Back to Journals » International Journal of Nanomedicine » Volume 12
Reductive nanocomplex encapsulation of cRGD-siRNA conjugates for enhanced targeting to cancer cells
Authors Zhou Z, Liu S, Zhang Y, Yang X, Ma Y, Guan Z, Wu Y, Zhang L, Yang ZJ
Received 11 March 2017
Accepted for publication 13 June 2017
Published 4 October 2017 Volume 2017:12 Pages 7255—7272
DOI https://doi.org/10.2147/IJN.S136726
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Linlin Sun
Zhaoxiu Zhou,* Shuang Liu,* Yanfen Zhang, Xiantao Yang, Yuan Ma, Zhu Guan, Yun Wu, Lihe Zhang, Zhenjun Yang
State Key Laboratory of Natural and Biomimetic Drugs, School of Pharmaceutical Sciences, Peking University, Beijing, People’s Republic of China
*These authors contributed equally to this work
Abstract: In this study, through covalent conjugation and lipid material entrapment, a combined modification strategy was established for effective delivery of small interfering RNA (siRNA). Single strands of siRNA targeting to BRAFV600E gene (siMB3) conjugated with cRGD peptide at 3'-terminus or 5'-terminus via cleavable disulfide bond was synthesized and then annealed with corresponding strands to obtain single and bis-cRGD-siRNA conjugates. A cationic lipid material (CLD) developed by our laboratory was mixed with the conjugates to generate nanocomplexes; their uniformity and electrical property were revealed by particle size and zeta potential measurement. Compared with CLD/siBraf, CLD/cRGD-siBraf achieved higher cell uptake and more excellent tumor-targeting ability, especially 21 (sense-5′/antisense-3″-cRGD-congjugate) nanocomplex. Moreover, they can regulate multiple pathways to varying degree and reduce acidification of endosome. Compared with the gene silencing of different conjugates, single or bis-cRGD-conjugated siRNA showed little differences except 22 (5/5) which cRGD was conjugated at 5'-terminus of antisense strand and sense strand. However bis-cRGD conjugate 21 nanocomplex exhibited better specific target gene silencing at multiple time points. Furthermore, the serum stabilities of the bis-cRGD conjugates were higher than those of the single-cRGD conjugates. In conclusion, all these data suggested that CLD/bis-conjugates, especially CLD/21, can be an effective system for delivery of siRNA to target BRAFV600E gene for therapy of melanoma.
Keywords: cRGD-siRNA conjugates, cationic lipids, targeting, silencing, intracellular pathways, CLD
Introduction
Melanoma is a kind of highly dangerous malignant tumor, and it is difficult to be cured clinically. More than 90% of malignant melanoma tissues have N-RAS (15%) or BRAF (60%–80%) variants, and BRAFV600E mutant is an ideal small interfering RNA (siRNA) target for gene therapy of melanoma. RNAi is a popular and promising treatment strategy for cancer therapy.1,2 Compared with traditional small molecule chemotherapeutic agents, siRNA can specifically target intracellular mRNA and genetically prevent the occurrence and development of diseases by silencing the disease-related gene.3 In fact, several siRNA drug candidates have been in clinical trial, including anti-tumor and anti-viral siRNAs.4,5 However, siRNAs are still facing many challenges such as system administration, target tissue delivery and, most importantly, cellular uptake and lysosome escape.6
At present, there are two main modification strategies for siRNA delivery: delivery vectors and covalent conjugation. In recent years, a variety of chemical molecules have been covalently conjugated to siRNA to improve its stability, specificity and transmembrane ability such as PEG, cholesterol, phospholipids, peptides, quantum dots, aptamer and antibody.7 Different siRNA conjugates possess their unique biological properties and characteristics, and cell-targeting peptides which specifically bind to overexpressed target proteins on the surface of certain cells can achieve specific endocytosis.8 A common cell-targeting peptide is Arg-Gly-Asp peptide sequence (cRGD), which implement endocytosis by binding to overexpressed integrin αvβ3 receptors on tumor cell surface.9 Many reports have demonstrated that the transport capacity of siRNAs was significantly improved when siRNAs were covalently conjugated with cRGD peptides.9–11 For example, Alam et al conjugated the divalent, trivalent and tetravalent cRGD to the 3′-terminus of the sense strand of siRNA via thioether bonds.10 Although the intracellular uptake efficiency of cRGD-siRNA conjugates with different multivalent values was not significantly different, the trivalent/tetravalent cRGD-siRNA conjugate showed enhanced luciferase silencing effect. It was found that trivalent RGD-siRNA single conjugate can mainly enter into cells through caveolae-mediated endocytosis (CvME) and present better silencing activity. However, in current studies of siRNA conjugates, more attention has been paid to the 3′-terminus of sense strand. Meanwhile, there are a few literatures reporting bis-conjugated siRNAs, such as 3′,3″-bis-phospholipid-siRNA conjugate,12 3′,3″-bis-polymer-siRNA conjugate13 and 3′,3″-bis-peptide-siRNA conjugate.14
In our previous research, we used a peptide derivative, CPG, to develop a relatively simple strategy to prepare peptide-siRNA conjugates in which hydrophobic hexapeptide (KALLAL) was conjugated at 3′-terminus of single oligonucleotide.15–17 Serum stability results showed that the peptide modified at the 3′-terminus of sense strand affected the nucleases attack and could significantly reduce the off-target effect caused by incorporation of sense strand into RNA-induced silencing complex (RISC), thereby improving the serum stability and silencing activity. In addition, 3′,3″-bis-peptide-siRNA conjugate showed better serum stability and nuclease resistance while maintaining longer term gene-silencing activity.14
Low weight siRNA conjugates are easily eliminated by the kidneys due to their small size. Studies on delivery vectors such as engineered viruses, inorganic nanoparticles, organic nanoparticles and hybrid nanoparticles have made progress.18 Engineered viruses were the most efficient delivery vectors for different cells, but they caused immune system response and high toxicity, which limited their application. In contrast with the viral methods, lipid-based nanoparticles, such as cationic lipids and neutral lipids that have favorable biocompatible and biodegradable properties, are widely used for RNA delivery.19 The cationic lipid material (CLD; Figure S1) designed by our group has a disulfide bond that can be cleaved in reductive environment of cell, and it has good biodegradation property, low toxicity and efficient delivery ability.20,21 Due to the ability to reduce immune response and highly improve RNA’ gene-silencing activity in vivo, disulfide bond has been widely used in siRNA conjugates.22,23 Bis-peptide-siRNA conjugates have been shown to exhibit good biological effects. We have also used CLD in specific delivery of 3′,3″-bis-peptide-siRNA conjugate, and obtained regular and homogeneous nanolipid complexes with dual actions (electrostatic and van der Waals force). The experiments showed that terminal peptide can be inserted into hydrophobic region in bilayers of CLD, which could improve the binding ability of siRNAs with CLD to reduce the amount of the carrier. MT-pp-siRNA/CLDs (MT means mixed two-phase) could regulate the pathway of siRNA endocytosis and intracellular distribution, making it enter into the cell mainly through caveolin mediated endocytosis (CvME) and macropinocytosis to reduce the degradation of lysosome, thereby further improving its silencing activity. However, the experiment did not design contrast experiment (single conjugates vs double conjugates); moreover, the prepared nanoparticles did not target tumor cells and may damage healthy tissues causing adverse side effects.
In the modification strategy of siRNA, covalent conjugation can decrease the shortage of siRNA itself, while the carrier delivery can help the siRNA to achieve system administration. To improve the silencing effect of siRNA, the combination of covalent conjugation and vector encapsulation has become a trend. It is reported that HA-siRNA/PEI nanocomplexes were successfully internalized into the cells by receptor-mediated endocytosis and then downregulated the target gene expression in CD44 cancer cells. The nanocomplex can be specifically delivered to the liver and achieve target gene silencing after systemic administration.24 In addition, Chen et al, based on studies of the SPACE-siRNA conjugates, further embedded it in cationic alcohol plastids with additional free SPACE peptides to facilitate the delivery of SPACE-siRNA conjugates, which showed a dramatic increase in cellular gene-silencing activity.25
In this study, we used a combined modification strategy for siRNA delivery. To improve the gene silencing and the targeting of siRNA, and compare the biological mechanism of different conjugates, cRGD was conjugated to siRNA at four different sites (3′- or 5′-terminus of sense strand, or 3″-, 5″-terminus of antisense strand) via cleavable linkages to comparatively evaluate their effects on gene silencing and cellular uptake. Five cRGD-siRNA conjugates including single-conjugated ones and bis-conjugated ones were mixed with CLD to form nanocomplexes through the MT method. The siMB3 sequence effectively target BRAFV600E in melanoma was selected as our siRNA sequence. We hypothesized that two identical target peptides, conjugated at the same end of siRNA, allow more siRNA conjugates to bind receptors on the cell surface. Meanwhile, we constructed 5′,3″-cRGD-siRNA conjugates, whose cRGD may also be easily exposed to the surface of the CLD/5′,3″-cRGD-siRNA nanocomplex and promote the targeted cellular uptake. On the other hand, CLD/cRGD-siRNA nanocomplex could also regulate the pathway of siRNA endocytosis because of the dual actions, electrostatic and van der Waals force. Therefore, the cRGD-siRNA conjugates and the CLD/cRGD-siRNA nanocomplexes will be used to inhibit the expression of BRAFV600E in melanoma cells. Cellular uptake and intracellular pathways were investigated through flow cytometry analysis, and specific degradation of target mRNA by cleavable cRGD-siRNA conjugates was verified by quantitative polymerase chain reaction (qPCR) and western blot analysis.
Materials and methods
Reagents
The cyclo(Arg-Gly-Asp-D-Phe-Lys) peptide (cRGD) was obtained from GL Biochem (Shanghai) Ltd. N-{6-[3-(2-Pyridyldithio)propionamido]hexanoyloxy}-sulfosuccinimide, sodium salt (Sulfo-AC5-SPDP), was purchased from Dojindo Laboratories (Kumamoto, Japan). The thiol-modifier C6 S-S CE phosphoramidite was purchased from Glen Research (Sterling, VA, USA). 6-5′-fluorescein phosphoramidite (FAM) was obtained from Lumiprobe (Hallandale Beach, FL, USA). The novel Gemini-like cationic lipid (CLD) that was utilized for the delivery of siRNA was synthesized by previously reported method.22 The negative control siRNA (siNC) was purchased from Guangzhou RiboBio Co., Ltd. All other chemicals and reagents were of analytical grade.
Synthesis of cRGD-siRNA Conjugates
Solid-phase synthesis of oligonucleotides
All oligonucleotides used in this study were synthesized on an ABI 394 DNA synthesizer using standard phosphoramidite chemistry with commercially available phosphoramidite monomers. The 3′- or 5′-terminus C6 thiol function groups were introduced to siRNA using commercially available thiol-modifier C6 S-S CE phosphoramidite to obtain thiol-modified oligonucleotides 4–7 (Figure 1). The FAM was used to label the 5′-terminus of siRNA antisense or sense strands on solid phase to obtain fluorescence-labeled oligonucleotides 15–17 (Table 1). Cleavage of the oligonucleotides were performed in concentrated methylamine aqueous solution and methylamino alcohol (v:v =1:1) at 60°C for 90 min; 2′-TBDMS group was removed by treating with TEA·3HF at 65°C for 90 min. The crude oligonucleotides were purified by anion-exchange high-performance liquid chromatography (HPLC) and then desalted.
  | Table 1 Single-strand siRNAs |
The HPLC condition was used as follows: an anion-exchange column (Dionex DNA Pac PA200, 9×250 mm) equipped with a UV detector at 260 nm using a linear gradient of 15%–40% eluent A within 35 min. Solutions of 0.02 M tris(hydroxymethyl)aminomethane (Tris)-HClO4 containing 10% CH3CN (pH 8) was used as eluent B, and 0.4 M NaClO4 in eluent B was used as eluent A. Then the purified oligonucleotides were desalted by using Sephadex G25 column. The purity and identity of the oligonucleotides were monitored by anion-exchange chromatography and electrospray ionization mass spectrometry (ESI-MS). Finally, the pure oligonucleotides were lyophilized and stored at −80°C.
Synthesis of cRGD-conjugated oligonucleotides linked with a disulfide bond
The HPLC-purified disulfide-modified oligonucleotides (50 nmol) were reduced by using 0.1 M dithiothreitol (DTT) in phosphate-buffered saline (PBS; 0.02 M Tris) (0.5 mL) at pH =8.4 for 3 h at room temperature under argon to provide the corresponding thiol-modified oligonucleotides.26
Conjugation of cyclic RGD with Sulfo-AC5-SPDP was performed as follows: RGD peptide (386 μM) was mixed with Sulfo-AC5-SPDP in PBS and incubated for 3 h at room temperature. The resulting product, cRGD-PDP, was analyzed and purified by using a HPLC with a C18 column (Venusil XBP C18 (2), 4.6×250 mm, 150 Å, 5 μm) equipped with a UV detector at 220 nm. Liquid chromatography was carried out in the following conditions: mobile phase A, 0.5% trifluoroaceticacid in sterile water; mobile phase B, acetonitrile; a gradient elution from 5% B to 65% B in 30 min; flow rate 1.0 mL/min; column temperature 25°C. The cRGD-PDP was concentrated to obtain a working stock solution by lyophilizing twice with DEPC water.
Then, 10 mol equivalent of the HPLC-purified product was added to the thiol-modified oligonucleotides in PBS-ethylenediaminetetraacetic acid buffer to yield cRGD-conjugated oligonucleotides at room temperature overnight.27 The resultant crude product was purified by anion-exchange HPLC under the same conditions as already mentioned.
Preparation of fluorescently labeled cRGD-siRNA conjugates
The 6-FAM was used to label the fluorescent dye to oligonucleotides at 5′-terminus of antisense. cRGD-siRNA conjugates with and without 6-FAM were prepared for the present study. Annealing of single oligonucleotides 11–19 (Table 1) individually with equimolar amount of each other at 85°C for 5 min in sterile water followed by slow cooling afforded the siRNAs 20–30 (Table 2).
  | Table 2 Double-strand siRNAs |
Preparation of CLD/siRNA nanocomplexes
CLD and siMB3 were mixed at charge radio (N/P =5:1). The lipid materials were added to naked siRNA solution at the concentration of 1 μM, then vortexed for 1 min and put in ultrasonic instrument for 10 min at 70°C to form CLD/siRNA nanocomplexes.
Characterization of CLD/siRNA nanocomplexes
The particle size and the surface charge (zeta potential, mV) of nanocomplexes were measured at room temperature with photon correlation spectroscopy at a scattering angle of 90°C using a Zetasizer Nano ZS90 instrument (Malvern Instruments, Worcestershire, UK). Samples were diluted with ddH2O to obtain appropriate viscosities. Data were analyzed using a software package (ELS-8000 software) supplied by the manufacturer, and the data were obtained from three repeated measurements.
Cell culture
Human malignant melanoma cells (A375) and human cervical carcinoma cells (HeLa) were obtained from the Institute of Basic Medical Science and Chinese Academy of Medical Sciences respectively (Beijing, People’s Republic of China). It has been previously demonstrated that A375 cell lines express integrin αvβ3 on their cell surface.26,28 Therefore, A375 cell line was selected to explore the gene-silencing ability of CLD/cRGD-siRNA nanocomplexes in the following in vitro studies. The cells were cultured in DMEM in a humidified incubator supplied with 5% CO2 and maintained at 37°C. All media were supplemented with 10% heat-inactivated fetal bovine serum (FBS).
Serum stability
Nine microliter of 20 pmol siRNA 25 or cRGD-siRNA 20–24 was mixed with 1 μL FBS and incubated at 37°C for 0, 1, 2, 4 and 6 h. Aliquots were taken at each of the time points, and then the solution was immediately stored at −80°C until analysis. The control assay was performed for each siRNA without FBS as well. Together with 2 μL of 6× RNA loading buffer, all the samples were resolved in non-denature polyacrylamide gels stained with SYBR Gold, and the electrophoresis was conducted at 110 V for 150 min.
Anti-proliferation and cytotoxicity analysis
The A375 cells were seeded into 96-well plates at a density of 3,000 cells per well in 200 μL DMEM supplemented with 10% FBS in 5% CO2 incubator at 37°C. After 18–24 h of incubation, cells were transfected with 100 nM (final concentration) CLD/cRGD-siRNA 20–24, CLD/siRNA 25, CLD/NC or siNC and non-conjugated siRNA 25 using Lipofectamine® 2000 (Life Technologies, Camarillo, CA USA) according to the manufacturer’s protocols. After continued incubation for 72 h, the effect of different treatments on cell viability was assessed by a cell counting kit-8 (CCK-8) assay (Dojindo Laboratories). Aliquots were drawn from each well, and the absorbance at 450 nm was determined by a microplate reader (Molecular Devices, California, USA). Untreated cells were used as a control with 100% viability. The relative cell viability (%) compared to control cells was calculated as follows:
|
RA, RB, RE were defined as the absorbance of experimental samples, untreated samples and blank controls, respectively. All treatments were done in triplicates.
Flow cytometry
For quantitative comparison of transfection efficiency, CLD/FAM-siRNAs’ uptake was measured by flow cytometry. A375 cells or HeLa cells (30×104 cells/2 mL/well) were seeded into 6-well plates and precultured overnight. A375 cells were transfected with 100 nM of CLD/cRGD-siRNA 26–29, CLD/siRNA 30 for 6 h in DMEM medium. For competitive study, A375 cells were incubated with cRGD peptide (15 μg/mL) at 37°C or cRAD peptide (15 μg/mL) for 30 min, and subsequently transfected with 100 nM of CLD/FAM-labeled cRGD-siRNA 26, 30. Afterwards, medium was removed and the cells were washed three times with cold PBS, and cells were fixed by 4% paraformaldehyde at 4°C for later flow cytometry analysis.
Endocytotic pathway assay
A375 cells or HeLa cells (30×104 cells/2 mL/well) were seeded into 6-well plates and precultured overnight. For investigating the endocytotic pathway of CLD/cRGD-siRNA nanocomplexes, cells were incubated with the inhibitors (genistein 40 μg/mL; chlorpromazine 5 μg/mL and amiloride 0.5 mM) to inhibit corresponding endocytotic pathway of CvME, clathrin-mediated endocytosis (CME) and micropinocytosis 30 min prior to transfection.23 Then, the cells were transfected with different CLD/siRNA nanocomplexes at the fixed final concentration of 50 nM for 4 h, including 26, 28, 29, 30. Afterwards, medium was removed and the cells were washed three times with cold PBS. The cells were collected for later flow cytometry analysis.
RT-qPCR
To verify the silencing efficiency of BARFV600E gene in A375 cells, the cells were transfected with different CLD/siRNA nanocomplexes, including 20–25. The cells were seeded into 6-well plates (2×105 cells/well) and were incubated in DMEM at 37°C under 5% CO2/95% air atmosphere overnight. Cells were transfected using the lipid material CLD with siNC, modified and unmodified siMB3 (siBraf-mu: targeting to BRAFV600E, antisense: 5′-AUC GAG AUU UCU CUG UAG Cdtdt-3′; sense: 5′-GCU ACA GAG AAA UCU CGA Udtdt-3′). SiNC (RibioBio, Guangzhou, People’s Republic of China) was used as negative control and 25 (siMB3) was transfected with Lipofectamine 2000 as positive control. Total RNA was extracted by using Trizol® Reagent (Life Technologies) following the protocol suggested by the manufacturer and quantified after 24 h of transfection. cDNA was synthesized using the Reverse Transcription System (Promega, Madison, WI, USA). The real-time PCR was performed with SYBR Green. PCR reaction was run by using a modified three-step amplification protocol followed by a melting curve measurement to confirm the production. The primers are as follows: β-actin, forward primer, 5′-CCA ACC GCG AGA AGA TGA-3′, reverse primer, 5′-CCA GAG GCG TAC AGG GAT AG-3′; Braf-mu, forward primer, 5′-TGG TGT GAG GGC TCC AGC TTG T-3′, reverse primer, 5′-ATG GGA CCC ACT CCA TCG AGA TTT CT-3′. The threshold cycle number (Ct) was calculated for BRAFV600E and β-actin (an inner control) using the Mx3005P QPCR instrument (Agilent Technologies, Santa Clara, CA, USA). The expression of BRAFV600E and β-actin mRNA was analyzed by the 2−ΔΔCt relative quantitative method as described previously.
Western blot analysis
The melanoma cells A375 were seeded into 6-well plates (2×105 cells/well) 1 day before transfection. Cells were transfected using the lipid material CLD with modified and unmodified siMB3. The cells were collected and lysed after 24 h of transfection. The lysates were centrifuged at 12,000 rpm for 10 min. The supernatant was analyzed for protein concentration using the Pierce BSA protein assay kit (Thermo Scientific, Waltham, MA, USA). Equal amount (16 μg) of protein was subjected to electrophoresis on sodium dodecyl sulfate polyacrylamide gels and then transferred to polyvinylidene fluoride membrane (Millipore, Billerica, MA, USA). The blotted membranes were immune stained with antibodies specific for BRAF antigens (Abcam, Cambridge, MA, USA). The signals were developed by a standard electrochemiluminescence method according to the manufacturer’s protocol (ChemiDocXRS, Bio-Rad Laboratories Inc., Hercules, CA, USA).
Cleavage of single-strand cRGD-siRNA conjugates in a reductive environment
According to the reported concentrations of DTT (10–300 mM);35–40 we used 100 mM of DTT to detect the cleavage of disulfide bond. The reaction was allowed to proceed at 37°C for 2 h, which was stopped by immediately storing the conjugates at −80°C until analysis. The cleaved siRNAs were visualized by electrophoresis.
Statistical analysis
All experimental data were described as mean ± standard deviation. Student’s t-test and one-way analysis of variance were used to analyze the results. A P-value <0.05 was considered to be significant.
Results and discussion
Synthesis and characterization
As shown in Figure 1, the amine group of the cRGD peptide containing a lysine residue was firstly reacted with an active acibenzolar group of a hetero-functional coupling reagent, Sulfo-AC5-SPDP, to produce cRGD-PDP 10. The pure cRGD-PDP was obtained after HPLC purification (Figure S2). The oligonucleotides derivatized with a sulfhydryl group at the end were then added to obtain cRGD-s-s-siRNA conjugates via a sulfhydryl exchange reaction. The resulting products were purified by anion-exchange HPLC (Figure S3). The molecular weight of conjugate molecules composed of peptides and siRNA strands was determined by ESI-MS (Figure S4), as listed in Table 1.
Sequences of siRNAs are listed in Table 2. For visualization of cellular uptake, the tested cRGD-siRNA molecules were labeled with 6-FAM, and fluorescence was monitored using flow cytometry. Due to structural and synthesis-related constraints, fluorescent peptide-siRNA conjugates described previously had to be prepared by conjugating peptide moiety with 3′ or 5′-terminus of sense strands, 3′-terminus of antisense strands and linking 6-FAM to the 5″-terminus of antisense strands of molecules (26–30). However, 5″-terminus phosphate of antisense strands is critical for siRNA’s ability to activate RISC.29 To ensure RISC-activating abilities, corresponding sequences (20–25) without FAM conjugation at the 5″-terminus of antisense strands were used in the following experiments.
The choice of cross-linker, however, may influence the ability of siRNA cargo to interact with RISC. The interior of the cells is a reductive environment, which would lead to cleavage of the disulfide bond in the cRGD-siRNA conjugates to free the siRNA. In general, 5″-terminus of siRNA antisense strand is not suggested to be modified, but if siRNA can be released from the disulfide bond through reductive environment, the siRNA conjugates would exert gene-silencing ability. To minimize RNase degradation of siRNA, gloves and sterile materials were used during the purification steps and DEPC-treated water was used. Purified conjugates were characterized by ESI-MS (Figure S4), and the purity of all single strands was above 90%.
The size and zeta potential of CLD/siRNA nanocomplexes
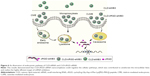
In accordance with the previous method of operation, we mixed the lipid material CLD with siRNA (siMB3) in N/P =5:1 ratio to get CLD/cRGD-siRNA nanocomplexes. On the basis of previous studies, a possible combination model of 21 (5/3) and 20 (3/3) with CLD is shown in Figure 2. The particle size and the surface charge (zeta potential, mV) of nanocomplexes were measured using a Zetasizer Nano ZS90 instrument (Malvern Instruments) (Table 3 and Figure 3). The results demonstrated that particle sizes of CLD/cRGD-siRNAs had little differences, and 24 (5/As) nanocomplex was slightly larger. On the whole, the double conjugates, such as 20 (3/3), 21 (5/3), 22 (5/5) nanocomplexes, had a smaller size than single conjugates. The particle sizes of 20 (3/3) and 22 (5/5) nanocomplexes were almost identical, indicating that they have the same binding pattern of siRNA conjugates with lipid material CLD. Size of 21 (5/3) was the smallest, which is partially due to the higher binding capacity between cRGD-siRNA and the carrier material CLD. The zeta potential of CLD/cRGD-siRNA nanocomplexes was higher than that of CLD/siRNA, which is possibly attributed to the cRGD, which contains a certain amount of positive charge.
  | Figure 2 The combination model of CLD/cRGD-siRNA nanocomplexes (5/3, 3/3). |
  | Table 3 The characteristics of CLD/siRNA nanocomplexes |
  | Figure 3 Size and zeta potential of CLD/siRNA nanocomplexes. (A) Particle size and distribution of CLD/siRNA nanocomplexes. (B) Sizes and zeta potentials of CLD/siRNA nanocomplexes. |
Serum stability of siRNA
The non-denaturing gels were run to study serum stability of siRNA 20–25 (Figure 4). The corresponding time curve is shown in Figure 4. Drawn from the experimental results, it is also not difficult to find that serum stability of siRNA-terminal conjugation was improved to some extent, such as 23 (3/As) and 24 (5/As). This implied that sense strand 3′ or 5′-terminus modified with cRGD peptide can improve siRNAs’ ability of resistance to cleavage. Their corresponding sequences 20 (3/3) and 21 (5/3), which were further modified with cRGD at antisense 3″-terminus, showed increased serum stability, indicating that modification at 3″-terminus of antisense can also improve serum stability of siRNA. In summary, the results indicated that 3′,5′-terminus of sense strand and 3″-terminus of antisense strand were readily attacked by nucleases, whereas our modification improved its enzyme-resistant ability. All these data indicated that the serum stability of siMB3 modified with cRGD peptide was improved, especially 20 and 21.
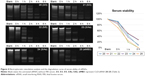  | Figure 4 Electrophoretic retardation analysis and the degradation curve of serum ability of siRNAs. |
Cellular uptake of CLD/siRNA nanocomplexes
Flow cytometry technique was used for the detection of cell uptake of CLD/siRNA 26–30. A375 cells were incubated with 100 nM CLD/FAM-siRNA for 6 h, and the corresponding results are shown in Figure 5A. After administration at the same concentrations, the uptake rates of cRGD-conjugated siRNA were higher than that of the non-targeting siRNA, wherein 23 (3/As) group uptake was increased by 2.6 times and 21 (5/3) group uptake was increased by 5.4 times. The uptake of 21 (5/3) group increased much more than others, which implied that it may form different assembly ways with the CLD, leading to more cRGD exposure on the surface of CLD/siRNA 21 (5/3) nanocomplex. Alternatively, A375 cells were incubated with cRGD peptide (15 μg/mL) or cRAD peptide (15 μg/mL) at 37°C for 30 min, and then transfected with 100 nM of CLD/FAM-labeled cRGD-siRNAs 26, 30; the corresponding results are shown in Figure 5B. These data demonstrate the ability of cRGD to effectively and specifically assist transportation of conjugated CLD/siRNA nanocomplexes into the cells expressing integrin αvβ3.
  | Figure 5 Uptake of CLD/siRNA nanocomplexes by A375 cells and HeLa cells. (A and C) Flow cytometry analysis of fluorescence intensity cells treated with CLD/FAM-siRNA at 100 nM for 6 h. (B) A375 cells were incubated with cRGD peptide (15 μg/mL) or cRAD peptide (15 μg/mL) at 37°C for 30 min in the beginning, and then cellular uptake levels was measured by flow cytometry after cells treated with CLD/FAM-siRNA at 100 nM for 6 h. (D) Intracellular pathway investigation for four nanocomplexes in A375 cells. Channel ratio image of four nanocomplexes in cellular uptake (n=3). |
Integrin αvβ3-negative HeLa cells9 were selected to investigate the effect of the interaction between pepetide and CLD to form nanocomplexes on cell uptake. HeLa cells were incubated with 100 nM CLD/FAM-siRNA for 6 h, and the corresponding results are shown in Figure 5C. The results showed that 20 (3/3), 21 (5/3) and 23 (3/As) have higher uptake rates than the non-targeting siRNA, wherein 20 (3/3) and 21 (5/3) were higher than 23 (3/As) illustrating that double siRNA conjugates have advantages over single siRNA conjugates.
These results indicated that CLD/cRGD-siRNA nanocomplexes achieved higher efficiency of cellular uptake in A375 cells and HeLa cells, wherein 23 (3/As) showed higher cellular uptake than non-targeting siRNA, and 21 (5/3) showed advantages in A375 cells and 20 (3/3) in HeLa cells. CLD/cRGD-siRNA nanocomplexes’ targeting ability and the dual actions, electrostatic and van der Waals force, both led to the increase of cellular uptake.
In fact, we have studied the cellular uptake of cRGD-siRNA conjugates without carrier or transfection reagent at cell level. The study demonstrated that U87 cell lines expressed more integrin αvβ3 than A375 cell lines;28 so, Glioma cells U87 were incubated with FAM-siRNA to evaluate the transmembrane ability of cRGD-siRNA conjugates. U87 cells were incubated with 100, 200, 300 and 400 nM CLD/FAM-labeled cRGD-siRNA 26 for 6 h without transfection reagent and we found that the cellular uptake was increased in a concentration-dependent manner. However, the cellular uptake in 400 nM siRNA concentration was very low in comparison with CLD/cRGD-siRNA nanocomplexes in 100 nM siRNA concentration (data not shown). A375 cells were chosen to evaluate their gene-silencing effect due to lack of target gene in U87 cells, and it was found that they have no noticeable gene-silencing effect in A375 cells (data not shown).
Endocytotic pathway assay
To calculate the ratio of three main uptake channels of siRNA, we chose inhibitors (genistein 40 μg/mL; chlorpromazine 5 μg/mL and amiloride 0.5 mM) to inhibit corresponding endocytotic pathway of CvME, CME and macropinocytosis, and the corresponding results are shown in Figure 5D. An illustration of endocytotic pathway is shown in Figure 6. CME is a classical cellular endocytosis mechanism. In the early intracellular bodies stage, quite a small part of drug content such as siRNA is released into the cytoplasm with the reduction of pH value, and the contents are hydrolyzed by lysosomes or exocytosed by exocytosis, which is not conducive to the escape of siRNA.30,31 CvME is formed by inward hollows of the bottle-shaped form of cell membrane. This endocytosis pathway does not merge with lysosome, thereby avoiding degradation of the hydrolytic enzymes in the lysosomes.32,33 The giant endocytic vesicles, formed by macrophage, are called macropinosomes and can fuse rapidly with lysosomal vesicles.34 Since this pathway shortens the time for vesicle acidification, it shortens the period of siRNA degradation in vesicles, facilitating lysosomal escape of siRNA. Compared with 25 (siMB3) group, 23 (3/As) group reduced the uptake of the CME by 15% and 20 (3/3) group and 21 (5/3) group reduced the uptake by 20%. Moreover, they all increased CvME uptake, especially 21 (5/3) and 20 (3/3) groups, which is partially due to the dual force (electrostatic attraction and Van der Waals forces) between CLD and cRGD-siRNA in nanocomplexes, it indicated that CLD/cRGD-siRNAs have more tighter structure in comparison with CLD/siRNA, and the higher zeta potential might also enable them to improve the route of non-acid-sensitive CvME pathway. All these results suggested that CLD/cRGD-siRNA nanocomplexes regulate multiple pathways to varying degrees. What is more, they all reduced acidification of CME pathway, which implied that they may promote efficient release of siRNA into the cytoplasm, leading to efficient silencing of target mRNA.
Anti-proliferation and cytotoxicity analysis
As we know, cationic lipid carriers are toxic to normal tissue. CCK-8 method was used to test the cytotoxicity of the CLD/siRNA nanocomplexes (Figure 7B). siMB3 and siNC were administered at the same concentration (100 nM). Meanwhile, cells were transfected with siNC and siMB3 with Lipofectamine 2000 as control. The results showed that the cytotoxicity of Lipofectamine 2000 was much higher than that of lipid material CLD. Compared with the blank group, the cytotoxicity of NC nanocomplex (siNC mixed with CLD lipid) showed few differences, which suggested that CLD was less toxic to cells. All CLD/siRNA nanocomplexes exhibited significantly high ability of inhibiting cell proliferation. 21 (5/3) group showed the strongest inhibition effect, while 22 (5/5) group was on the contrary. This is probably because 5″-terminus modification affected binding ability of the antisense strand of siMB3 to RISC complex, which leads to the loss of its gene-silencing activity. However, the ability of 22 (5/5) group to inhibit cell proliferation is slightly stronger than NC group, which is probably caused by cRGD peptide, which has inhibition ability of cell proliferation,28 and the residue part also probably exerted inhibit action when cRGD departed from the cRGD-siRNA. These results indicated that the predominant siRNA-induced target mRNA silencing inhibits cell proliferation, in accordance with the RT-PCR results (Figure 7A).
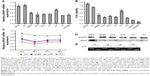  | Figure 7 Efficacy of BRAFV600E gene silencing, downregulation of BRAF protein expression, anti-proliferation with CLD/cRGD-siRNA nanocomplexes in vitro and gel electrophoretic analysis of disulfide cleavage of single-strand cRGD-siRNA conjugates in a reductive environment. (A) Cells were incubated with 100 nM siMB3 using Lipofectamine 2000, CLD/NC and CLD/siRNA 20–25 (Table 2) for 24 h, NC as a negative control, cells were then collected for mRNA isolation. BRAFV600E mRNA was quantified using RT-qPCR and normalized to internal control. Data were plotted relative to untreated cells. (B) Cells were incubated with 100 nM siNC and siMB3 using Lipofectamine 2000, CLD/NC and CLD/siRNA 20–25 (Table 2) for 72 h; cellular viability was measured using the CCK-8 assay. Blank states the untreated cells. (C) Cells were incubated with 100 nM CLD/siNC and CLD/siRNA 21, 23, 25 (Table 2) for 24, 48, 72 and 96 h. BRAFV600E mRNA was quantified using RT-qPCR and normalized to internal control. Data were plotted relative to untreated cells. (D) Downregulation of BRAF protein expression by CLD/siRNA. (E) 20 pmol single-strand cRGD-siRNAs 11, 12, 13, 14 (Table 1) incubated in the buffer containing 0.1 M DTT at 37°C for 2 h, and then analyzed by gel electrophoretic. |
Inhibition of BRAF expression
To confirm the gene-silencing effect of CLD/siRNA nanocomplexes, real-time PCR test was conducted to detect the mRNA level of BRAFV600E gene in A375 cell (Figure 7). Cells treated without siRNA (Blank) or treated with CLD/siNC were used as negative control, and cells treated with 25 using Lipofectamine 2000 were used as positive control. Real-time PCR (Figure 7A) and western blot results (Figure 7D) showed that CLD/siRNA nanocomplexes specifically and efficiently downregulate the expression of targeted gene and protein. 21(5/3) decreased slightly more gene expression than lipoMB3 (47%, 50% respectively), which was probably due to the modification at 5′-terminus of sense strand resulting in decrease of the off-target effect caused by incorporation of sense strand into RISC. Similarly, 23 (3/As) achieved 50% gene silencing, and 25 (siMB3) achieved 39% gene silencing, but 22 (5/5) showed the worst silencing, because 5″-terminus modification affected siRNA antisense strand’s binding to RISC complex as previously discussed. All results also showed that CLD has a similar transfection potency with Lipofectamine 2000.
To test the dynamics and longevity of gene knockdown with CLD/cRGD-siRNA, A375 cells were treated with 100 nM of CLD/siNC, CLD/siRNA 21, 23, 25; the levels of target mRNA were measured at multiple time points after the treatment. The resulting time curve (Figure 7C) demonstrated that maximum knockdowns of mRNA occurred at 48 h, and the silencing activity was sustained (for mRNA) for up to 96 h after the initial treatment.
In conclusion, both CLD/siRNA and CLD/cRGD-siRNAs can activate RNAi machinery and produce efficient, specific and sustainable silencing activity of targeted genes over 24 h. Above all, 21 (5/3) exhibited superior specific target gene silencing activity at multiple time points.
siRNA release from cRGD-siRNAs
The non-denaturing gels were run to verify the cleavage of disulfide linkage in cRGD-s-s-siRNA conjugates. The results (Figure 7E) showed that the disulfide band of single-strand cRGD-siRNAs 11–14 (Table 1) was indeed cleaved in highly reductive condition, and the top bands were the bands of single-strand cRGD-siRNAs, and the following bonds were the residual nucleic acid bands after disulfide bonds were cleaved. The results illustrated that cRGD-siRNA conjugates can release the naked siRNAs in highly reductive environment like cytoplasm. The cleavage of disulfide bond probably reduced the immune response,22 and the released siRNAs could be readily bound to the RISC for targeting mRNA degradation. In summary, cRGD-siRNA conjugates could be applied safely to siRNA therapy.
Conclusion
In conclusion, five cRGD-siRNA conjugates, including single-conjugated ones and bis-conjugated ones mixed with CLD to generate corresponding nanocomplexes, have been evaluated. Compared with other groups, 21 (5/3) exhibited a significantly enhanced uptake. Meanwhile, cRGD-siRNA conjugates 20, 21, 23, 24 also showed better gene-silencing effect than 25. What is more, we have developed the covalent modification strategy in combination with vector delivery, which demonstrated once again that the mode of assembly, formed by the interaction of covalent conjugates with the carrier, can affect cellular uptake and the endocytic pathway. Especially, the mode of assembly can reduce clathrin-mediated transmembrane and lysosomal degradation. All these results showed that CLD/21 nanocomplex was endowed with improved stability, cellular uptake and tumor cell specificity, which could be a meaningful supplement for siRNA drug development.
Acknowledgment
This work was supported by the National Natural Science Foundation of China (Grant No 81302626, 20932001), the Ministry of Science and Technology of China (Grant No 2012AA022501, 2012CB720604).
Disclosure
The authors report no conflicts of interest in this work.
References
Seyhan AA. RNAi: a potential new class of therapeutic for human genetic disease. Hum Genet. 2011;130(5):583–605. | ||
Perrimon N, Ni JQ, Perkins L. In vivo RNAi: today and tomorrow. Cold Spring Harb Perspect Biol. 2010;2(8):a003640. | ||
Wilson RC, Doudna JA. Molecular mechanisms of RNA interference. Annu Rev Biophys. 2013;42:217–239. | ||
Ozpolat B, Sood AK, Lopez-Berestein G. Liposomal siRNA nanocarriers for cancer therapy. Adv Drug Deliv Rev. 2014;66:110–116. | ||
Ming X, Laing B. Bioconjugates for targeted delivery of therapeutic oligonucleotides. Adv Drug Deliv Rev. 2015;87:81–89. | ||
Ku SH, Jo SD, Lee YK, Kim K, Kim SH. Chemical and structural modifications of RNAi therapeutics. Adv Drug Deliv Rev. 2016;104:16–28. | ||
Jeong JH, Mok H, Oh YK, Park TG. siRNA conjugate delivery systems. Bioconjug Chem. 2009;20(1):5–14. | ||
Juliano RL, Carver K, Cao C, Ming X. Receptors, endocytosis, and trafficking: the biological basis of targeted delivery of antisense and siRNA oligonucleotides. J Drug Target. 2013;21(1):27–43. | ||
Liu X, Wang W, Samarsky D, et al. Tumor-targeted in vivo gene silencing via systemic delivery of cRGD-conjugated siRNA. Nucleic Acids Res. 2014;42(18):11805–11817. | ||
Alam MR, Ming X, Fisher M, et al. Multivalent cyclic RGD conjugates for targeted delivery of small interfering RNA. Bioconjug Chem. 2011;22(8):1673–1681. | ||
Alam MR, Ming X, Nakagawa O, Jin J, Juliano RL. Covalent conjugation of oligonucleotides with cell-targeting ligands. Bioorg Med Chem. 2013;21(20):6217–6223. | ||
Liu H, Li Y, Mozhi A, et al. SiRNA-phospholipid conjugates for gene and drug delivery in cancer treatment. Biomaterials. 2014;35(24):6519–6533. | ||
Averick SE, Paredes E, Dey SK, et al. Autotransfecting short interfering RNA through facile covalent polymer escorts. J Am Chem Soc. 2013;135(34):12508–12511. | ||
Fan X, Zhang Y, Liu X, et al. Biological properties of a 3′,3″-bis-peptide-siRNA conjugate in vitro and in vivo. Bioconjug Chem. 2016;27(4):1131–1142. | ||
Wang X, Huang Y, Liu Y, et al. Synthesis and biological evaluation of peptide-siRNA conjugates with phosphodiester unit as linker. Sci China Chem. 2013;56(11):1542–1549. | ||
Liu Y, Wang XF, Chen Y, Zhang LH, Yang ZJ. A solid-phase method for peptide-siRNA covalent conjugates based on click chemistry. Med Chem Comm. 2012;3(4):506–511. | ||
Zou L, Huang Y, Wang X, et al. Serum stability enhancement of siRNA caused by peptide conjugation at 3′ terminus of sense strand. J Chin Pharm Sci. 2014;23(4):215–219. | ||
Young SW, Stenzel M, Yang JL. Nanoparticle-siRNA: a potential cancer therapy? Crit Rev Oncol Hematol. 2016;98:159–169. | ||
Lin Q, Chen J, Zhang Z, Zheng G. Lipid-based nanoparticles in the systemic delivery of siRNA. Nanomedicine (Lond). 2014;9(1):105–120. | ||
Zheng Y, Guo Y, Li Y, Wu Y, Zhang L, Yang Z. A novel gemini-like cationic lipid for the efficient delivery of siRNA. New J Chem. 2014;38(10):4952–4962. | ||
Ma XF, Sun J, Qiu C, et al. The role of disulfide-bridge on the activities of H-shape gemini-like cationic lipid based siRNA delivery. J Control Release. 2016;235:99–111. | ||
Jung S, Lee SH, Mok H, Chung HJ, Park TG. Gene silencing efficiency of siRNA-PEG conjugates: effect of PEGylation site and PEG molecular weight. J Control Release. 2010;144(3):306–313. | ||
Lu K, Duan QP, Ma L, Zhao DX. Chemical strategies for the synthesis of peptide-oligonucleotide conjugates. Bioconjug Chem. 2010;21(2):187–202. | ||
Jang YL, Ku SH, Jin S, et al. Hyaluronic acid-siRNA conjugate/reducible polyethylenimine complexes for targeted siRNA delivery. J Nanosci Nanotechnol. 2014;14(10):7388–7394. | ||
Chen M, Zakrewsky M, Gupta V, et al. Topical delivery of siRNA into skin using SPACE-peptide carriers. J Control Release. 2014;179:33–41. | ||
Alam MR, Dixit V, Kang H, et al. Intracellular delivery of an anionic antisense oligonucleotide via receptor-mediated endocytosis. Nucleic Acids Res. 2008;36(8):2764–2776. | ||
Waite CL, Roth CM. PAMAM-RGD conjugates enhance siRNA delivery through a multicellular spheroid model of malignant glioma. Bioconjug Chem. 2009;20(10):1908–1916. | ||
Hou J, Diao Y, Li W, et al. RGD peptide conjugation results in enhanced antitumor activity of PD0325901 against glioblastoma by both tumor-targeting delivery and combination therapy. Int J Pharm. 2016;505(1–2):329–340. | ||
Martinez J, Tuschl T. RISC is a 5′ phosphomonoester-producing RNA endonuclease. Genes Dev. 2004;18(9):975–980. | ||
Bareford LM, Swaan PW. Endocytic mechanisms for targeted drug delivery. Adv Drug Deliv Rev. 2007;59(8):748–758. | ||
Sahay G, Alakhova DY, Kabanov AV. Endocytosis of nanomedicines. J Control Release. 2010;145(3):182–195. | ||
McLendon PM, Fichter KM, Reineke TM. Poly(glycoamidoamine) vehicles promote pDNA uptake through multiple routes and efficient gene expression via caveolae-mediated endocytosis. Mol Pharm. 2010;7(3):738–750. | ||
Zhu XD, Zhuang Y, Ben JJ, et al. Caveolae-dependent endocytosis is required for class A macrophage scavenger receptor-mediated apoptosis in macrophages. J Biol Chem. 2011;286(10):8231–8239. | ||
Thurn KT, Arora H, Paunesku T, et al. Endocytosis of titanium dioxide nanoparticles in prostate cancer PC-3M cells. Nanomedicine. 2011;7(2): 123–130. | ||
Pichon C, LeCam E, Guérin B, Coulaud D, Delain E, Midoux P. Poly[Lys-(AEDTP)]: a cationic polymer that allows dissociation of pDNA/cationic polymer complexes in a reductive medium and enhances polyfection. Bioconjug Chem. 2002;13(1):76–82. | ||
Mo RH, Zaro JL, Shen WC. Comparison of cationic and amphipathic cell penetrating peptides for siRNA delivery and efficacy. Mol Pharm. 2012;9(2):299–309. | ||
Shirazi RS, Ewert KK, Leal C, Majzoub RN, Bouxsein NF, Safinya CR. Synthesis and characterization of degradable multivalent cationic lipids with disulfide-bond spacers for gene delivery. Biochim Biophys Acta. 2011;1808(9):2156–2166. | ||
Novo L, van Gaal EV, Mastrobattista E, van Nostrum CF, Hennink WE. Decationized crosslinked polyplexes for redox-triggered gene delivery. J Control Release. 2013;169(3):246–256. | ||
Zhao F, Yin H, Zhang Z, Li J. Folic acid modified cationic γ-cyclodextrin-oligoethylenimine star polymer with bioreducible disulfide linker for efficient targeted gene delivery. Biomacromolecules. 2013;14(2):476–484. | ||
Zhu Y, Tang GP, Xu FJ. Efficient poly(N-3-hydroxypropyl)aspartamide-based carriers via ATRP for gene delivery. ACS Appl Mater Interfaces. 2013;5(5):1840–1848. |
Supplementary materials
  | Figure S4 ESI-MS of the purified single oligonucleotide strand. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.