT1.59 Further, E2F transcription factor 1 (E2F1) transactivates SLC16A1 and SLC16A1-AS1 due to the shared response promoter. Reciprocally, SLC16A1-AS1 binds with E2F1 and creates a lncRNA–protein complex to co-activate SLC16A1/MCT1. This feed-forward loop leads to enhanced aerobic glycolysis and tumorigenesis in bladder cancer.59
LncRNAs-Mediated Regulatory Mechanisms of Glycolytic Enzymes
Glycolysis is the process that decomposes glucose into lactate to produce ATP. It contains the steps, catalyzed by corresponding enzymes. During this process, three rate-limiting enzymes work together to convert glucose into pyruvate.60 Hexokinase (HK) is the first rate-limiting enzyme of the glycolytic metabolism, which converts glucose into glucose-6-phosphate.61 Phosphofructokinase (PFK) 1 is the second rate-limiting enzyme that catalyzes fructose-6-phosphate (F6P) into fructose-1,6-biphosphate.62 Pyruvate Kinase (PK) is the last glycolytic rate-limiting enzyme that converts phosphoenolpyruvate (PEP) into pyruvate.63 Then, lactate dehydrogenase A (LDHA) converts pyruvate into lactate.64 Moreover, glyceraldehyde 3-phosphate dehydrogenase also plays a vital role in glycolysis. It catalyzes glyceraldehyde 3-phosphate into 1,3-bisphosphoglycerate accompanied by the formation of NADHs.60
In addition to affecting GLUT expression or translocation, lncRNAs modulate glycolytic enzymes (Figure 1).
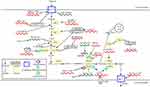 |
Figure 1 LncRNAs regulate cancer glycolysis by modulating metabolic enzymes and transporters. |
LncRNAs Directly Regulate Glycolytic Enzymes at Multiple Regulatory Levels
At the transcriptional level, lncRNAs activate gene expression in aerobic glycolysis. For instance, lncRNA Hox transcript antisense RNA (HOTAIR) is highly expressed in pancreatic adenocarcinoma and associated with poor prognosis. This lncRNA promotes cell proliferation and glycolysis through transcriptional activation of HK2.65 In ovarian cancer, lncRNA ceruloplasmin (NRCP) promotes aerobic glycolysis and tumor growth. Mechanistically, NRCP, as an intermediate binding partner, promotes an interaction between RNA polymerase II and signal transducer and activator of transcription 1 (STAT1) in the nucleus. This interaction results in increased expression of several glycolytic enzymes, including glucose-6-phosphate isomerase, aldolase A (ALDOA) and ALDOC.66 Under glucose starvation, overexpression of lncRNA insulin-like growth factor binding protein 4–1 (IGFBP4-1) upregulates the expression of HK2 and LDHA at the transcriptional level, thus promoting aerobic glycolysis and tumor growth.67
At the post-transcriptional level, lncRNAs directly regulate mRNA splicing, protein localization, and act as ceRNAs for miRNAs. In aerobic glycolysis, lncRNAs mediate the post-transcriptional regulation of enzymes, thus affecting their expression and functional diversifications.
Alternative splicing is essential for proteomic diversity and contributes to spatial and temporal gene expression.68 Alternative splicing of pyruvate kinase type M (PKM) precursor mRNA by heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1) is an important example.69 PKM exists in two isoforms, PKM1 and PKM2. PKM2-mediated aerobic glycolysis may be advantageous for tumorigenesis, and the ratio of PKM2/PKM1 is commonly upregulated in cancers.69 Lan et al found that lncRNA small nucleolar RNA host gene (SNHG) 6 is associated with hnRNPA1-mediated splicing of PKM in colorectal cancer (CRC). SNHG6 directly binds to the precursor mRNA of PKM and induces hnRNPA1 to bind to flanking exon 9. These actions increase the proportion of PKM2/PKM1 and enhance aerobic glycolysis and promote tumor growth. Thus, the role of SNHG6 in carcinogenesis might be exerted by promoting aerobic glycolysis by alternative splicing of PKM.70
LncRNAs are often considered as dark matter without encoded peptides.71 However, one current investigation has reversed this concept. The NR_033201.2 variant of HOXB cluster antisense RNA 3 (HOXB-AS3) is downregulated in colon cancer and encodes a small endogenous peptide that acts as a tumor suppressor. This peptide selectively binds to the hnRNPA1 binding domain. This binding targeted sequences adjacent to PKM exon 9 results in downregulation of PKM2 expression. This process reduces lactate production and inhibits glycolytic flux.72
DLX6 Antisense RNA 1 (DLX6-AS1), an upregulated lncRNA in gastric cancer, facilitates aerobic glycolysis and tumor development by acting as a ceRNA for miR-4290. This interaction upregulates pyruvate dehydrogenase kinase 1 (PDK1), a gatekeeper enzyme that inhibits pyruvate entry into the tricarboxylic acid (TCA) cycle. The enzyme inhibits the formation of acetyl-coA from pyruvate.73 In osteosarcoma, lncRNA potassium voltage-gated channel subfamily Q member 1 opposite strand 1 (KCNQ1OT1) can sponge miR-34c-5p to enhance expression of ALDOA, thus facilitating aerobic glycolysis and tumor growth.74
LncRNAs modulate glycolytic enzyme activity by affecting their localization. Guan et al found that nuclear receptor subfamily four group A member 1 (Nur77) activates transcription of lncRNA WAP four-disulfide core domain 21 pseudogene (WFDC21P). Transcription of the lncRNA attenuates the Warburg effect and hepatocarcinogenesis. Mechanistically, WFDC21P inhibits tetramer formation of phosphofructokinase, platelet type (PFKP) and inhibits its catalytic activity. It also blocks nuclear localization of PKM2, thus suppressing its transcriptional activation of oncogenic factors, including c-Myc and hypoxia-inducible factor (HIF)-1α.75,76 Thus, the Nur77/WFDC21P axis may be a promising therapeutic target for HCC, though its regulation remains elusive.
LncRNAs mediate post-translational regulation of several enzymes in aerobic glycolysis, thus influencing the formation and degradation of proteins. The main post-transcriptional modifications are phosphorylation and ubiquitination. For example, lncRNA highly up-regulated in liver cancer (HULC) directly binds to LDHA and PKM2 and enhances their binding with fibroblast growth factor receptor type 1 (FGFR1). These interactions result in elevated phosphorylation of the two enzymes, thus promoting glycolysis and cell proliferation.77 Additionally, lncRNA LINC00538 (YIYA) facilitates glycolytic flux and breast cancer growth through regulating cyclin-dependent kinase 6 (CDK6) dependent phosphorylation of fructose bisphosphatase PFK2 (PFKFB3).78 This enzyme converts F-6-P into fructose-2,6-bisphosphate (F-2,6-BP).79 Fez family zinc finger protein 1 antisense ribonucleic acid 1 (FEZF1-AS1), an upregulated lncRNA in CRC, accelerates metastasis and proliferation of cancer cells. FEZF1-AS1 increases the stability of PKM2 protein through inhibiting ubiquitin-mediated degradation. These actions lead to enhanced expression of PKM2 in the cytoplasm and nucleus and increased aerobic glycolysis.80
Liu et al reported that LINC00470 directly binds with a ternary complex composed of RNA-binding protein fused in sarcoma (FUS) and V-akt murine thymoma viral oncogene homolog (AKT). This binding leads to AKT phosphorylation. Elevated AKT phosphorylation prevents HK1 from ubiquitinate-mediated degradation, thus enhancing aerobic glycolysis and carcinogenesis.81
Interestingly, the post-translational modification of PKM2 can trigger Warburg effect. For example, HIF-1α antisense lncRNA (HIFAL) recruits proline hydroxylase domain (PHD) 3 to PKM2 and promotes prolyl hydroxylation at proline 408. Hydroxylated PKM2 drives nuclear translocation of hydroxyl-PKM2/PHD3 complex through binding with heterogeneous nuclear ribonucleoprotein F (hnRNPF). This process promotes HIF-1α transactivation, resulting in enhanced expression of LDHA, PDK1, GLUT1, and HK2. Further, activated HIF-1α potentiates HIFAL transcription, inducing the positive HIF-1α- HIFAL- HIF-1α regulation and promoting aerobic glycolysis and tumor growth.82
In conclusion, lncRNA may control the function of glycolytic enzymes at the post-translational level through adding or removing functional groups. These changes regulate glycolytic flux and cancer development.
LncRNAs Regulate Glycolytic Enzymes Through Signaling Pathways
An interaction between dysregulated lncRNAs and signal transduction pathways significantly contributes to alteration of cancer glycolysis, thereby promoting or inhibiting carcinogenesis and cancer progression (Figure 2).
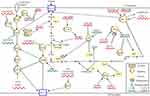 |
Figure 2 LncRNAs regulate cancer glycolysis by modulating metabolic pathways. |
The aerobic glycolysis is closely associated with hypoxic tumor microenvironment.18 HIF serves as an important transcription factor expressed by malignant cell adapted to anoxic environments.83 Activating HIF-1α enhances a glycolytic phenotype through promoting transcription of GLUTs or glycolysis-related enzymes.84 LncRNAs play critical regulatory roles in HIF signaling. LncRNA PCED1B antisense RNA 1 (PCED1B-AS1) supports the translation of HIF-1α mRNA by directly binding to its 5’-UTR, leading to enhanced HIF-1α expression. Increased HIF-1α expression potentiates aerobic glycolysis and carcinogenesis in glioblastoma.85 In triple-negative breast cancer (TNBC), overexpression of lncRNA miR210 host gene (MIR210HG) increases HIF-1α protein levels and upregulates the expression of glycolysis target genes such as GLUT1, PKM2, LDHA. These alterations enhance glucose consumption and tumor growth.86
Under normoxia, the PHD-dependent hydroxylation of HIF-1α at Proline 402 or Proline 564 leads to destabilization of HIF-1α via the von Hippel-Lindau protein (VHL)-mediated ubiquitination process. Ubiquitination leads to rapid degradation in the proteasome pathway.87 Thus, inhibition of hydroxylation and subsequent ubiquitin-mediated degradation of HIF-1α is a critical aspect of regulating its function in aerobic glycolysis. For example, one study showed that accumulation of HIF-1α leads to enhanced transcription of long intergenic noncoding RNA (lincRNA)-p21 under hypoxic conditions.88 Moreover, overexpressed lincRNA-p21 competitively binds with HIF-1α to disrupt the interaction between HIF-1α and VHL. Thus, HIF-1α is stabilized by inhibition of ubiquitin-mediated degradation. This reciprocal regulation ensures a high level of active HIF-1α during hypoxia. In turn, increased HIF-1α expression leads to activation of GLUT1 and LDHA, which facilitates the Warburg effect.88 Lin and colleagues revealed that a cytoplasmic lncRNA, long intergenic noncoding RNA for kinase activation (LINK-A), promotes reprogramming of glucose metabolism and tumorigenesis in TNBC.89 LINK-A recruited breast tumor kinase (BRK) binds to EGFR- heterodimer and transmembrane glycoprotein NMB (GPNMB) complex and subsequently activates BRK kinase. Activated BRK kinase phosphorylates HIF-1α at tyrosine 565. This phosphorylation results in inhibition of HIF-1α hydroxylation at proline 564 and prevents its subsequent degradation. Moreover, LINK-A also recruits leucine-rich repeat kinase 2 (LRRK2) to mediate phosphorylation of HIF-1α at Ser797, thereby activating its target genes.89
The MYC gene family is a canonical oncogene that is widely downregulated in tumors.90 The MYC oncogene encodes c-Myc, a transcription factor that promotes glycolysis through activating glycolytic enzymes.91 For instance, lncRNA prostate cancer gene expression marker 1 (PCGEM1) is overexpressed in prostate cancer and is advantageous for cancer cell growth via metabolic reprogramming. PCGEM1 promotes transcriptional activation of metabolic genes through enhancing histone H3 and H4 acetylation of c-Myc. Moreover, silencing PCGEM1 inhibits the expression of several genes and attenuates glycolysis. Thus, PCGEM1 plays a critical role in prostate cancer metabolism.92 In CRC, glycolysis-associated lncRNA of colorectal cancer (GLCC1) is highly expressed during energy stress and is associated with poor prognosis. Mechanistically, GLCC1 strengthens the interaction between heat shock protein 90 (HSP90) and ubiquitin-specific protease 22 (USP22), an important deubiquitinating enzyme. Consequently, c-Myc, the target of HSP90, is stabilized by abolishing proteasome-mediated degradation. c-Myc can then support transcriptional activation of LDHA and promote glycolysis and cell proliferation.93
Existing evidence has revealed that ROS plays a vital role in aerobic glycolysis.94 Moreover, Xiang et al found that c-Myc is the regulator of ROS. c-Myc transcriptionally represses lncRNA isocitrate dehydrogenase 1 (IDH1) antisense RNA1 (IDH1-AS1). IDH1-AS1 increases the activity of IDH1 by potentiating its dimerization. This process enhances the production of α-ketoglutarate (α-KG) and reduces ROS under normoxia.95 α-KG, a component of the TCA cycle, is involved in the degradation of HIF-1α.96 Thus, lncRNA IDH1-AS1 regulated by c-Myc destabilizes HIF-1α through activating the IDH1-αKG/ROS-HIF1α cascade, leading to inhibition of Warburg effect.95
Growth factors stimulate phosphoinositide 3-kinase (PI3K) and activate the AKT/mechanistic target of rapamycin (mTOR) axis. This axis plays an important role in numerous physiological processes, such as tumorigenesis and metabolic reprogramming.97,98 Notably, AKT activates mTORC1 by inhibiting tuberous sclerosis complex 2 (TSC2) activity. This inhibition leads to repression of Ras homolog enriched in brain guanosine triphosphatase (Rheb GTPase).99 In this process, phosphatase and tensin homolog on chromosome 10 (PTEN) negatively controls PI3K signaling by inactivating AKT.100 For example, overexpression of LINC00184 recruits DNA methyltransferase 1 (DNMT1) to the promoter of PTEN, thus inhibiting its expression. Decreased PTEN removes its repression of the PI3K/AKT cascade and leads to enhanced glycolysis in esophageal cancer.101
Additionally, AKT activates mTOR, which in turn upregulates GLUTs or glycolytic enzymes. Antisense non-coding RNA in the INK4 locus (ANRIL) activates AKT/mTOR signaling to upregulate GLUT-1, resulting in enhanced glycolysis in nasopharyngeal carcinoma.102 Li et al reported that urothelial cancer-associated 1 (UCA1) activates mTOR to upregulate HK2 via both activating STAT3 and inhibiting miR-143, thereby uncovering a novel metabolic cascade in bladder cancer.103 Zheng et al revealed that LINC01554 inactivates AKT/mTOR signaling pathway to abolish tumorigenicity and glycolysis in HCC.104
Insulin receptor substrate (IRS) is a cytoplasmic adaptor protein that amplifies the PI3K signal and plays a critical role in cancer metabolism.105 Cheng et al demonstrated that lncRNA X–inactive specific transcript (XIST) is highly expressed in glioblastoma and directly sponges miR-126. This activity promotes glycolysis via the IRS1/PI3K/Akt/GLUT cascade.106 Thus, IRS1/PI3K/AKT may provide one mechanism for metabolism reprogramming. The complex combinatorial possibilities of the IRS/PI3K/AKT pathway may affect cancer metabolic remodeling through multiple dimensions.107 More studies are needed to explore this linkage.
AMPK is an energy sensor. Cancer cells typically downregulate AMPK to evade confining influences on proliferation and develop their glycolytic phenotype.108 Activated AMPK phosphorylates the TSC2 complex, inhibiting the activity of Rheb. Rheb inhibition results in the inactivation of mTOR.109 Under energy stress, activated TSC2 mediated by AMPK reduces cell apoptosis.110
Liver kinase B1 (LKB1), a critical tumor suppressor, modulates cell growth and metabolism through regulating mTOR activity.111 As a primary upstream modulator of AMPK, LKB1 activates AMPK in response to low intracellular levels of ATP. Low ATP levels inhibit cell proliferation.112 Knockdown of LKB1 facilitates tumor growth, with elevated uptake of glucose, upregulated ATP production, and synthesis of macromolecules.113 Zeng et al revealed that LKB1 inhibits human papillomavirus (HPV)-induced glycolysis. Loss of LKB1 facilitates HPV-stimulated tumor metastasis through upregulating the expression of HK2.114 LKB1 deficiency thus promotes glycolysis and tumorigenesis.
Nuclear lncRNA LINC00473 interacts with non-POU domain-containing octamer-binding protein (NONO), a regulator of cancer glycolytic metabolism.115,116 LINC00473 is induced by LKB1 loss and promotes lung tumor growth.115 Under energy stress, the LKB1/AMPK pathway activates the transcription of lncRNA neighbor of BRCA1 gene 2 (NBR2), which potentiates AMPK activation. Depletion of NBR2 counteracts glucose-starvation-induced AMPK activation, leading to alterations in the apoptosis/autophagy responsive ratio and enhanced breast cancer development. These findings suggest the modulation of the NBR2/AMPK feed-forward loop mechanism.117 Additionally, NBR2 attenuates thyroid cancer epithelial-mesenchymal transition and GLUT-1 expression by potentiating AMPK activation.118 Therefore, interactions between lncRNAs and LKB1/AMPK signaling pathway play a critical role in cancer glycolysis.
p53, a well-known tumor suppressor, is deregulated in many cancers.119 Recently, several studies have revealed that p53 is crucial in cancer glycolysis. The cellular absence of p53 increases glycolysis.119 p53 directly inhibits the transcription of GLUT1 and GLUT4 to decrease glucose uptake.120 p53 also participates in glucose metabolism via modulating expression of multiple glycolytic enzymes. Activated p53 enhances ubiquitinate-mediated degradation of phosphoglycerate mutase (PGM), which inhibits the production of pyruvate.121 p53 also controls LDHA transcriptional activity through an interaction with the LDHA promoter region. This action attenuates glycolysis in breast cancer.122
LncRNAs interact with p53 signaling directly or indirectly. In HepG2 cells, lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) enhances deacetylases sirtuin 1 (SIRT1)- deacetylated-p53 and inhibits the transcription of its target genes. These actions promote cell growth.123 Overexpression of maternally expressed gene 3 (MEG3) increases p53 expression and activates its target genes.124 MEG3 promotes p53-mediated transactivation, thereby inhibiting breast cancer development.125 Accordingly, MALAT1 and MEG3 are critical to the p53 pathway, but detailed mechanisms need further investigation.
Overall, the role of p53 pathway in cancer development is partially manifested in inhibition of glucose metabolic reprogramming, and lncRNAs may play a crucial role in these processes.
Lee et al demonstrated that wingless (Wnt) facilitates glycolysis via upregulating pyruvate carboxylase (PC). This enzyme is critical for the conversion of pyruvate to oxaloacetate.126 Pate et al then reported that inhibition of the Wnt/β‐catenin pathway leads to reduced glycolysis in colon cancer cells. Mechanistically, Wnt regulated glycolytic metabolism, in part, through controlling PDK1.127 These findings reveal that Wnt/β‐catenin signaling pathway promotes aerobic glycolysis through regulating PC and PDK1, enzymes that switch metabolic energy production from oxidative phosphorylation to aerobic glycolysis.
LncRNAs are implicated in glycolytic metabolism and cancer progression via the Wnt/β-catenin pathway. For example, Zhang et al showed that lncRNA SNHG9 is involved in aerobic glycolysis of glioblastoma. They suggested that overexpression of SNHG9 promotes glycolytic metabolism and cell growth by downregulating miR-199a-5p and upregulating Wnt2.128 In laryngeal squamous cell carcinoma, knockdown of SNHG3 inhibits glycolysis and tumor growth by eliminating the Wnt pathway via modulating miR-340-5p/yes-associated protein 1 (YAP1) axis.129 However, whether lncRNAs-mediated Wnt/glycolysis signaling correlates with alterations in PC and PDK1 awaits further investigation.
The Hippo pathway plays an important role in organ development and tissue homeostasis by suppressing YAP/PDZ-binding motif (TAZ) transcription co-activators. The deregulated Hippo pathway usually causes tumorigenesis in model organisms.130 Currently, several studies have revealed that Hippo pathway is a key regulator of cancer glycolysis. For example, LINC00857 promotes ovarian cancer glycolysis through upregulating YAP1.131 Additionally, YAP enhances the expression of lncRNA Breast Cancer Antiestrogen Resistance 4 (BCAR4), which coordinates the Hedgehog pathway to upregulate HK2 and PFKFB3 in breast cancer.132
Transforming growth factor beta (TGF-β) pathway is considered as a pro-metastatic signaling by inducing EMT.133 However, TGF-β also cooperates with Hippo signaling in cancer glycolysis. Mota et al reported that loss of tumor suppressor Merlin results in the loss of small mothers against decapentaplegic 7, which activates TGF-β and enables the co-operative engagement of TGF-β-Hippo signaling. This process upregulates lncRNA UCA1 that enhances the expression of HK2, leading to increased aerobic glycolysis in breast cancer cells.134
Kirsten rat sarcoma viral oncogene homolog (KRAS), an important member of RAS family, is mutated in various cancers.135 Mutated KRAS has been reported to promote aerobic glycolysis through upregulating GLUT1 and glycolytic enzymes.136 LINC01420 promotes pancreatic cancer progression through upregulating KRAS.137 Long noncoding RNA activated by TGF-beta (lncRNA-ATB) has been reported to enhance cell proliferation and invasion of bladder cancer through miR-126/KRAS axis.138 Moreover, lncRNA H19 promotes colon cancer progression by activating RAS/mitogen-activated protein kinase (MAPK) signaling pathway.139 In conclusion, LINC01420, lncRNA-ATB, H19 may be critical regulators of RAS. More studies are needed to understand their functions in aerobic glycolysis.
Deregulated Mechanisms of LncRNAs in Cancer Glycolysis
Epigenetic Regulation
LINC00261 is downregulated in pancreatic cancer cell lines and overexpressed LINC00261 inhibits glycolysis and proliferation of pancreatic cancer cells. Mechanistically, LINC00261 is downregulated by enhancer of zeste homolog 2 (EZH2)-mediated histone H3 lysine 27 trimethylation and high methylated levels in the promoter region. This finding reveals novel epigenetic regulatory mechanisms of LINC00261 in pancreatic cancer glycolysis and provides a promising target for pancreatic cancer targeted therapy.140
Transcriptional Regulation
Transcription factor AP2alpha (TFAP2A) plays an essential role in regulating cell proliferation and differentiation.141 Moreover, Zhang et al found that TFAP2A directly binds to the promoter region of lncRNA SNHG16 and activates its transcription. Activated SNHG16 promotes the proliferation and glycolysis of endometrial carcinoma.142 Moreover, lncRNAs can be regulated by c-Myc. In NSCLC, c-Myc activates LINC01123 to promote aerobic glycolysis.143 On the contrary, c-Myc transcriptionally represses IDH1-AS1.95 Additionally, lncRNAs may also be transcriptional targets of p53. LncRNA actin gamma 1 pseudogene (AGPG) stabilizes PFKFB3 by inhibiting its ubiquitination and subsequent degradation. Upregulated PFKFB3 stimulates glycolytic flux and cell proliferation. Moreover, AGPG is repressed by p53, which indicates a p53-AGPG-PFKFB3 axis may be a promising target for cancer treatment.144 Liao et al reported that lncRNA EPB41L4A antisense RNA 1 (EPB41L4A-AS1), a p53 inducible lncRNA, is downregulated in multiple cancers. Knockdown of EPB41L4A-AS1 enhances the localization of histone deacetylase 2 (HDAC2) in the nucleoplasm, promotes HDAC2 occupation on the VHL promoter region, stabilizes HIF-1α, and thus promotes glycolysis.145 This finding links two critical glycolytic pathways, reflecting cancer metabolism complexity and the need for further study.
It has been reported that Fusobacterium nucleatum (F. nucleatum) is highly abundant in CRC tissues and correlated with the poor prognosis in patients with CRC.146 But the detail molecular mechanism is unknown. Hong et al found that F. nucleatum supports carcinogenesis by promoting CRC cell glycolysis. Mechanistically, F. nucleatum activates lncRNA enolase1-intronic transcript 1 (ENO1-IT1) transcription through recruiting transcription factor specificity protein 1 (SP1) to the promoter region of ENO1-IT1. Upregulated ENO1-IT activates the target genes including ENO1, resulting in elevated glycolysis and enhanced tumor growth.147
HIF-1α induces upregulation of lncRNA AC020978 in NSCLC. Elevated AC020978 stabilizes PKM2 by protecting it from degradation. Stabilized PKM2 augments HIF-1α transcriptional activity. This positive AC020978/PKM2/HIF-1α loop activates Warburg effect.148 Moreover, other lncRNAs such as HIFAL and linc-p21 form positive feedback loops as well.82,88 On the one hand, hypoxia induces the upregulation of lncRNAs. On the other hand, lncRNAs can promote the transcriptional activity of HIF-1α, thus facilitating the glycolysis.
Overall, during aerobic glycolysis, lncRNAs expression can be regulated by epigenetic mechanisms. Additionally, several transcriptional factors regulate lncRNAs expression as well (Table 2 and Figure 3).
 |
Table 2 The Deregulated Mechanisms of lncRNAs in Aerobic Glycolysis |
 |
Figure 3 The deregulated mechanisms of lncRNAs in cancer glycolysis. |
MiRNAs and CircRNAs in Cancer Glycolysis
MiRNAs and circRNAs are important ncRNAs in tumorigenesis. Moreover, recent studies have shown the involvement of miRNAs and circRNAs in cancer glycolysis.149 Several miRNAs have been shown to regulate the expression of GLUT1 and GLUT3. For example, miR-148b downregulates GLUT1, thus impending gastric cancer glycolysis.150 Moreover, miR-10a enhances cancer cell glycolysis and proliferation in OSCC via upregulating GLUT1.151 In bladder cancer, miR-195-5p inhibits glycolysis and cell proliferation by downregulating GLUT3.152 Additionally, miRNAs also regulate several glycolytic enzymes. In NSCLC, miR-214 upregulates HK2 and PKM2 through PTEN/Akt/mTOR signaling pathway.153 Wang et al reported that LDHA is targeted by several miRNAs including miR-34a, miR-34c, miR-369-3p, miR-374a, and miR-4524a/b, thereby altering glycolysis in CRC.154
Mounting evidence has shown that circRNAs regulate glycolysis through multiple mechanisms: (i) circRNAs act as ceRNAs for miRNAs. (ii) circRNAs regulate gene transcription. (iii) circRNAs encode peptides. For example, circRNA MAT2B facilitates HCC glycolysis via miR-338-3p/PKM2 axis.155 CircMDM2 accelerates the glycolysis of OSCC through targeting miR-532-3p/HK2 axis.156 CircFNDC3B-218aa, a novel peptide encoded by circFNDC3B, can inhibit glucose uptake, lactate production and cell invasion in colon cancer.157
Obviously, miRNAs, circRNAs, and lncRNAs are all important regulators in cancer glycolysis. Therefore, understanding their regulatory mechanisms in aerobic glycolysis may provide efficient strategies for cancer treatment.
Perspectives: The Potential Therapeutic Role of LncRNAs
Cancer is the primary cause of human mortality, and resistance of tumors to therapeutic interventions significantly impedes cancer treatment. More efficient strategies to evade resistance and successfully treat cancer have been sought for decades. Liu et al noted the role of LINC00518 in radio-resistance of cutaneous malignant melanoma (CMM) from the perspective of aerobic glycolysis and proposed LINC00518 as a promising target for improving the treatment outcomes of patients receiving radiotherapy. Knockdown of LINC00518 inhibits glycolysis through downregulating LDHA protein levels via the miR-33a-3p/HIF-1α axis. This inhibition enhances the radiosensitivity of CMM cells. Moreover, treatment with an inhibitor of glycolysis, 2-deoxy-D-glucose, reverses the positive effect of overexpression of LINC00518 on LDHA and HIF-1α expression and radio-resistance in CMM cells. The discovery of the LIN00518/miR-33a-3p/HIF-1α/LDHA/axis in the radiotherapy of CMM identifies glycolysis as an efficient target to increase the therapeutic efficiency of radiotherapy.15 Moreover, targeting the UCA1/HK2/glycolysis pathway can also be a method to increase the radiosensitivity of cervical cancer.158
Aberrant energy metabolism is a hallmark of cancer, and biguanides are potential anticancer drugs for their abilities to regulate glycolysis.159 Phenformin is a kind of biguanide used as a chemotherapeutic agent because it impedes cancer development.160 Liu et al found that lncRNA NBR2 is a likely regulator in cancer cells for the adaptive response under energy stress. Enhanced NBR2 expression inhibits apoptosis induced by phenformin and increases glucose uptake through elevating GLUT1 expression. Upregulation of NBR2 also promotes tumor growth.161 Several lncRNAs, such as SNHG7,16 HOTAIR,162 XIST163 are also involved in resistance to cisplatin, temozolomide and 5-fluorouracil via altering glycolysis in gastric cancer, glioblastoma and CRC, respectively. Interestingly, Fu et al reported that lncRNA plasmacytoma variant translocation 1 (PVT1) maintains Myc expression in Sjögren’s syndrome. This expression regulates proliferation and immune effects of CD4+ T cells by controlling glycolytic metabolism.164 Investigating the role of the PVT1-T cell glycolysis axis in tumor models may provide a novel insight for improving T cell-based immunotherapy.
The important role of lncRNAs-mediated glucose metabolic reprogramming in cancer adjuvant therapy, as discussed above, makes them promising targets for treating cancers. Several studies have revealed that siRNA, antisense oligonucleotides (ASOs) and clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9 (CRISPR/Cas9) may be promising methods of targeting lncRNAs.165 However, the application of lncRNAs-based targeted treatment faces several challenges. For example, both the gene network triggering aerobic glycolysis and the intricate molecular mechanisms linking metabolic reprogramming with therapeutic resistance remain to be fully investigated. Only a small number of lncRNAs are clearly understood. Moreover, several studies have revealed that the same lncRNA may play diametrically opposite functional roles in different types of cancers. For example, HOXB-AS3 plays an oncogenic role in lung cancer, acute myeloid leukemia (AML), hepatoma, and epithelial ovarian cancer (EOC).166–169 Xu et al reported that HOXB-AS3 promotes EOC glycolysis by altering LDHA expression.166 However, Huang et al reported that HOXB-AS3 is downregulated in colon cancer and encodes a peptide to inhibit glycolysis.72 Papaioannou et al proposed that the HOXB-AS3 transcript variant in AML cells presents limited overlap with its variant that is downregulated in colon cancer cells. HOXB-AS3, which promotes cell growth in AML, shows high concentrations in the nucleus and a low association with polysomal fractions.168 It may be a reasonable explanation for the differential functions of HOXB-AS3 in different cancers. Additionally, NF-kappa B interacting long noncoding RNA (NKILA) exerts antitumor activity in various cancers,170 but stimulates aerobic glycolysis and angiogenesis in gliomas.171 This activity awaits further investigation.
Further, inhibiting oncogenic lncRNAs by siRNAs requires an engineered delivering system to maintain serum stability, prevent immune destruction, and act specifically on tumor cells. Poor membrane permeability of ASOs172 makes ASOs-based therapy difficult to employ for nuclear lncRNA as a target. Further technological advancement is needed to address this problem. Also, CRISPR/Cas9 system that contains a single guide RNA (sgRNA) and a Cas9 enzyme is now a prevalent approach for targeting lncRNAs. Potent off-target actions are the major obstacles for the clinical application of CRISPR/Cas9. Modifying Cas9 or sgRNA173 and sgRNA truncation174,175 are strategies for reducing these off-target effects. Further research is needed to identify stable and efficient means to overcome off-target effects and allow clinical application of CRISPR/Cas9. Finally, lncRNAs display notable high tissue specificity. LncRNAs expression may vary even within distinct tumor cells and in different individuals due to tumor heterogeneity. Such variability may interject uncertainty into evaluation and verification at the preclinical stage. Developing advanced sequencing and analytical techniques can ensure precise screening of differentially expressed lncRNAs in cancers. Such study will improve understanding of underlying molecular mechanisms in cancer glycolysis and promote the application of lncRNA-based clinical therapy.
Conclusion
Cancer cells exhibit distinct metabolic characteristics that employ glycolysis to provide energy and intermediary metabolites. This specific metabolic phenomenon is advantageous for cancer cell survival. We highlight regulatory mechanisms of lncRNAs in cancer glycolysis in this review. LncRNAs interact with metabolic enzymes or transporters involved in glycolysis and significantly influence glycolytic metabolism and cancer progression. Additionally, deregulated signaling pathways are vital for cancer glycolysis. LncRNAs, the crucial regulators of glycolysis, are the target of substantial researches into possible therapeutic targets for cancer treatment. More comprehensive investigations are required to explore underlying mechanisms by which lncRNAs regulate aerobic glycolysis. Understanding these mechanisms will promote the development of lncRNA-based strategies to control aberrant metabolic reprogramming and cancer development. Ultimately, such effort will lead to successful clinical application.
Ethics Approval and Consent to Participate
This article does not contain any studies with human participants/animals performed by any of the authors.
Funding
The work was supported by Foundation of Science and Technology Department of Sichuan Province (2020YJ0147); Foundation of “apricot grove scholar” of Chengdu University of Traditional Chinese Medicine (2019yky09); Postdoctoral Science Foundation of Chengdu University of Traditional Chinese Medicine (030054080).
Disclosure
All the authors declare that they have no conflicts of interest.
References
1. Judge A, Dodd MS. Metabolism. Essays Biochem. 2020;64(4):607–647. doi:10.1042/ebc20190041
2. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi:10.1016/j.cell.2011.02.013
3. Sun L, Suo C, Li ST, Zhang H, Gao P. Metabolic reprogramming for cancer cells and their microenvironment: beyond the Warburg effect. Biochim Biophys Acta. 2018;1870(1):51–66. doi:10.1016/j.bbcan.2018.06.005
4. Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314. doi:10.1126/science.123.3191.309
5. Icard P, Shulman S, Farhat D, Steyaert JM, Alifano M, Lincet H. How the Warburg effect supports aggressiveness and drug resistance of cancer cells? Drug Resist Updat. 2018;38:1–11. doi:10.1016/j.drup.2018.03.001
6. Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi:10.1126/science.1160809
7. Yang J, Ren B, Yang G, et al. The enhancement of glycolysis regulates pancreatic cancer metastasis. Cell Mol Life Sci. 2020;77(2):305–321. doi:10.1007/s00018-019-03278-z
8. Lin J, Xia L, Liang J, et al. The roles of glucose metabolic reprogramming in chemo- and radio-resistance. J Exp Clin Cancer Res. 2019;38(1):218. doi:10.1186/s13046-019-1214-z
9. Hombach S, Non-coding KM. RNAs: classification, biology and functioning. Adv Exp Med Biol. 2016;937:3–17. doi:10.1007/978-3-319-42059-2_1
10. Lin S, Zhen Y, Guan Y, Yi H. Roles of wnt/β-catenin signaling pathway regulatory long non-coding RNAs in the pathogenesis of non-small cell lung cancer. Cancer Manag Res. 2020;12:4181–4191. doi:10.2147/cmar.S241519
11. Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388. doi:10.1080/15476286.2015.1020271
12. Zhang X, Wang W, Zhu W, et al. Mechanisms and functions of long non-coding RNAs at multiple regulatory levels. Int J Mol Sci. 2019;20(22). doi:10.3390/ijms20225573
13. Zhang Z, Fang E, Rong Y, et al. Hypoxia-induced lncRNA CASC9 enhances glycolysis and the epithelial-mesenchymal transition of pancreatic cancer by a positive feedback loop with AKT/HIF-1α signaling. Am J Cancer Res. 2021;11(1):123–137.
14. Xu Z, Zhang D, Zhang Z, et al. MicroRNA-505, suppressed by oncogenic long non-coding RNA LINC01448, acts as a novel suppressor of glycolysis and tumor progression through inhibiting HK2 expression in pancreatic cancer. Front Cell Dev Biol. 2020;8:625056. doi:10.3389/fcell.2020.625056
15. Liu Y, He D, Xiao M, Zhu Y, Zhou J, Cao K. Long noncoding RNA LINC00518 induces radioresistance by regulating glycolysis through an miR-33a-3p/HIF-1α negative feedback loop in melanoma. Cell Death Dis. 2021;12(3):245. doi:10.1038/s41419-021-03523-z
16. Pei LJ, Sun PJ, Ma K, Guo YY, Wang LY, Liu FD. LncRNA-SNHG7 interferes with miR-34a to de-sensitize gastric cancer cells to cisplatin. Cancer Biomark. 2020. doi:10.3233/cbm-201621
17. Hinshaw DC, Shevde LA. The tumor microenvironment innately modulates cancer progression. Cancer Res. 2019;79(18):4557–4566. doi:10.1158/0008-5472.Can-18-3962
18. Avagliano A, Fiume G, Pelagalli A, et al. Metabolic plasticity of melanoma cells and their crosstalk with tumor microenvironment. Front Oncol. 2020;10:722. doi:10.3389/fonc.2020.00722
19. Xu S, Herschman HR. A tumor agnostic therapeutic strategy for hexokinase 1-null/hexokinase 2-positive cancers. Cancer Res. 2019;79(23):5907–5914. doi:10.1158/0008-5472.Can-19-1789
20. Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis?. Nat Rev Cancer. 2004;4(11):891–899. doi:10.1038/nrc1478
21. Ganeshan K, Chawla A. Metabolic regulation of immune responses. Annu Rev Immunol. 2014;32:609–634. doi:10.1146/annurev-immunol-032713-120236
22. Lu J, Tan M, Cai Q. The Warburg effect in tumor progression: mitochondrial oxidative metabolism as an anti-metastasis mechanism. Cancer Lett. 2015;356(2 Pt A):156–164. doi:10.1016/j.canlet.2014.04.001
23. Wang G, Wang JJ, Yin PH, et al. New strategies for targeting glucose metabolism-mediated acidosis for colorectal cancer therapy. J Cell Physiol. 2018;234(1):348–368. doi:10.1002/jcp.26917
24. Turpin A, El Amrani M, Bachet JB, Pietrasz D, Schwarz L, Hammel P. Adjuvant pancreatic cancer management: towards new perspectives in 2021. Cancers. 2020;12(12):3866. doi:10.3390/cancers12123866
25. Afonso J, Santos LL, Morais A, Amaro T, Longatto-Filho A, Baltazar F. Metabolic coupling in urothelial bladder cancer compartments and its correlation to tumor aggressiveness. Cell Cycle. 2016;15(3):368–380. doi:10.1080/15384101.2015.1121329
26. Zhang Z, Tan X, Luo J, Yao H, Si Z, Tong JS. The miR-30a-5p/CLCF1 axis regulates sorafenib resistance and aerobic glycolysis in hepatocellular carcinoma. Cell Death Dis. 2020;11(10):902. doi:10.1038/s41419-020-03123-3
27. Ng SP, Leong T. Role of radiation therapy in gastric cancer. Ann Surg Oncol. 2021. doi:10.1245/s10434-021-09639-y
28. Zhao F, Ming J, Zhou Y, Fan L. Inhibition of Glut1 by WZB117 sensitizes radioresistant breast cancer cells to irradiation. Cancer Chemother Pharmacol. 2016;77(5):963–972. doi:10.1007/s00280-016-3007-9
29. Tian RF, Li XF, Xu C, et al. SiRNA targeting PFK1 inhibits proliferation and migration and enhances radiosensitivity by suppressing glycolysis in colorectal cancer. Am J Transl Res. 2020;12(9):4923–4940.
30. Zhao H, Jiang H, Li Z, et al. 2-Methoxyestradiol enhances radiosensitivity in radioresistant melanoma MDA-MB-435R cells by regulating glycolysis via HIF-1α/PDK1 axis. Int J Oncol. 2017;50(5):1531–1540. doi:10.3892/ijo.2017.3924
31. Kumari N, Das A, Bhatt AN. Interleukin-6 confers radio-resistance by inducing Akt-mediated glycolysis and reducing mitochondrial damage in cells. J Biochem. 2020;167(3):303–314. doi:10.1093/jb/mvz091
32. Krishna S, Lowery FJ, Copeland AR, et al. Stem-like CD8 T cells mediate response of adoptive cell immunotherapy against human cancer. Science. 2020;370(6522):1328–1334. doi:10.1126/science.abb9847
33. O’Donnell JS, Teng MWL, Smyth MJ. Cancer immunoediting and resistance to T cell-based immunotherapy. Nat Rev Clin Oncol. 2019;16(3):151–167. doi:10.1038/s41571-018-0142-8
34. Cascone T, McKenzie JA, Mbofung RM, et al. Increased tumor glycolysis characterizes immune resistance to adoptive T cell therapy. Cell Metab. 2018;27(5):977–87.e4. doi:10.1016/j.cmet.2018.02.024
35. Rowshanravan B, Halliday N, Sansom DM. CTLA-4: a moving target in immunotherapy. Blood. 2018;131(1):58–67. doi:10.1182/blood-2017-06-741033
36. Chikuma S. CTLA-4, an essential immune-checkpoint for T-cell activation. Curr Top Microbiol Immunol. 2017;410:99–126. doi:10.1007/82_2017_61
37. Zappasodi R, Serganova I, Cohen IJ, et al. CTLA-4 blockade drives loss of T(reg) stability in glycolysis-low tumours. Nature. 2021;591(7851):652–658. doi:10.1038/s41586-021-03326-4
38. Pascale RM, Calvisi DF, Simile MM, Feo CF, Feo F. The Warburg effect 97 years after its discovery. Cancers. 2020;12(10):2819. doi:10.3390/cancers12102819
39. Chen LQ, Cheung LS, Feng L, Tanner W, Frommer WB. Transport of sugars. Annu Rev Biochem. 2015;84:865–894. doi:10.1146/annurev-biochem-060614-033904
40. Ancey PB, Contat C, Meylan E. Glucose transporters in cancer - from tumor cells to the tumor microenvironment. FEBS J. 2018;285(16):2926–2943. doi:10.1111/febs.14577
41. Zhou D, Jiang L, Jin L, Yao Y, Wang P, Zhu X. Glucose transporter-1 cooperating with AKT signaling promote gastric cancer progression. Cancer Manag Res. 2020;12:4151–4160. doi:10.2147/cmar.S251596
42. Deng D, Xu C, Sun P, et al. Crystal structure of the human glucose transporter GLUT1. Nature. 2014;510(7503):121–125. doi:10.1038/nature13306
43. Mueckler M, Thorens B. The SLC2 (GLUT) family of membrane transporters. Mol Aspects Med. 2013;34(2–3):121–138. doi:10.1016/j.mam.2012.07.001
44. Leto D, Saltiel AR. Regulation of glucose transport by insulin: traffic control of GLUT4. Nat Rev Mol Cell Biol. 2012;13(6):383–396. doi:10.1038/nrm3351
45. Tsai TH, Yang CC, Kou TC, et al. Overexpression of GLUT3 promotes metastasis of triple-negative breast cancer by modulating the inflammatory tumor microenvironment. J Cell Physiol. 2021;236(6):4669–4680. doi:10.1002/jcp.30189
46. Zambrano A, Molt M, Uribe E, Salas M. Glut 1 in cancer cells and the inhibitory action of resveratrol as a potential therapeutic strategy. Int J Mol Sci. 2019;20(13):3374. doi:10.3390/ijms20133374
47. Mao A, Zhou X, Liu Y, Ding J, Miao A, Pan G. KLF8 is associated with poor prognosis and regulates glycolysis by targeting GLUT4 in gastric cancer. J Cell Mol Med. 2019;23(8):5087–5097. doi:10.1111/jcmm.14378
48. Garrido P, Osorio FG, Morán J, et al. Loss of GLUT4 induces metabolic reprogramming and impairs viability of breast cancer cells. J Cell Physiol. 2015;230(1):191–198. doi:10.1002/jcp.24698
49. Ellis BC, Graham LD, Molloy PL. CRNDE, a long non-coding RNA responsive to insulin/IGF signaling, regulates genes involved in central metabolism. Biochim Biophys Acta. 2014;1843(2):372–386. doi:10.1016/j.bbamcr.2013.10.016
50. Wang Y, Zhang X, Wang Z, et al. LncRNA-p23154 promotes the invasion-metastasis potential of oral squamous cell carcinoma by regulating Glut1-mediated glycolysis. Cancer Lett. 2018;434:172–183. doi:10.1016/j.canlet.2018.07.016
51. Chang L, Xu W, Zhang Y, Gong F. Long non-coding RNA-NEF targets glucose transportation to inhibit the proliferation of non-small-cell lung cancer cells. Oncol Lett. 2019;17(3):2795–2801. doi:10.3892/ol.2019.9919
52. Shi Z, Guo F, Jia D, et al. Long non-coding RNA mortal obligate RNA transcript suppresses tumor cell proliferation in prostate carcinoma by inhibiting glucose uptake. Oncol Lett. 2019;18(4):3787–3791. doi:10.3892/ol.2019.10711
53. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi:10.1016/s0092-8674(04)00045-5
54. Han TS, Hur K, Cho HS, Ban HS. Epigenetic associations between lncRNA/circRNA and miRNA in hepatocellular carcinoma. Cancers. 2020;12(9):2622. doi:10.3390/cancers12092622
55. Chen XX, Zhang N, Fu XF, Jiang Y, Wang MY. LncRNA DBH-AS1 facilitates the tumorigenesis of melanoma by targeting miR-233-3p via IGF-1R/Akt signaling. Eur Rev Med Pharmacol Sci. 2020;24(14):7698–7708. doi:10.26355/eurrev_202007_22272
56. Zhang C, Liu J, Wu R, et al. Tumor suppressor p53 negatively regulates glycolysis stimulated by hypoxia through its target RRAD. Oncotarget. 2014;5(14):5535–5546. doi:10.18632/oncotarget.2137
57. Zhao Y, Liu Y, Lin L, et al. The lncRNA MACC1-AS1 promotes gastric cancer cell metabolic plasticity via AMPK/Lin28 mediated mRNA stability of MACC1. Mol Cancer. 2018;17(1):69. doi:10.1186/s12943-018-0820-2
58. Payen VL, Mina E, Van Hée VF, Porporato PE, Sonveaux P. Monocarboxylate transporters in cancer. Mol Metab. 2020;33:48–66. doi:10.1016/j.molmet.2019.07.006
59. Logotheti S, Marquardt S, Gupta SK, et al. LncRNA-SLC16A1-AS1 induces metabolic reprogramming during bladder cancer progression as target and co-activator of E2F1. Theranostics. 2020;10(21):9620–9643. doi:10.7150/thno.44176
60. Akram M. Mini-review on glycolysis and cancer. J Cancer Educ. 2013;28(3):454–457. doi:10.1007/s13187-013-0486-9
61. Wilson JE. Isozymes of mammalian hexokinase: structure, subcellular localization and metabolic function. J Exp Biol. 2003;206(Pt 12):2049–2057. doi:10.1242/jeb.00241
62. Al Hasawi N, Alkandari MF, Luqmani YA. Phosphofructokinase: a mediator of glycolytic flux in cancer progression. Crit Rev Oncol Hematol. 2014;92(3):312–321. doi:10.1016/j.critrevonc.2014.05.007
63. Israelsen WJ, Vander Heiden MG. Pyruvate kinase: function, regulation and role in cancer. Semin Cell Dev Biol. 2015;43:43–51. doi:10.1016/j.semcdb.2015.08.004
64. Yin D, Hua L, Wang J, Liu Y, Li X. Long non-coding RNA DUXAP8 facilitates cell viability, migration, and glycolysis in non-small-cell lung cancer via regulating HK2 and LDHA by inhibition of miR-409-3p. Onco Targets Ther. 2020;13:7111–7123. doi:10.2147/ott.S243542
65. Ma Y, Hu M, Zhou L, et al. Long non-coding RNA HOTAIR promotes cancer cell energy metabolism in pancreatic adenocarcinoma by upregulating hexokinase-2. Oncol Lett. 2019;18(3):2212–2219. doi:10.3892/ol.2019.10551
66. Rupaimoole R, Lee J, Haemmerle M, et al. Long noncoding RNA ceruloplasmin promotes cancer growth by altering glycolysis. Cell Rep. 2015;13(11):2395–2402. doi:10.1016/j.celrep.2015.11.047
67. Yang B, Zhang L, Cao Y, et al. Overexpression of lncRNA IGFBP4-1 reprograms energy metabolism to promote lung cancer progression. Mol Cancer. 2017;16(1):154. doi:10.1186/s12943-017-0722-8
68. Ule J, Blencowe BJ. Alternative splicing regulatory networks: functions, mechanisms, and evolution. Mol Cell. 2019;76(2):329–345. doi:10.1016/j.molcel.2019.09.017
69. Gu Z, Xia J, Xu H, Frech I, Tricot G, Zhan F. NEK2 promotes aerobic glycolysis in multiple myeloma through regulating splicing of pyruvate kinase. J Hematol Oncol. 2017;10(1):17. doi:10.1186/s13045-017-0392-4
70. Lan Z, Yao X, Sun K, Li A, Liu S, Wang X. The interaction between lncRNA SNHG6 and hnRNPA1 contributes to the growth of colorectal cancer by enhancing aerobic glycolysis through the regulation of alternative splicing of PKM. Front Oncol. 2020;10:363. doi:10.3389/fonc.2020.00363
71. Grillone K, Riillo C, Scionti F, et al. Non-coding RNAs in cancer: platforms and strategies for investigating the genomic “dark matter”. J Exp Clin Cancer Res. 2020;39(1):117. doi:10.1186/s13046-020-01622-x
72. Huang JZ, Chen M, Chen D, et al. A peptide encoded by a putative lncRNA HOXB-AS3 suppresses colon cancer growth. Mol Cell. 2017;68(1):171–84.e6. doi:10.1016/j.molcel.2017.09.015
73. Qian Y, Song W, Wu X, et al. DLX6 antisense RNA 1 modulates glucose metabolism and cell growth in gastric cancer by targeting microRNA-4290. Dig Dis Sci. 2021;66(2):460–473. doi:10.1007/s10620-020-06223-4
74. Shen Y, Xu J, Pan X, et al. LncRNA KCNQ1OT1 sponges miR-34c-5p to promote osteosarcoma growth via ALDOA enhanced aerobic glycolysis. Cell Death Dis. 2020;11(4):278. doi:10.1038/s41419-020-2485-1
75. Yang W, Zheng Y, Xia Y, et al. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat Cell Biol. 2012;14(12):1295–1304. doi:10.1038/ncb2629
76. Guan YF, Huang QL, Ai YL, et al. Nur77-activated lncRNA WFDC21P attenuates hepatocarcinogenesis via modulating glycolysis. Oncogene. 2020;39(11):2408–2423. doi:10.1038/s41388-020-1158-y
77. Wang C, Li Y, Yan S, et al. Interactome analysis reveals that lncRNA HULC promotes aerobic glycolysis through LDHA and PKM2. Nat Commun. 2020;11(1):3162. doi:10.1038/s41467-020-16966-3
78. Xing Z, Zhang Y, Liang K, et al. Expression of long noncoding RNA YIYA promotes glycolysis in breast cancer. Cancer Res. 2018;78(16):4524–4532. doi:10.1158/0008-5472.Can-17-0385
79. Shi L, Pan H, Liu Z, Xie J, Han W. Roles of PFKFB3 in cancer. Signal Transduct Target Ther. 2017;2:17044. doi:10.1038/sigtrans.2017.44
80. Bian Z, Zhang J, Li M, et al. LncRNA-FEZF1-AS1 promotes tumor proliferation and metastasis in colorectal cancer by regulating PKM2 signaling. Clin Cancer Res. 2018;24(19):4808–4819. doi:10.1158/1078-0432.Ccr-17-2967
81. Liu C, Zhang Y, She X, et al. A cytoplasmic long noncoding RNA LINC00470 as a new AKT activator to mediate glioblastoma cell autophagy. J Hematol Oncol. 2018;11(1):77. doi:10.1186/s13045-018-0619-z
82. Zheng F, Chen J, Zhang X, et al. The HIF-1α antisense long non-coding RNA drives a positive feedback loop of HIF-1α mediated transactivation and glycolysis. Nat Commun. 2021;12(1):1341. doi:10.1038/s41467-021-21535-3
83. Schito L, Semenza GL. Hypoxia-inducible factors: master regulators of cancer progression. Trends Cancer. 2016;2(12):758–770. doi:10.1016/j.trecan.2016.10.016
84. Nagao A, Kobayashi M, Koyasu S, Chow CCT, Harada H. HIF-1-dependent reprogramming of glucose metabolic pathway of cancer cells and its therapeutic significance. Int J Mol Sci. 2019;20(2). doi:10.3390/ijms20020238
85. Yao Z, Zhang Q, Guo F, et al. Long noncoding RNA PCED1B-AS1 promotes the Warburg effect and tumorigenesis by upregulating HIF-1α in glioblastoma. Cell Transplant. 2020;29:963689720906777. doi:10.1177/0963689720906777
86. Du Y, Wei N, Ma R, Jiang SH, Song D. Long noncoding RNA MIR210HG promotes the Warburg effect and tumor growth by enhancing HIF-1α translation in triple-negative breast cancer. Front Oncol. 2020;10:580176. doi:10.3389/fonc.2020.580176
87. Strowitzki MJ, Cummins EP, Taylor CT. Protein hydroxylation by hypoxia-inducible factor (HIF) hydroxylases: unique or ubiquitous? Cells. 2019;8(5):384. doi:10.3390/cells8050384
88. Yang F, Zhang H, Mei Y, Wu M. Reciprocal regulation of HIF-1α and lincRNA-p21 modulates the Warburg effect. Mol Cell. 2014;53(1):88–100. doi:10.1016/j.molcel.2013.11.004
89. Lin A, Li C, Xing Z, et al. The LINK-A lncRNA activates normoxic HIF1α signalling in triple-negative breast cancer. Nat Cell Biol. 2016;18(2):213–224. doi:10.1038/ncb3295
90. Baluapuri A, Wolf E, Eilers M. Target gene-independent functions of MYC oncoproteins. Nat Rev Mol Cell Biol. 2020;21(5):255–267. doi:10.1038/s41580-020-0215-2
91. Stine ZE, Walton ZE, Altman BJ, Hsieh AL, Dang CV. MYC, metabolism, and cancer. Cancer Discov. 2015;5(10):1024–1039. doi:10.1158/2159-8290.Cd-15-0507
92. Hung CL, Wang LY, Yu YL, et al. A long noncoding RNA connects c-Myc to tumor metabolism. Proc Natl Acad Sci U S A. 2014;111(52):18697–18702. doi:10.1073/pnas.1415669112
93. Tang J, Yan T, Bao Y, et al. LncRNA GLCC1 promotes colorectal carcinogenesis and glucose metabolism by stabilizing c-Myc. Nat Commun. 2019;10(1):3499. doi:10.1038/s41467-019-11447-8
94. Rodic S, Vincent MD. Reactive oxygen species (ROS) are a key determinant of cancer’s metabolic phenotype. Int J Cancer. 2018;142(3):440–448. doi:10.1002/ijc.31069
95. Xiang S, Gu H, Jin L, Thorne RF, Zhang XD, Wu M. LncRNA IDH1-AS1 links the functions of c-Myc and HIF1α via IDH1 to regulate the Warburg effect. Proc Natl Acad Sci U S A. 2018;115(7):E1465–e74. doi:10.1073/pnas.1711257115
96. Pan Y, Mansfield KD, Bertozzi CC, et al. Multiple factors affecting cellular redox status and energy metabolism modulate hypoxia-inducible factor prolyl hydroxylase activity in vivo and in vitro. Mol Cell Biol. 2007;27(3):912–925. doi:10.1128/mcb.01223-06
97. Hoxhaj G, Manning BD. The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat Rev Cancer. 2020;20(2):74–88. doi:10.1038/s41568-019-0216-7
98. Wang B, Shen Y, Zou Y, et al. TOP2A promotes cell migration, invasion and epithelial-mesenchymal transition in cervical cancer via activating the PI3K/AKT signaling. Cancer Manag Res. 2020;12:3807–3814. doi:10.2147/cmar.S240577
99. Menon S, Dibble CC, Talbott G, et al. Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell. 2014;156(4):771–785. doi:10.1016/j.cell.2013.11.049
100. Parsons R. Discovery of the PTEN tumor suppressor and its connection to the PI3K and AKT oncogenes. Cold Spring Harb Perspect Med. 2020;10(8):a036129. doi:10.1101/cshperspect.a036129
101. Li W, Huang K, Wen F, et al. LINC00184 silencing inhibits glycolysis and restores mitochondrial oxidative phosphorylation in esophageal cancer through demethylation of PTEN. EBioMedicine. 2019;44:298–310. doi:10.1016/j.ebiom.2019.05.055
102. Zou ZW, Ma C, Medoro L, et al. LncRNA ANRIL is up-regulated in nasopharyngeal carcinoma and promotes the cancer progression via increasing proliferation, reprograming cell glucose metabolism and inducing side-population stem-like cancer cells. Oncotarget. 2016;7(38):61741–61754. doi:10.18632/oncotarget.11437
103. Li Z, Li X, Wu S, Xue M, Chen W. Long non-coding RNA UCA1 promotes glycolysis by upregulating hexokinase 2 through the mTOR-STAT3/microRNA143 pathway. Cancer Sci. 2014;105(8):951–955. doi:10.1111/cas.12461
104. Zheng YL, Li L, Jia YX, et al. LINC01554-mediated glucose metabolism reprogramming suppresses tumorigenicity in hepatocellular carcinoma via downregulating PKM2 expression and inhibiting Akt/mTOR signaling pathway. Theranostics. 2019;9(3):796–810. doi:10.7150/thno.28992
105. Shaw LM. The insulin receptor substrate (IRS) proteins: at the intersection of metabolism and cancer. Cell Cycle. 2011;10(11):1750–1756. doi:10.4161/cc.10.11.15824
106. Cheng Z, Luo C, Guo Z. LncRNA-XIST/microRNA-126 sponge mediates cell proliferation and glucose metabolism through the IRS1/PI3K/Akt pathway in glioma. J Cell Biochem. 2020;121(3):2170–2183. doi:10.1002/jcb.29440
107. Taniguchi CM, Emanuelli B, Kahn CR. Critical nodes in signalling pathways: insights into insulin action. Nat Rev Mol Cell Biol. 2006;7(2):85–96. doi:10.1038/nrm1837
108. Faubert B, Boily G, Izreig S, et al. AMPK is a negative regulator of the Warburg effect and suppresses tumor growth in vivo. Cell Metab. 2013;17(1):113–124. doi:10.1016/j.cmet.2012.12.001
109. Herzig S, Shaw RJ. AMPK: guardian of metabolism and mitochondrial homeostasis. Nat Rev Mol Cell Biol. 2018;19(2):121–135. doi:10.1038/nrm.2017.95
110. Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115(5):577–590. doi:10.1016/s0092-8674(03)00929-2
111. Nguyen HB, Babcock JT, Wells CD, Quilliam LA. LKB1 tumor suppressor regulates AMP kinase/mTOR-independent cell growth and proliferation via the phosphorylation of Yap. Oncogene. 2013;32(35):4100–4109. doi:10.1038/onc.2012.431
112. Hezel AF, Bardeesy N. LKB1; linking cell structure and tumor suppression. Oncogene. 2008;27(55):6908–6919. doi:10.1038/onc.2008.342
113. Shackelford DB, Vasquez DS, Corbeil J, et al. mTOR and HIF-1alpha-mediated tumor metabolism in an LKB1 mouse model of Peutz-Jeghers syndrome. Proc Natl Acad Sci U S A. 2009;106(27):11137–11142. doi:10.1073/pnas.0900465106
114. Zeng Q, Chen J, Li Y, et al. LKB1 inhibits HPV-associated cancer progression by targeting cellular metabolism. Oncogene. 2017;36(9):1245–1255. doi:10.1038/onc.2016.290
115. Chen Z, Li JL, Lin S, et al. cAMP/CREB-regulated LINC00473 marks LKB1-inactivated lung cancer and mediates tumor growth. J Clin Invest. 2016;126(6):2267–2279. doi:10.1172/jci85250
116. Kim SJ, Ju JS, Park SS, et al. An RNA-binding-protein, NONO governs energy metabolism by regulating NAMPT in lung cancer. Biochem Biophys Res Commun. 2020;528(2):376–382. doi:10.1016/j.bbrc.2020.01.011
117. Liu X, Xiao ZD, Han L, et al. LncRNA NBR2 engages a metabolic checkpoint by regulating AMPK under energy stress. Nat Cell Biol. 2016;18(4):431–442. doi:10.1038/ncb3328
118. Yang W, Zheng Z, Yi P, et al. LncRNA NBR2 inhibits the malignancy of thyroid cancer, associated with enhancing the AMPK signaling. Front Oncol. 2020;10:956. doi:10.3389/fonc.2020.00956
119. Bensaad K, Vousden KH. p53: new roles in metabolism. Trends Cell Biol. 2007;17(6):286–291. doi:10.1016/j.tcb.2007.04.004
120. Schwartzenberg-Bar-Yoseph F, Armoni M, Karnieli E. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res. 2004;64(7):2627–2633. doi:10.1158/0008-5472.can-03-0846
121. Kondoh H, Lleonart ME, Gil J, et al. Glycolytic enzymes can modulate cellular life span. Cancer Res. 2005;65(1):177–185.
122. Zhou Y, Niu W, Luo Y, et al. p53/Lactate dehydrogenase A axis negatively regulates aerobic glycolysis and tumor progression in breast cancer expressing wild-type p53. Cancer Sci. 2019;110(3):939–949. doi:10.1111/cas.13928
123. Chen R, Liu Y, Zhuang H, et al. Quantitative proteomics reveals that long non-coding RNA MALAT1 interacts with DBC1 to regulate p53 acetylation. Nucleic Acids Res. 2017;45(17):9947–9959. doi:10.1093/nar/gkx600
124. Uroda T, Anastasakou E, Rossi A, et al. Conserved pseudoknots in lncRNA MEG3 are essential for stimulation of the p53 pathway. Mol Cell. 2019;75(5):982–95.e9. doi:10.1016/j.molcel.2019.07.025
125. Zhang Y, Wu J, Jing H, Huang G, Sun Z, Xu S. Long noncoding RNA MEG3 inhibits breast cancer growth via upregulating endoplasmic reticulum stress and activating NF-κB and p53. J Cell Biochem. 2019;120(4):6789–6797. doi:10.1002/jcb.27982
126. Lee SY, Jeon HM, Ju MK, et al. Wnt/Snail signaling regulates cytochrome C oxidase and glucose metabolism. Cancer Res. 2012;72(14):3607–3617. doi:10.1158/0008-5472.Can-12-0006
127. Pate KT, Stringari C, Sprowl-Tanio S, et al. Wnt signaling directs a metabolic program of glycolysis and angiogenesis in colon cancer. EMBO J. 2014;33(13):1454–1473. doi:10.15252/embj.201488598
128. Zhang H, Qin D, Jiang Z, Zhang J. SNHG9/miR-199a-5p/Wnt2 axis regulates cell growth and aerobic glycolysis in glioblastoma. J Neuropathol Exp Neurol. 2019;78(10):939–948. doi:10.1093/jnen/nlz078
129. Kang R, Yao DF, Xu GZ, Zhou YH. The knockdown of SNHG3 inhibits the progression of laryngeal squamous cell carcinoma by miR-340-5p/YAP1 axis and Wnt/β-catenin pathway. Neoplasma. 2020;67(5):1094–1105. doi:10.4149/neo_2020_191022N1073
130. Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13(4):246–257. doi:10.1038/nrc3458
131. Lin X, Feng D, Li P, Lv Y. LncRNA LINC00857 regulates the progression and glycolysis in ovarian cancer by modulating the Hippo signaling pathway. Cancer Med. 2020;9(21):8122–8132. doi:10.1002/cam4.3322
132. Zheng X, Han H, Liu GP, et al. LncRNA wires up Hippo and Hedgehog signaling to reprogramme glucose metabolism. EMBO J. 2017;36(22):3325–3335. doi:10.15252/embj.201797609
133. Hao Y, Baker D, Ten Dijke P. TGF-β-mediated epithelial-mesenchymal transition and cancer metastasis. Int J Mol Sci. 2019;20(11):2767. doi:10.3390/ijms20112767
134. Mota MSV, Jackson WP, Bailey SK, et al. Deficiency of tumor suppressor Merlin facilitates metabolic adaptation by co-operative engagement of SMAD-Hippo signaling in breast cancer. Carcinogenesis. 2018;39(9):1165–1175. doi:10.1093/carcin/bgy078
135. Wu HZ, Xiao JQ, Xiao SS, Cheng Y. KRAS: a promising therapeutic target for cancer treatment. Curr Top Med Chem. 2019;19(23):2081–2097. doi:10.2174/1568026619666190905164144
136. Pupo E, Avanzato D, Middonti E, Bussolino F, Lanzetti L. KRAS-driven metabolic rewiring reveals novel actionable targets in cancer. Front Oncol. 2019;9:848. doi:10.3389/fonc.2019.00848
137. Zhai H, Zhang X, Sun X, Zhang D, Ma S. Long non-coding RNA LINC01420 contributes to pancreatic cancer progression through targeting KRAS proto-oncogene. Dig Dis Sci. 2020;65(4):1042–1052. doi:10.1007/s10620-019-05829-7
138. Zhai X, Xu W. Long noncoding RNA ATB promotes proliferation, migration, and invasion in bladder cancer by suppressing microRNA-126. Oncol Res. 2018;26(7):1063–1072. doi:10.3727/096504018x15152072098476
139. Yang W, Redpath RE, Zhang C, Ning N. Long non-coding RNA H19 promotes the migration and invasion of colon cancer cells via MAPK signaling pathway. Oncol Lett. 2018;16(3):3365–3372. doi:10.3892/ol.2018.9052
140. Zhai S, Xu Z, Xie J, et al. Epigenetic silencing of LncRNA LINC00261 promotes c-myc-mediated aerobic glycolysis by regulating miR-222-3p/HIPK2/ERK axis and sequestering IGF2BP1. Oncogene. 2021;40(2):277–291. doi:10.1038/s41388-020-01525-3
141. Schulte JH, Kirfel J, Lim S, et al. Transcription factor AP2alpha (TFAP2a) regulates differentiation and proliferation of neuroblastoma cells. Cancer Lett. 2008;271(1):56–63. doi:10.1016/j.canlet.2008.05.039
142. Zhang G, Ma A, Jin Y, Pan G, Wang C. LncRNA SNHG16 induced by TFAP2A modulates glycolysis and proliferation of endometrial carcinoma through miR-490-3p/HK2 axis. Am J Transl Res. 2019;11(11):7137–7145.
143. Hua Q, Jin M, Mi B, et al. LINC01123, a c-Myc-activated long non-coding RNA, promotes proliferation and aerobic glycolysis of non-small cell lung cancer through miR-199a-5p/c-Myc axis. J Hematol Oncol. 2019;12(1):91. doi:10.1186/s13045-019-0773-y
144. Liu J, Liu ZX, Wu QN, et al. Long noncoding RNA AGPG regulates PFKFB3-mediated tumor glycolytic reprogramming. Nat Commun. 2020;11(1):1507. doi:10.1038/s41467-020-15112-3
145. Liao M, Liao W, Xu N, et al. LncRNA EPB41L4A-AS1 regulates glycolysis and glutaminolysis by mediating nucleolar translocation of HDAC2. EBioMedicine. 2019;41:200–213. doi:10.1016/j.ebiom.2019.01.035
146. Yu T, Guo F, Yu Y, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. 2017;170(3):548–63.e16. doi:10.1016/j.cell.2017.07.008
147. Hong J, Guo F, Lu SY, et al. F. nucleatum targets lncRNA ENO1-IT1 to promote glycolysis and oncogenesis in colorectal cancer. Gut. 2020. doi:10.1136/gutjnl-2020-322780
148. Hua Q, Mi B, Xu F, et al. Hypoxia-induced lncRNA-AC020978 promotes proliferation and glycolytic metabolism of non-small cell lung cancer by regulating PKM2/HIF-1α axis. Theranostics. 2020;10(11):4762–4778. doi:10.7150/thno.43839
149. Mirzaei H, Hamblin MR. Regulation of glycolysis by non-coding RNAs in cancer: switching on the Warburg effect. Mol Ther Oncolytics. 2020;19:218–239. doi:10.1016/j.omto.2020.10.003
150. Ding X, Liu J, Liu T, Ma Z, Wen D, Zhu J. miR-148b inhibits glycolysis in gastric cancer through targeting SLC2A1. Cancer Med. 2017;6(6):1301–1310. doi:10.1002/cam4.1008
151. Chen YH, Song Y, Yu YL, Cheng W, Tong X. miRNA-10a promotes cancer cell proliferation in oral squamous cell carcinoma by upregulating GLUT1 and promoting glucose metabolism. Oncol Lett. 2019;17(6):5441–5446. doi:10.3892/ol.2019.10257
152. Fei X, Qi M, Wu B, Song Y, Wang Y, Li T. MicroRNA-195-5p suppresses glucose uptake and proliferation of human bladder cancer T24 cells by regulating GLUT3 expression. FEBS Lett. 2012;586(4):392–397. doi:10.1016/j.febslet.2012.01.006
153. Zhang K, Zhang M, Jiang H, Liu F, Liu H, Li Y. Down-regulation of miR-214 inhibits proliferation and glycolysis in non-small-cell lung cancer cells via down-regulating the expression of hexokinase 2 and pyruvate kinase isozyme M2. Biomed Pharmacother. 2018;105:545–552. doi:10.1016/j.biopha.2018.06.009
154. Wang J, Wang H, Liu A, Fang C, Hao J, Wang Z. Lactate dehydrogenase A negatively regulated by miRNAs promotes aerobic glycolysis and is increased in colorectal cancer. Oncotarget. 2015;6(23):19456–19468. doi:10.18632/oncotarget.3318
155. Li Q, Pan X, Zhu D, Deng Z, Jiang R, Wang X. Circular RNA MAT2B promotes glycolysis and malignancy of hepatocellular carcinoma through the miR-338-3p/PKM2 axis under hypoxic stress. Hepatology. 2019;70(4):1298–1316. doi:10.1002/hep.30671
156. Zheng Z, Ma X, Li H. Circular RNA circMDM2 accelerates the glycolysis of oral squamous cell carcinoma by targeting miR-532-3p/HK2. J Cell Mol Med. 2020;24(13):7531–7537. doi:10.1111/jcmm.15380
157. Pan Z, Cai J, Lin J, et al. A novel protein encoded by circFNDC3B inhibits tumor progression and EMT through regulating Snail in colon cancer. Mol Cancer. 2020;19(1):71. doi:10.1186/s12943-020-01179-5
158. Fan L, Huang C, Li J, Gao T, Lin Z, Yao T. Long non‑coding RNA urothelial cancer associated 1 regulates radioresistance via the hexokinase 2/glycolytic pathway in cervical cancer. Int J Mol Med. 2018;42(4):2247–2259. doi:10.3892/ijmm.2018.3778
159. Sakai T, Matsuo Y, Okuda K, et al. Development of antitumor biguanides targeting energy metabolism and stress responses in the tumor microenvironment. Sci Rep. 2021;11(1):4852. doi:10.1038/s41598-021-83708-w
160. García Rubiño ME, Carrillo E, Ruiz Alcalá G, et al. Phenformin as an anticancer agent: challenges and prospects. Int J Mol Sci. 2019;20(13):3316. doi:10.3390/ijms2013
161. Liu X, Gan B. lncRNA NBR2 modulates cancer cell sensitivity to phenformin through GLUT1. Cell Cycle. 2016;15(24):3471–3481. doi:10.1080/15384101.2016.1249545
162. Zhang J, Chen G, Gao Y, Liang H. HOTAIR/miR-125 axis-mediated Hexokinase 2 expression promotes chemoresistance in human glioblastoma. J Cell Mol Med. 2020;24(10):5707–5717. doi:10.1111/jcmm.15233
163. Zheng H, Zhang M, Ke X, et al. LncRNA XIST/miR-137 axis strengthens chemo-resistance and glycolysis of colorectal cancer cells by hindering transformation from PKM2 to PKM1. Cancer Biomark. 2020. doi:10.3233/cbm-201740
164. Fu J, Shi H, Wang B, et al. LncRNA PVT1 links Myc to glycolytic metabolism upon CD4(+) T cell activation and Sjögren’s syndrome-like autoimmune response. J Autoimmun. 2020;107:102358. doi:10.1016/j.jaut.2019.102358
165. Chen Y, Li Z, Chen X, Zhang S. Long non-coding RNAs: from disease code to drug role. Acta Pharm Sin B. 2021;11(2):340–354. doi:10.1016/j.apsb.2020.10.001
166. Xu S, Jia G, Zhang H, et al. LncRNA HOXB-AS3 promotes growth, invasion and migration of epithelial ovarian cancer by altering glycolysis. Life Sci. 2021;264:118636. doi:10.1016/j.lfs.2020.118636
167. Jiang W, Kai J, Li D, Wei Z, Wang Y, Wang W. lncRNA HOXB-AS3 exacerbates proliferation, migration, and invasion of lung cancer via activating the PI3K-AKT pathway. J Cell Physiol. 2020;235(10):7194–7203. doi:10.1002/jcp.29618
168. Papaioannou D, Petri A, Dovey OM, et al. The long non-coding RNA HOXB-AS3 regulates ribosomal RNA transcription in NPM1-mutated acute myeloid leukemia. Nat Commun. 2019;10(1):5351. doi:10.1038/s41467-019-13259-2
169. Zhang XM, Chen H, Zhou B, et al. lncRNA HOXB-AS3 promotes hepatoma by inhibiting p53 expression. Eur Rev Med Pharmacol Sci. 2018;22(20):6784–6792. doi:10.26355/eurrev_201810_16145
170. Tian S, Yu Y, Huang H, Xu A, Xu H, Zhou Y. Expression level and clinical significance of NKILA in human cancers: a systematic review and meta-analysis. Biomed Res Int. 2020;2020:4540312. doi:10.1155/2020/4540312
171. Chen Z, Li S, Shen L, et al. NF-kappa B interacting long noncoding RNA enhances the Warburg effect and angiogenesis and is associated with decreased survival of patients with gliomas. Cell Death Dis. 2020;11(5):323. doi:10.1038/s41419-020-2520-2
172. Crooke ST. Molecular mechanisms of antisense oligonucleotides. Nucleic Acid Ther. 2017;27(2):70–77. doi:10.1089/nat.2016.0656
173. Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351(6268):84–88. doi:10.1126/science.aad5227
174. Kocak DD, Josephs EA, Bhandarkar V, Adkar SS, Kwon JB, Gersbach CA. Increasing the specificity of CRISPR systems with engineered RNA secondary structures. Nat Biotechnol. 2019;37(6):657–666. doi:10.1038/s41587-019-0095-1
175. Lee JK, Jeong E, Lee J, et al. Directed evolution of CRISPR-Cas9 to increase its specificity. Nat Commun. 2018;9(1):3048. doi:10.1038/s41467-018-05477-x
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
