Back to Journals » OncoTargets and Therapy » Volume 11
Regulation of cell proliferation and metastasis by microRNA-593-5p in human gastric cancer
Authors Yu H, Wei W, Cao W, Zhan Z, Yan L, Wu K, Xie D, Cai B, Xie Y, Xiao Q
Received 25 June 2018
Accepted for publication 5 September 2018
Published 24 October 2018 Volume 2018:11 Pages 7429—7440
DOI https://doi.org/10.2147/OTT.S178151
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Yao Dai
Han Yu,1,* Weiyuan Wei,1,2,* Wenlong Cao,1 Zexu Zhan,1 Linhai Yan,2 Kun Wu,1 Dongyi Xie,1 Bin Cai,1 Yubo Xie,3 Qiang Xiao1
1Department of Surgery, The First Affiliated Hospital of Guangxi Medical University, Nanning, China; 2Department of Gastrointestinal Surgery, Affiliated Tumor Hospital of Guangxi Medical University, Nanning, China; 3Department of Anesthesiology, The First Affiliated Hospital of Guangxi Medical University, Nanning, China
*These authors contributed equally to this work
Background: MicroRNA (miRNA) array analysis has reported that the expression of miR-593-5p is associated with lymph node metastasis in gastric cancer (GC); however, the function and mechanism of miR-593-5p in GC have not been described yet. miR-593-5p has also not been elucidated widely in other cancers.
Methods: miR-593-5p expression was detected by quantitative RT-PCR (qRT-PCR) in human GC tissues and cell lines. Cell proliferation was investigated using CCK-8 assays, cell cycle was detected by flow cytometric method, and cell migration and invasion abilities were evaluated by wound-healing and transwell assays. miR-593-5p-influenced gene expression profiles were detected by total gene expression chip method in MGC-803 cells, and miR-593-5p candidate target genes were predicted using bioinformatics methods. The candidate target gene and downstream of miR-593-5p were determined by qRT-PCR, Western blot, and dual-luciferase reporter assays. The effects of miR-593-5p on the growth and metastasis of GC were evaluated by tumor xenograft experiment in vivo.
Results: miR-593-5p was frequently downregulated in GC patients and GC cell lines. miR-593-5p was significantly correlated with tumor size and distant metastasis in GC patients. miR-593-5p inhibited cell proliferation, migration, and invasion and also arrested cell cycle at the G0/G1 phase in SGC-7901 and MGC-803 cells in vitro. miR-593-5p also suppressed tumor growth and metastasis in vivo. miR-593-5p influenced gene expression profile in MGC-803 cells. MST4 was indirectly targeted by miR-593-5p. miR-593-5p also downregulated FAK, MMP12, and JUN protein expression.
Conclusion: Our study suggests that miR-593-5p may function as a tumor suppressor in GC through a mechanism that regulates JUN pathway via indirectly targeting the MST4 gene.
Keywords: miR-593-5p, gastric cancer, MST4, JUN pathway
Introduction
Gastric cancer (GC) is one of the most common malignancies worldwide. MicroRNAs (miRNAs) are about 22 nt small endogenous nucleotide RNAs that play an important role in most biological processes in GC, such as carcinogenesis, apoptosis, and invasion.1,2 Mature miRNA is incorporated into an RNA-induced silencing complex that binds to a target messenger RNA (mRNA).3,4 The location is 3′-untranslated region (UTR) of the target gene.5 Yang et al6 had initially reported that the expression of miR-593-5p was lower in lymph node metastasis-positive tissue than in lymph node metastasis-negative tissue in miRNA chip expression profile in GC patients. This indicates that miR-593-5p may play a role in metastasis in GC.
Serine/threonine protein kinase 26 (STK26) is a member of the GCK group III family of kinases, which are a subset of the Ste20-like kinases. STK26 is also known as MST4. MST4 can promote cell proliferation in tumors. The expression of MST4 is very high in colon cancer and liver cancer cell lines.7,8 Interestingly, MST4 was found to be a crucial downstream target gene of miR-593-5p using bioinformatics tools (TargetScan: https://www.targetscan.org, miRDB: http://www.mirdb.org, and miRNA: http://www.microrna.org). These indicate that MST4 may be closely involved in tumor and can be regulated by miRNAs. Thus, we posited that MST4 may be directly regulated by miR-593-5p. To date, the function of miR-593-5p has not been reported.
In this study, we investigated the differential expression of miR-593-5p and its clinicopathological implication in GC. We further discussed the function of miR-593-5p in GC in vitro and in vivo and investigated whether miR-593-5p directly targets the MST4 gene. The data show that miR-593-5p suppresses GC progression through indirectly targeting the MST4 gene, which suggests that miR-593-5p can serve as a new potential therapeutic target for GC.
Materials and methods
Tissue samples and cell lines
Seventy-three pairs of GC tissues and their corresponding nonmalignant tissues from gastrectomy were obtained from the First Affiliated Hospital of Guangxi Medical University (Nanning, China). Written informed consents were obtained from the patients before collecting the tissues. The samples were divided into two parts: one part was immediately frozen in liquid nitrogen and stored at −80°C until use, and the other part was fixed in 10% formalin and analyzed by H&E staining. The subjects were informed of the study design and purpose as per the Declaration of Helsinki. This study was approved by the ethics committee of the First Affiliated Hospital of Guangxi Medical University (approval no 2013[KY-E-003]). The pathological diagnoses were reconfirmed according to the WHO 2007 classification of GC.
The GC cell lines AGS, GES-1, HGC-27, MGC-803, and SGC-7901 were obtained from Cell Bank of Shanghai Institute of Cell Biology, Chinese Academy of Sciences (Shanghai, China). The cells were maintained at 37°C and 5% CO2 in RPMI 1640 medium (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% FBS (Thermo Fisher Scientific), 100 U/mL penicillin, and 100 μg/mL streptomycin.
Quantitative RT-PCR analysis of mRNA and miRNA expression
Total RNA was extracted from fresh tissues and the cells using TRIzol reagent (Thermo Fisher Scientific). Reverse transcription was performed using an RNA PCR Kit (AMV) (TaKaRa, Otsu, Japan). Quantitation of miRNAs and mRNA was carried out by qRT-PCR using SYBR Premix Ex Taq™ (TaKaRa) according to the manufacturer’s instructions. U6 was used as the internal control for miR-593-5p expression levels. GAPDH was used as an internal control for other genes’ mRNA expression. The primers (Table S1) for the qRT-PCR detection of genes were synthesized by TaKaRa. The fold changes in expression were calculated using the 2−ΔΔCt method.
Establishment of stable miR-593-5p-expressing cell lines
Lentivirus expressing hsa-miR-593-5p and control vector were structured and provided by Genechem (Shanghai, China). Transfection of MGC-803 and SGC-7901 cells was carried out by Lipofectamine 2000 reagent (Thermo Fisher Scientific) according to the manufacturer’s protocol. After 24 hours of transfection, culture medium was replenished. At 48 hours post-transfection, puromycin (Sigma-Aldrich Co., St Louis, MO, USA) was added to the medium (0.002 mg/mL), and puromycin-resistant colonies were selected and separately expanded. When cells became confluent, they were harvested for the subsequent experiment.
Cell proliferation assay
To measure the effect of miR-593-5p on cellular proliferation rates, cells were incubated in 10% CCK-8 (Dojindo, Kumamoto, Japan) diluted in normal culture media at 37°C until visual color conversion was observed. Proliferation rates were determined at 0, 24, 48, 72, and 96 hours post-transfection, and the absorbance was determined at wavelength 450 nm by a microplate reader.
In vitro assays of cell cycle
To analyze the effect of miR-593-5p on cell cycle, the transfected cells were detached and fixed in 1 mL of cold 70% ethanol in PBS for 24 hours. The cells were washed once with 3 mL of PBS. Fixed cells were resuspended in 1 mg/mL propidium iodide (BD Biosciences, San Jose, CA, USA) and 0.5 mg/mL RNaseA (Keygen Biotech, Nanjing, China) in PBS for 30 minutes at 37°C. Approximately 1.5×105 cells were analyzed by a CyFlow® flow cytometer (BD Biosciences).
In vitro assays of migration and invasion
To analyze the effect of miR-593-5p on cell invasion, the transfected cells were detected by the assays of invasion. The top of the transwell chambers was covered with 0.5 mg/mL matrigel (BD Biosciences). The gastric cell lines MGC-803 and SGC-7901 were transferred to the top of the transwell chambers in serum-free RPMI 1640 medium (1×105 cells/pore). RPMI 1640 medium containing 5% FBS was added to the lower chambers as a cell-inducible factor. In the cultured system, cells were cultured after 48 hours, and unpenetrated cells were removed from the top wells by using a cotton ball. Cells on the lower membrane surface of the transwell chambers were fixed in 4% formaldehyde and stained with 0.2% crystal violet. Invading cells were manually counted in eight randomly chosen fields under a microscope, and photographs were taken.
To analyze the effect of miR-593-5p on cell migration, the transfected cells were detected by wound-healing assay. Cell migration was measured by culturing the GC cells in 6-well plates, and a straight line scratch wound was created with a 200 μL sterile pipette tip once the cells attained 100% confluency. The wound conditions were monitored at 0, 48, and 72 hours microscopically and the relative motility was calculated using the following formula: relative motility = (initial distance − a time point distance)/initial distance ×100%.
Microarray hybridization of whole genome expression profile
Total RNA was extracted from fresh tissues and the cells using TRIzol reagent (Thermo Fisher Scientific). After RNA extraction, all quantitation and microarray experiments were performed at the Phalanx Biotech Group Laboratory (Taiwan, China). Concentration and quality of RNA were analyzed by a NanoDrop™ (Thermo Fisher Scientific) and an Agilent Bioanalyzer 2100 system (Agilent Technologies, Palo Alto, CA, USA). Experimental procedures were carried out according to the manufacturer’s instructions. The arrays were washed and stained with streptavidin phycoerythrin in black chip box and then scanned using an Agilent Microarray Scanner (G2505C). Primary microarray imaging data were captured by GenePix™, and the data were saved as .GPR files. All the files were integrated into a .GPR file. Gene expression spectrum detection data of the chip were analyzed by Rosetta Resolver® System (Rosetta Biosoftware, Kirkland, WA, USA). Differentially expressed genes were selected at |log2(Ratio)|≥1 and P-value (differentially expressed) <0.05. PCA and clustering analyses were performed using Heartbeat 3.0.3.
Prediction of miRNA target genes
miR-593-5p target genes were predicted using the following algorithms as described previously: TargetScan, miRDB, and miRNA.
Western blot analysis
Specific antibodies (rabbit anti-human antibodies) to MST4, FAK, MMP12, JUN, and GAPDH were provided by Cell Signaling Technology (Beverly, MA, USA) and Abcam (Cambridge, MA, USA). Infrared-labeled secondary antibodies (goat anti-rabbit antibodies) to IRDye 800 were obtained from Li-Cor Biosciences (Lincoln, NE, USA). Cells were washed twice with PBS and lysed in a cell lysis buffer for the reagent of Western blotting and immunoprecipitation (Beyotime, Beijing, China) and protease inhibitor phenylmethanesulfonyl fluoride (Beyotime). Equal amounts of protein were heated to 100°C for 5 minutes with Laemmli sample buffer, and then protein samples were separated using 12% SDS-PAGE and transferred onto polyvinylidene fluoride membranes. The membranes were incubated with the primary antibodies (1:1,000) at 4°C overnight, washed three times in PBST, and incubated with peroxidase-conjugated secondary antibodies (1:10,000) for 1 hour at 25°C. The membranes were scanned and the net intensities of the bands were quantified using Odyssey Software Version 3.0 system (Li-Cor Biosciences). Then, the protein expression levels of each gene were measured with GAPDH protein as a reference at membrane.
Dual luciferase reporter assay
In brief, the 3′-UTR of MST4 mRNA containing the miR-593-5p binding site was PCR amplified. Wild-type and mutant MST4 3′-UTRs containing the putative binding region of miR-593-5p were constructed and cloned into the pmiR-REPORT vector (Ambion, Foster City, CA, USA). Site-directed Gene Mutagenesis Kit (Roche, Basle, Switzerland) was used to produce the mutations of the miR-593-5p targeting site. The luciferase activities were measured at 48 hours after cotransfection with Dual-Luciferase Reporter Assay system (Promega, Madison, WI, USA). Then, vector containing Renilla luciferase was co-transfected as a reference control. The experiments were performed in triplicate and repeated three times.
Tumor xenograft experiment
Athymic nude male BALBC/c mice, aged 4–5 weeks, were purchased from Guangxi Animal Center (Nanning, China). The mice were maintained in specific pathogen-free, temperature-controlled isolation conditions and fed with sterilized food and autoclaved water according to the experimental animal guidelines. All animal experiments were conducted as per the provisions of the ethics committee of Guangxi Medical University. Then, the mice were subcutaneously injected with an inoculation dosage of 1×107 cells in 200 μL of PBS per mouse. The number of nude male BALBC/c mice was 12 in each group. Group 1 was injected with negative control vectors transfected MGC-803-LV-miR-NC cells (803-NC), Group 2 was injected with MGC-803-LV-hsa-miR-593-5p cells stably expressing miR-593-5p (803-593-5p), Group 3 was injected with MGC-803 cells (803-Ctrl), Group 4 was injected with negative control vectors transfected SGC-7901-LV-miR-NC cells (7901-NC), Group 5 was injected with SGC-7901-LV-hsa-miR-593-5p cells stably expressing miR-593-5p (7901-593-5p), and Group 6 was injected with MGC-7901 cells (7901-Ctrl). Tumor size (V) was calculated every 3 days by a digital caliper using the following formula: V (mm3) =1/2×a (length)×b (width).2 After 28 days of treatment, nude mice were killed and tumors were assessed. Liver tissue, lung tissue, and the formed tumors were removed, fixed, and embedded in paraffin. Paraffin-embedded sections were prepared for H&E staining.
Statistical analysis
SPSS 17.0 software was used for statistical analysis. Data were shown as mean ± SD unless otherwise noted. Chi-squared analysis and Student’s t-test were used for statistical analysis. In all experiments, P<0.05 was considered significant.
Results
miR-593-5p is frequently downregulated in GC
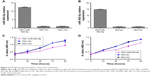
We performed qRT-PCR to examine the expression level of miR-593-5p in GC. We first examined the expression level of miR-593-5p in 73 pairs of GC tissues and adjacent normal gastric tissues. The PCR results showed that 42 of 73 (57.53%) cases had reduced levels of miR-593-5p in GC tissues compared with the corresponding normal tissues; in contrast, only 31 of 73 (42.47%) cases had increased levels of miR-593-5p in GC tissues compared with the adjacent normal tissues (P<0.05) (Figure 1A). Subsequently, we examined the expression of mature miR-593-5p in four human GC cell lines (AGS, HGC-27, MGC-803, and SGC-7901) and a human gastric epithelium cell line (GES-1). The expression level of miR-593-5p in the four GC cell lines was significantly lower than that in GES-1, with the expression in MGC-803 cells being the lowest (Figure 1B). Collectively, the data indicated that miR-593-5p was significantly attenuated in tumor tissues compared with adjacent normal tissues and might act as a tumor suppressor in GC.
Low-level expression of miR-593-5p is associated with GC metastasis
We analyzed the correlation between positive miR-593-5p expression and various clinicopathological characteristics that may affect the prognosis of GC patients. We found that miR-593-5p expression was correlated with tumor size and distant metastasis (P<0.05). Age, sex, T classification, and other clinical variables were not correlated with miR-593-5p expression in GC patients (P>0.05) (Table 1).
  | Table 1 Clinicopathological variables and the expression of miR-593-5p in gastric cancer patients |
miR-593-5p regulates cell proliferation and cell cycle in vitro
MGC-803 and SGC-7901 cell lines were chosen for transfection with lentiviral vectors. Lentiviral vectors contained overexpression of miR-593-5p sequences. miR-593-5p was successfully overexpressed in these two cell lines as confirmed by qPCR (Figure 2A and B). To evaluate the potential functions of miR-593-5p in regulating tumor cell growth and cell cycle, CCK-8 assay and flow cytometry were performed to detect cell proliferation and cell cycle. In both MGC-803 and SGC-7901 cells, the CCK-8 assay results showed that exogenous expression of miR-593-5p inhibited cell proliferation (Figure 2C and D). Flow cytometry results showed that exogenous expression of miR-593-5p inhibited cell transition in G1 phase (Figure 3). These results suggest that miR-593-5p inhibits cell proliferation and cell cycle G1/S transition.
Upregulation of miR-593-5p can inhibit GC cell migration and invasion
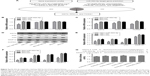
The effects of miR-593-5p on the invasion and migration of MGC-803 and SGC-7901 cell lines were investigated by transwell methods and wound-healing assay. Compared with the scramble control and untreated cells, the transwell assay results showed that overexpression of miR-593-5p dramatically decreased the invasion and migration of the two GC cell lines (Figures 4 and 5). These results indicated that miR-593-5p played important roles in regulating cell migration and invasion in GC.
miR-593-5p indirectly targets MST4 in GC cells
We further investigated the mechanism by which miR-593-5p functioned as a tumor suppressor in GC. However, miR-593-5p has not been reported to directly target any gene in carcinoma. Therefore, total gene expression chip of Homo sapiens was taken to find significantly differential expression of genes in MGC-803 cells infected with lentivirus containing overexpression of miR-593-5p. The results showed that 475 genes were regulated and 263 genes were downregulated in stably expressing miR-593-5p cells (803-593-5p) compared with the negative control transfected MGC-803-LV-miR-NC cells (803-NC) (Table S2). In addition, miR-593-5p targets were predicted by the following algorithms: TargetScan, miRDB, and miRNA. The total number of bioinformatics website software-predicted target genes of miR-593-5p was 163 (Table S3). Intersection genes were taken between the results of gene expression chip (Table S2) and the results of bioinformatics website software-predicted target genes (Table S3). The intersection genes were MST4, PPM1A, and TWSG1, which were candidate target genes of miR-593-5p. To confirm the target genes of miR-593-5p, qRT-PCR was performed to detect whether the expression of MST4, PPM1A, and TWSG1 was regulated by miR-593-5p in MGC-803 and SGC-7901 cells infected with miR-593-5p or scramble lentivirus. The results of the qRT-PCR and Western blot analyses showed a notable reduction of the mRNA and protein levels of MST4 in the cells infected with miR-593-5p compared with those infected with scramble lentivirus. However, the results of luciferase activity assay showed that miR-593-5p could not directly regulate MST4. The results displayed that the protein levels of FAK, MMP12, and JUN were lower in the miR-593-5p group (P<0.05) (Figure 6). Taken together, these results indicated that miR-593-5p can indirectly target MST4, FAK, MMP12, and JUN in GC cells in vitro.
miR-593-5p suppressed tumor growth and metastasis in a xenograft model
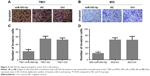
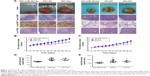
To directly evaluate the role of miR-593-5p in tumor formation and growth in vivo, the xenograft model of human GC MGC-803 and SGC-7901 cells in nude mice was adopted. Briefly, MGC-803 and SGC-7901 cells infected with miR-593-5p or scramble lentivirus were injected subcutaneously into each flank of nude mice. After the cells were injected, the tumor volume was monitored every 4 days, and the growth curves of the tumors were plotted accordingly. Finally, all the mice were killed to harvest the xenograft. It is obvious that the mean volume of the tumors generated from the miR-593-5p overexpression group was significantly lower compared with the negative and control groups (Figure 7). We then studied the effect of miR-593-5p on tumor metastasis in vivo. The lungs and livers of the mice were dissected, and H&E staining was performed to evaluate the tissue morphology (Figure 7). As shown in Table 2, a significantly lower number of macroscopic liver metastases could be observed in cells infected with miR-593-5p than the negative and control groups. These results indicate that miR-593-5p may repress GC proliferation and metastasis.
Discussion
Many miRNAs have been suggested to play an important role in GC progression. miRNA dysregulation is regarded as a potentially disastrous event that leads to carcinogenesis.9,10 Therefore, abnormity of miRNA expression in GC may have a relationship with the disease. Chen et al had initially reported that the expression of miR-593-5p is abnormal in GC patients,6 indicating that an abnormal expression of miR-593-5p may have a significant effect in the development of GC. However, clinicopathological characteristics and function of miR-593-5p have not been concretely reported yet. To further understand miR-593-5p, we studied the role of miR-593-5p in GC in vitro and vivo.
In this study, we expanded the expression analysis in 73 pairs of GC tissues and adjacent normal gastric tissues, GC cell lines, and human gastric epithelium cell line. Our results demonstrated that miR-593-5p was frequently downregulated in GC tissues and GC cell line (Figure 1), and expression level of miR-593-5p was lower in GC distant metastasis tissues than no distant metastasis tissues (Table 1). This result suggests that miR-593-5p may act as a tumor suppressor gene. Lentivirus expressing miRNAs are transfected in cancer cells to observe its function. This experimental method has been widely adopted in the research field.11,12 To assess whether miR-593-5p influences GC metastasis, transwell chambers and wound-healing assay method were used to detect the abilities of cell invasion and migration, and SGC-7901 and MGC-803 cell lines were transfected with miR-593-5p precursor and miR-593-5p over-expressed stably in the two cell lines. The results show that overexpression of miR-593-5p inhibits GC cell migration and invasion in vitro (Figures 4 and 5). Similarly, in vivo, SGC-7901 and MGC-803 cells overexpressing miR-593-5p grew significantly slower than the control group xenografts (Figure 7). Tumor liver metastasis rate was lower in overexpressing miR-593-5p of mouse xenograft model than those of the control group (Table 2). Collectively, our results reveal that miR-593-5p inhibits GC growth and metastasis.
Many research studies have confirmed that functions of miRNAs are dependent on their main downstream target genes; miRNAs bind to the 3′-UTRs of transcripts and may also target ORFs.13,14 However, the direct target gene of miR-593-5p has not been reported yet. To date, target genes of miRNAs were mainly predicted by bioinformatics websites such as TargetScan, miRDB, and miRNA. However, the bioinformatics website predicted hundreds of miRNA target genes, as shown in Table S3. In order to avoid blind choice, total gene expression chip method was used to find genes that had expressions affected by overexpression of miR-593-5p in GC, as shown in Table S2. The intersection genes of Tables S2 and S3 were MST4, PPM1A, and TWSG1, which were candidate target genes of miR-593-5p. To confirm which gene was the target gene of miR-593-5p, first, qRT-PCR was conducted to verify MST4, PPM1A, and TWSG1 genes. The results showed that miR-593-5p was downregulated in transfected cells SGC-7901 and MGC-803. Furthermore, the results showed that miR-593-5p is found to indirectly target MST4 by dual luciferase reporter assay and Western blotting (Figure 6).
Accordingly, miR-593-5p inhibits GC cell growth and metastasis by indirectly targeting the MST4 gene, which regulates the levels of MMP12, JUN, and FAK, as shown in Figure 6. Furthermore, MST4 facilitates the EMT process via regulating the activation of ERK in hepatocellular carcinoma,15 and downexpression of MST4 could suppress cancer cell migration and invasion.7,16 Additionally, ERK-c-Jun system could modulate cell viability, and inhibition of ERK-c-JUN levels reduces proliferation and apoptosis,17,18 which is inconsistent with our study results showing that miR-593-5p inhibits GC cell growth. Downexpression of JUN can also reduce the expression of MMP protein.19 The level of FAK protein was downregulated in a study by Lee et al.20 FAK/Fyn signal also impacts the expression of MMP12. The level of MMP protein is positively correlated with the invasion ability of cancer.21,22 Downregulated MMP12 can inhibit GC cell invasion and migration.23 Thus, the JUN signal pathway is suppressed by indirect target gene MST4 of miR-593-5p in GC.
Conclusion
Our study reveals that miR-593-5p is frequently downregulated in GC. Our in vitro and in vivo data further prove that miR-593-5p may act as a tumor suppressor in GC and inhibit cell metastasis through indirectly targeting the MST4 gene.
Acknowledgment
This work was supported by grants from the National Natural Science Foundation of China (grant nos 81660511 and 81060201), the Natural Science Foundation of Guangxi (grant no 2015GXNSFDA227001), and the Guangxi Medical University Training Program for Distinguished Young Scholars.
Disclosure
The authors report no conflicts of interest in this work.
References
Karimi Kurdistani Z, Saberi S, Tsai KW, Mohammadi M. MicroRNA-21: Mechanisms of oncogenesis and its application in diagnosis and prognosis of gastric cancer. Arch Iran Med. 2015;18(8):524–536. | ||
Ni Q, Ji A, Yin J, Wang X, Liu X. Effects of Two Common Polymorphisms rs2910164 in miR-146a and rs11614913 in miR-196a2 on Gastric Cancer Susceptibility. Gastroenterol Res Pract. 2015;2015:764163. | ||
Shen CT, Qiu ZL, Song HJ, Wei WJ, Luo QY. miRNA-106a directly targeting RARB associates with the expression of Na(+)/I(−) symporter in thyroid cancer by regulating MAPK signaling pathway. J Exp Clin Cancer Res. 2016;35(1):101. | ||
Chen W, Wang J, Liu S, et al. MicroRNA-361-3p suppresses tumor cell proliferation and metastasis by directly targeting SH2B1 in NSCLC. J Exp Clin Cancer Res. 2016;35(1):76. | ||
Qu Y, Zhang H, Duan J, et al. MiR-17-5p regulates cell proliferation and migration by targeting transforming growth factor-β receptor 2 in gastric cancer. Oncotarget. 2016;7(22):33286–33296. | ||
Yang B, Jing C, Wang J, et al. Identification of microRNAs associated with lymphangiogenesis in human gastric cancer. Clin Transl Oncol. 2014;16(4):374–379. | ||
Xiong W, Matheson CJ, Xu M, et al. Structure-Based Screen Identification of a Mammalian Ste20-like Kinase 4 (MST4) Inhibitor with Therapeutic Potential for Pituitary Tumors. Mol Cancer Ther. 2016;15(3):412–420. | ||
Ma X, Zhao H, Shan J, et al. PDCD10 interacts with Ste20-related kinase MST4 to promote cell growth and transformation via modulation of the ERK pathway. Mol Biol Cell. 2007;18(6):1965–1978. | ||
Wu WK, Lee CW, Cho CH, et al. MicroRNA dysregulation in gastric cancer: a new player enters the game. Oncogene. 2010;29(43):5761–5771. | ||
Pan HW, Li SC, Tsai KW. MicroRNA dysregulation in gastric cancer. Curr Pharm Des. 2013;19(7):1273–1284. | ||
Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467(7311):86–90. | ||
Varambally S, Cao Q, Mani RS, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322(5908):1695–1699. | ||
Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. | ||
Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11(9):597–610. | ||
Lin ZH, Wang L, Zhang JB, et al. MST4 promotes hepatocellular carcinoma epithelial-mesenchymal transition and metastasis via activation of the p-ERK pathway. Int J Oncol. 2014;45(2):629–640. | ||
Xiong W, Knox AJ, Xu M, et al. Mammalian Ste20-like kinase 4 promotes pituitary cell proliferation and survival under hypoxia. Mol Endocrinol. 2015;29(3):460–472. | ||
Pathria G, Garg B, Garg K, Wagner C, Wagner SN. Dual JNK-Cyclin D1 and ERK-c-JUN Disjunction in Human Melanoma. Br J Dermatol. 2016;175(6):1221–1231. | ||
Park KH, Shin KS, Zhao TT, Park HJ, Lee KE, Lee MK. L-DOPA modulates cell viability through the ERK-c-Jun system in PC12 and dopaminergic neuronal cells. Neuropharmacology. 2016;101:87–97. | ||
Nishitani K, Ito H, Hiramitsu T, et al. PGE2 inhibits MMP expression by suppressing MKK4-JNK MAP kinase-c-JUN pathway via EP4 in human articular chondrocytes. J Cell Biochem. 2010;109(2):425–433. | ||
Lee SJ, Jung YH, Oh SY, Yong MS, Ryu JM, Han HJ. Netrin-1 induces MMP-12-dependent E-cadherin degradation via the distinct activation of PKCα and FAK/Fyn in promoting mesenchymal stem cell motility. Stem Cells Dev. 2014;23(16):1870–1882. | ||
Bradbury PA, Zhai R, Hopkins J, et al. Matrix metalloproteinase 1, 3 and 12 polymorphisms and esophageal adenocarcinoma risk and prognosis. Carcinogenesis. 2009;30(5):793–798. | ||
Yu D, Ye T, Xiang Y, et al. Quercetin inhibits epithelial-mesenchymal transition, decreases invasiveness and metastasis, and reverses IL-6 induced epithelial-mesenchymal transition, expression of MMP by inhibiting STAT3 signaling in pancreatic cancer cells. Onco Targets Ther. 2017;10:4719–4729. | ||
Kwon CH, Moon HJ, Park HJ, Choi JH, Park DY. S100A8 and S100A9 promotes invasion and migration through p38 mitogen-activated protein kinase-dependent NF-κB activation in gastric cancer cells. Mol Cells. 2013;35(3):226–234. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.