Back to Journals » International Journal of Nanomedicine » Volume 10 » Issue 1
Reducible chimeric polypeptide consisting of octa-d-arginine and tetra-l-histidine peptides as an efficient gene delivery vector
Authors Wang X, Tai Z , Tian J, Zhang W, Yao C, Zhang L, Gao Y, Zhu Q, Gao J , Gao S
Received 25 February 2015
Accepted for publication 12 May 2015
Published 22 July 2015 Volume 2015:10(1) Pages 4669—4690
DOI https://doi.org/10.2147/IJN.S83507
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Lei Yang
Xiaoyu Wang,1,2* Zongguang Tai,1* Jing Tian,1* Wei Zhang,3 Chong Yao,1 Lijuan Zhang,1 Yuan Gao,1 Quangang Zhu,1 Jing Gao,4 Shen Gao1
1Department of Pharmaceutics, Changhai Hospital, Second Military Medical University, Shanghai, 2Department of Pharmaceutics, ChengDu Military General Hospital, ChengDu, 3Department of Pharmaceutics, Shanghai Pulmonary Hospital, Tongji University, Shanghai, 4Department of Pharmaceutical Science, School of Pharmacy, Second Military Medical University, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Abstract: Cationic oligopeptide as a nonviral gene delivery vector has aroused much research interest recently, but its further application is limited by its low transfection efficiency. In the present study, we have created a high-efficiency gene vector by using octa-d-arginine and tetra-l-histidine to form a disulfide cross-linked chimeric polypeptide and used this vector to deliver the therapeutic gene tumor-necrosis-factor-related apoptosis-inducing ligand (TRAIL) to see whether the gene could be transferred and could exert antitumor effects in vitro and in vivo. The result showed that the newly designed vector was able to condense DNA into nanosized polyplexes effectively, thus facilitating its transmembrane transport, promoting its endosomal escape, and finally enabling degradation within the cell. Our study has demonstrated that this chimeric polypeptide is an effective gene carrier in cancer therapy.
Keywords: reducible polypeptide, disulfide bond, octa-d-arginine, tetra-l-histidine, TRAIL
Introduction
Gene therapy needs highly efficient and low-toxicity vectors to deliver nucleic acids into cells and release genes sufficiently. An ideal gene vector should possess ability for strong gene packing, efficient intracellular transportation, sufficient endosomal escape, and rapid gene release.1
Cationic peptides, such as lysine or arginine peptides, have been studied as gene vectors over the past decades owing to their excellent biocompatibility and nontoxicity.2 However, their poor stability due to their low molecular weights (MWs) greatly affects the efficiency of gene transfection.3
Recently, cross-linked cationic polymers based on the reduction-sensitive strategy have been developed to enhance transfection efficiency and decrease cytotoxicity.4,5 These disulfide-containing polymers can remain stable in the extracellular environment and degrade in the intracellular milieu.6 Several studies have reported disulfide cross-linked molecules for RNA or DNA delivery based on different types of frameworks such as linear and branched polyethylenimines (BPEIs) and their derivatives.7 In other words, disulfide cross-linking strategies are able to enhance the stability and efficiency of gene transfection by cationic peptides, along with low cytotoxicity.
After internalization, the complexes are mainly localized in endosomes and then these merge into lysosomes to eliminate exogenous substances.8 Therefore, efficient endosomal escape is a critical step for gene delivery.9 Histidine has been studied as a proton-buffering moiety that can swell and burst the endosomes to facilitate gene transfection.10,11
Although some studies have reported the combined use of histidine and lysine,12,13 the lysine peptide lacks cell-penetrating properties as compared with the arginine analog.14,15 Although Won et al16–18 reported reducible arginine polypeptides, they did not include the buffering moieties in these polypeptides. More importantly, all these homogeneous reducible polypeptides were prepared using fixed compositions of monomers.
In this study, we designed a novel reducible chimeric polypeptide as the gene vector, known as RHss. It contained two functional peptides: octa-D-arginine and tetra-L-histidine peptides. In RHss, octa-D-arginine worked as a DNA-binding moiety and cell penetration-facilitating group, while tetra-L-histidine peptide was responsible for the acquired endosomal buffering capability of the polypeptide. Two cysteines were present at both ends of the peptide to enable formation of intermolecular disulfide bonds under mild oxidative condition (in presence of H2O2) (Figure 1). It was easy to optimize the composition of polypeptides in our research using heterogeneously cross-linked functional peptides at different molar ratios.
Subsequently, we investigated the physiochemical characteristics of RHss/DNA polypeptides and evaluated the gene delivery efficiency with a reporter gene. In addition, the positive effect of histidine peptides on the efficiency of endosomal escape was studied in vitro. Finally, we applied the tumor-necrosis-factor-related apoptosis-inducing ligand (TRAIL) gene to the model drug and evaluated the therapeutic effect of RHss/pTRAIL (plasmid DNA [pDNA] encoding TRAIL) polyplexes on HeLa cells in vitro and in vivo.
Materials and methods
Materials
Materials used in this study were D/L-arginine, L-histidine hydrochloride, PEI (branched, 25 K), L-buthionine sulfoximine (L-BSO), 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI), and chloroquine phosphate (Sigma-Aldrich, St Louis, MO, USA); hydrogen peroxide (30% w/v, Shanghai Sangon Biotech Company, Shanghai, People’s Republic of China); bafilomycin A1 (Gene Operation Inc, Wuxi, People’s Republic of China); pDNA (pGL3, pEGFP, and pORF-hTRAIL) (Shanghai Innovation Biotechnology Co, Ltd, Shanghai, People’s Republic of China); luciferase assay kits (Promega, Beijing, People’s Republic of China); Cell Counting Kit-8 (CCK-8) (Dojindo Molecular Technologies, Inc, Shanghai, People’s Republic of China); Cy3 Label IT® Tracker™ (Mirus Bio LLC, Madison, WI, USA); enhanced bicinchoninic acid assay (BCA) kit and lactate dehydrogenase (LDH) cytotoxicity detection kit (Beyotime, Haimen, People’s Republic of China); propidium iodide (PI)/RNase staining buffer and fluorescein isothiocyanate (FITC)–Annexin V Apoptosis Detection Kit (BD Pharmingen™, Shanghai, People’s Republic of China); Dulbecco’s Modified Eagle’s Medium (DMEM), phosphate-buffered saline (PBS), trypsin, and fetal bovine serum (FBS) (Life Technologies Corporation, Shanghai, People’s Republic of China); HEK293T cells (human embryonic kidney cell line) and HeLa cells (human cervical cancer cell line) (Cell Culture Center of Shanghai Institutes for Biological Sciences of the Chinese Academy of Sciences, Shanghai, People’s Republic of China). All other reagents were of analytical grade. The study was approved by the Ethics Committee of Changhai Hospital.
Cell culture
HEK293T and HeLa cells were grown in DMEM with penicillin, streptomycin, and 10% FBS at 37°C in a humidified 5% CO2 and 95% air atmosphere.
Synthesis and characterization of reducible RHss polypeptides
Peptides Cys-(D-R8)-Cys (CRRRRRRRRC) and Cys-(L-H4)-Cys (CHHHHC) were synthesized using the solid-phase peptide synthesis method with Fmoc protection and purified by reverse-phase high-performance liquid chromatography. RHss was synthesized by hydrogen peroxide oxidation (Figure 2). Cys-(D-R8)-Cys and Cys-(L-H4)-Cys peptides were dissolved in PBS (pH =7.4). H2O2 with a final concentration of 0.1% (w/v) was added to the solution, and oxidative polymerization was conducted for 12 hours at room temperature (RT). To remove excess reactants, the products were purified for 24 hours in deionized water using a dialysis membrane (molecular weight cutoff =3,500 Da) and then lyophilized. To optimize the synthesized RHss, three molar ratios of H4 and R8 peptides (1:1, 1:2, and 1:4) were chosen. The products were termed as RHss1, RHss2, and RHss4, respectively. The single cross-linked R8 peptide was termed as Rss. The products were analyzed by H-spectra of nuclear magnetic resonance (1H-NMR, 600 MHz) and gel permeation chromatography (GPC).
To study the biodegradation property of the polypeptides, we applied GPC analysis to determine the MW of the vectors by incubation with or without dithiothreitol (DTT) for 1 hour. Briefly, 5 mg RHss4 was dissolved in 1 mL PBS containing 0.38 mg DTT. About 100 μL of solution was collected every 10 minutes and analyzed by GPC. RHss4 incubated in PBS was used as the control.
Formation and characterization of RHss/DNA polyplexes
Polyplexes comprising 0.8 μg pDNA and RHss with different N/P ratios (the ratios of moles of the amine groups of cationic polymers to those of the phosphate groups of DNA) self-assembled in 0.8 mL PBS (10 mM, pH 7.4) after 30 minutes of incubation at RT. The particle size and zeta potential were measured using Zetasizer Nano ZS90 (Malvern Instruments, Malvern, UK). The morphology of the polyplexes was measured at N/P =10 using an H-600 transmission electron microscope (Hitachi, Tokyo, Japan) under 75 kV acceleration voltage. The polyplex solution (concentration of pDNA =10 μg/mL) was placed on a carbon-coated copper grid. The excess solution was removed with a filter paper and the grid was dried in air.
Agarose gel electrophoresis of RHss/DNA polyplexes
The condensation ability of the polyplexes was determined by agarose gel electrophoresis. The RHss/DNA polyplexes were prepared at N/P ratios ranging from 0.25 to 2.0. After 30 minutes’ incubation, polyplexes containing 1 μg pDNA were loaded into the wells of 1.0% agarose gel containing Gelred™ and electrophoresis was conducted at 100 V for 30 minutes in Tris-acetate-EDTA buffer. The locations of the nucleic acid bands were visualized under ultraviolet light.
The protective effect of the polyplexes was evaluated by the DNase degradation test. RHss and pDNA were incubated at N/P ratio =2 for 30 minutes, followed by addition of 1 μL DNase (3 units [U]) to the polyplexes and incubation for 2 hours at 37°C. Next, 10 μL (500 U/mL) of heparin solution was added to release the condensed pDNA. All samples underwent agarose gel electrophoresis under the same conditions described earlier.
The DNA-releasing ability was evaluated using the salt dissociation assay. Briefly, polyplexes were prepared at N/P ratio =2, incubated in 25 mM DTT for 2 hours at 37°C, and mixed with NaCl solution (0–1 M). All samples underwent agarose gel electrophoresis under the same conditions described earlier.
Reporter gene transfection assay
Transfection experiments were conducted in 12-well plates. HEK293T cells were seeded at a density of 1×105 cells per well, and the cells were cultured for 24 hours. The medium was totally replaced by serum-free medium before transfection. Polyplexes containing 2 μg pDNA (pEGFP or pGL3) with different N/P ratios were added into the wells. After 4 hours of incubation, fresh medium with 10% serum was added to each well. For pEGFP transfection, gene expression was observed under a fluorescence microscope (Leica, Wetzlar, Germany). Flow cytometry was performed to quantitate the EGFP expression. After 48 hours of transfection, cells were digested and resuspended in 300 μL PBS. The fluorescence intensity of cells was analyzed with a flow cytometer (BD, Franklin Lakes, NJ, USA). For pGL3 transfection, cells were washed with PBS and treated with 200 μL lysis buffer reagent (Promega) for 30 minutes. Luciferase activity was measured with a luminometer (Promega) after addition of 100 μL substrate to 20 μL cell lysate. The luciferase activity was normalized with protein concentrations measured using an enhanced BCA protein assay kit according to the manufacturer’s protocol.
Cellular uptake and localization experiment
Cellular uptake of the polyplexes was analyzed by flow cytometry. The pGL3 plasmid was labeled using a Cy3 protein-labeling kit according to the manufacturer’s protocol. HEK293T cells were seeded onto 12-well plates at 2×105 cells per well and incubated for 24 hours. After replacing the culture medium, polyplexes containing 2 μg Cy3-pDNA were added into the wells. After 4-hour incubation, cells were digested and resuspended in 300 μL PBS. The fluorescence intensity of cells was analyzed by flow cytometry (BD).
Intracellular distribution of the polyplexes was measured with a confocal laser scanning microscope (Olympus, Tokyo, Japan). HEK293T cells were seeded onto glass-bottom 24-well plates at a density of 1.0×105 cells per well and incubated for 24 hours. After replacing the culture medium, polyplexes containing 2 μg Cy3-pDNA were added into the wells. After 1- or 4-hour incubation, cells were fixed in 4% paraformaldehyde solution and treated with DAPI for nucleus staining, washed with PBS three times, and sealed with mounting medium. The samples were imaged by confocal laser scanning microscopy (CLSM). For the 24-hour group, fresh medium with 10% serum was added to replace the polyplex-containing medium after 4 hours of incubation.
Cytotoxicity assay
Cytotoxicity of the polymers toward HEK293T and HeLa cells was evaluated by the CCK-8 and LDH release assays using the commercial kits. For the CCK-8 assay, cells were seeded onto 96-well plates at different densities (1×104 cells of HEK293T and 8×103 cells of HeLa cells) per well and incubated for 24 hours. The polymers were added into wells in a series of increasing concentrations and then incubated for 24 hours. The medium was replaced by fresh medium containing 10% CCK-8 solution and then incubated for 1 hour. The absorbance of each well was measured using a microplate reader at 450 nm. Untreated cells were used as control and the viability was set as 100%. The viability of other cells was expressed as the percentage relative to the absorbance of the untreated cells. The LDH release assay was carried out according to the manufacturer’s protocol. Briefly, 1×104 cells of HEK293T were seeded onto 96-well plates per well and incubated for 24 hours. The predetermined amount of polymers was added and incubated for 24 hours. About 100 μL LDH buffer was added into the collected supernatant and the mixture was incubated for another 0.5 hour. The absorbance of each well was measured with a microplate reader at 490 nm.
Efficacy of TRAIL gene-mediated apoptosis
HeLa cells were seeded onto 96-well plates at 8×103 cells per well and incubated for 24 hours. Polyplexes prepared at N/P ratio of 40:1 were studied to assess the TRAIL-induced cell apoptosis: RHss4/pTRAIL, Rss/pTRAIL, RHss4/pEGFP, and Rss/pEGFP. After 4-hour incubation, the polyplex medium was replaced by fresh FBS and incubated for 48 hours. Cell viability was measured by the CCK-8 assay as described earlier.
Cell apoptosis was detected by flow cytometry as previously described.19 Briefly, HeLa cells were seeded onto 12-well plates at 3×105 cells per well and incubated for 24 hours. The cells were assorted into six groups: 1) untreated group; 2) pTRAIL group; 3) RHss4/pTRAIL group; 4) RHss4/pGL3 group; 5) Rss/pTRAIL; and 6) Rss/pEGFP. The polyplexes were prepared at N/P ratio =40:1. After 4-hour incubation, the medium was replaced by fresh FBS. Cells were digested with trypsin and collected by centrifugation. Cell apoptosis was detected using the FITC–Annexin V kit according to manufacturer’s protocol without fixing treatment. Briefly, cells were resuspended in 300 μL 1× Annexin V binding buffer. Next, 5 μL FITC–Annexin V solution and 5 μL PI staining solution were added, followed by 15 minutes’ incubation. All cell samples were assayed by flow cytometry.
In vivo tumor transfection assay
A HeLa xenograft model was established for transfection assay and antitumor study. HeLa cells were harvested and resuspended in DMEM at a density of 1×107 cells/mL. The cell suspension and BD Matrigel™ were mixed in equal volumes and subcutaneously injected into 4-week-old female BALB/c nude mice at 0.2 mL. When the tumor volume reached nearly 50 mm3, the mice were intratumorally injected with 50 μL RHss4/plasmid polyplexes, as well as Rss/plasmid polyplexes containing 20 μg luciferase plasmid, and were prepared at N/P ratio of 40:1 in PBS solution. The injection was repeated daily for 2 weeks. Two days after the final injection, D-luciferin potassium salt (150 mg/kg weight of mice) was intraperitoneally injected into the mice. The image of luciferase expression was recorded using an in vivo imaging system. After the in vivo imaging assay, the mice were sacrificed, and tumors were homogenized in lysis buffer and centrifuged at 10,000× g for 5 minutes. The luciferase expression was detected as described previously.
In vivo antitumor assay
The therapeutic effect of polypeptides/pTRAIL polyplexes was evaluated in the HeLa xenograft model prepared as described above. When the tumor volume reached nearly 50 mm3, 20 mice were equally randomized into four groups: 1) PBS; 2) pTRAIL; 3) Rss/pTRAIL; and 4) RHss4/pTRAIL. The mice were intratumorally injected with 50 μL polyplex containing 20 μg pTRAIL and prepared at N/P ratio of 40:1 in PBS solution. The injection was performed twice a week for 3 weeks. The weight of the mice and the volume of the tumors were recorded every 2 days. The tumor volume was calculated according to the following equation: V = length × width2 ×0.5. Three weeks after treatment, the mice were sacrificed, and the dissected tumors were weighed and fixed in 4% formaldehyde.
Hematoxylin and eosin (HE) staining was carried out as described earlier. Briefly, the fixed tumor tissue was dehydrated using a gradient of ethanol and treated with dimethylbenzene to replace ethanol. The tumor tissue was embedded in paraffin, sliced into 5 μm sections, dewaxed using dimethylbenzene, and stained with HE according to the standard protocol.
Statistical analysis
For comparison of differences between groups, analysis of variance was conducted by SPSS 18.0. P-value <0.05 was considered statistically significant.
Results
Synthesis and characterization of RHss
To enhance the MW and the endosomal escape ability of the arginine peptides, RHss were prepared by H2O2-mediated oxidation between octa-D-arginine and tetra-L-histidine peptides in a near-neutral pH solution. It was found that the concentration of H2O2 (CH) and the oxidative reaction time (TOR) were two crucial factors for polypeptide synthesis. The transfection efficiency of RHss1 polypeptides prepared under various reaction conditions (four kinds of CHs, A: 0.05%, B: 0.1%, C: 0.2%, and D: 0.5%; and three TORs: 12 hours, 24 hours, and 48 hours) was evaluated. As shown in Figure S1 (Supplementary materials), transfection efficiency was the highest in group B-12. When CH was higher or lower than 0.1%, the transfection efficiency decreased significantly. In addition, when CH was >0.5%, the polyplexes exhibited significant cytotoxicity during incubation (data not shown) and EGFP expression was almost unobserved, indicating that the polypeptides were excessively cross-linked under this condition. Hence, the optimized reaction condition was 0.1% of CH and 12 hours of TOR, which was suitable for this polypeptide vector.
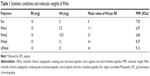
The structure of RHss4 was confirmed by 1H-NMR. As shown in Figure 2, the peaks at δ =1.51, 1.77, and 3.14 ppm were attributed to the –CH2– of the guanidine side chain of arginine (signals a, b, and e, respectively). The peaks at δ =4.18, 4.24, and 4.38 ppm were from the –CH– of arginine (signals g, c, and d, respectively). The peaks at δ =4.48 and 4.62 ppm were attributed to the tertiary carbons of histidine (signals k and j, respectively). The signal m (δ =3.15 ppm) was from the –CH2– of the imidazole side chain of histidine. The multiple peaks at δ =2.92 and δ =3.18 ppm were from the remaining –CH2– of arginine and histidine (signals f and L, respectively). The signals i and h (δ =7.13 and 8.31 ppm, respectively) were attributed to the protons of the imidazole in histidine. The MW of RHss was measured by GPC analysis and is listed in Table 1.
Changes in the MW of RHss4 in the reductive condition were also observed. DTT was applied to imitate the intracellular environment, and the degradation process in PBS was studied as control. As shown in Figure 3, RHss4 degraded rapidly in the reductive condition, and the polypeptide shrank to a peptide monomer after 1-hour incubation. However, in the physiological environment, the degradation rate was slow and the MW decreased by only 5.2%.
Characterization of the polyplexes
The ability of RHss4 to condense DNA was confirmed by the gel retardation assay. The RHss4/DNA polyplexes were prepared at different N/P ratios ranging from 0.25:1 to 2:1. As shown in Figure 4A, R8 completely condensed DNA at the N/P ratio of 1.25:1, while Rss polypeptides condensed DNA into polyplexes above the N/P ratio of 0.75:1. The RHss4/DNA complexes were completely inhibited above the N/P ratio of 1:1.
To determine the redox sensitivity of RHss4, the polyplex dissociation test was performed in NaCl solution, followed by DTT treatment. As shown in Figure 4B, RHss4 remained condensed even in the high-NaCl-concentration environment. Meanwhile, the polyplexes treated with 25 mM DTT released DNA gradually with increasing concentration of NaCl above 0.4 M.
The protection of DNA against DNase by RHss4 is shown in Figure 4C. Free DNA was prone to rapid degradation by DNase in serum. In line 3, naked DNA was degraded under DNase treatment, while RHss4 condensed DNA and subsequently released the DNA strand on addition of heparin (lines 4 and 5, respectively).
The particle size and zeta potential of the polyplexes are shown in Figure 5A: RHss4 could condense DNA into polyplexes, with the Z-average size around 200 nm at an N/P ratio of 2:1. As the N/P ratio increased, more condensed particles formed. The Z-average size decreased to 104.2 nm at an N/P ratio of 40:1. The polydispersity index (PDI) of the polyplexes was <0.3 at different N/P ratios, indicating a narrow size distribution. We next investigated the time-dependent changes in size of RHss4/DNA polyplexes in the presence or absence of the reductive environment. As shown in Figure 5B, the size of RHss4/DNA polyplexes prepared at N/P ratio of 5:1 increased from 117.2 nm to 482.3 nm after 25 mM DTT treatment, and the PDI increased correspondingly from 0.273 to 0.752 in 4 hours. The size of the polyplexes without DTT treatment remained around 110 nm with a low PDI (<0.3). In addition to RHss4/DNA, the size of the other polyplexes is shown in Figure 5C. The zeta potential of the polyplexes prepared at different N/P ratios is presented in Figure 5D. The morphological features of the polyplexes and size distribution are shown in Figure S2 (Supplementary materials). The RHss4/DNA polyplexes formed a spherical nanostructure, and the size was <200 nm at N/P ratio of 10:1.
Cellular uptake and intracellular localization of RHss4/DNA polyplexes
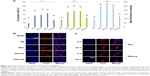
Cy3-labeled plasmid DNA was used to display cellular uptake of the polyplexes in HEK293T cells. The flow cytometry results are shown in Figure 6A. The cellular uptake value was time dependent in all groups. After 4-hour incubation, the polyplexes of Rss and RHss4 exhibited a significantly higher ratio of positive cells than R8 monomer group (P<0.01). In addition, the ratio of positive cells was 98.32% and 94.81% in RHss4 and Rss groups, respectively, vs 32.21% for R8 and 95.56% for BPEI. The mean fluorescence intensity also reflected the cellular uptake of RHss4 and Rss polyplexes. More importantly, the polypeptide LRHss4 prepared by L-R8 and H4 peptides in the same way displayed a significantly lower uptake ability as compared with the D-arginine analog group (P<0.01).
The cellular uptake and intracellular distribution of the polyplexes were also determined by CLSM. As shown in Figure 6B, the red fluorescence of Rss and RHss4 groups displayed stronger cellular uptake than the other groups after 4-hour incubation, indicating efficient cellular uptake, which is consistent with the flow cytometry data. The intracellular distribution of Rss and RHss4 groups after different incubation periods was further determined. As shown in Figure 6C, only the cytoplasm was filled with polyplexes in both groups after 4-hour incubation, and intranuclear distribution was observed after 24 hours’ treatment.
In vitro transfection efficiency of RHss
The transfection efficiency of RHss was measured using two reporter genes (EGFP, luciferase) in HEK293T and HeLa cells. As shown in Figures 7A and 8, after cross-linking with H4 peptide, the RHss polypeptides exhibited significantly higher EGFP transgene expression efficiency than the single Rss polypeptides and their monomers in HEK293T and HeLa cells. To investigate the effect of the ratio of H4 in polypeptides on gene transfection, three types of RHss molecules with different ratios were synthesized. With the ratio of H4 peptide increasing, the transfection efficiency increased significantly. RHss4 showed the best exogenous gene expression. When the molar ratio of H4 and R8 was >4, the polypeptides were prone to precipitation during preparation. Hence, the R8-to-H4 ratio of 1:4 was optimal for polypeptide vectors, and RHss4 was studied in detail for further transfection and cytotoxicity examination.
To quantitate the transfection efficiency, flow cytometry was used to determine the percentage and mean fluorescence value of EGFP gene-positive cells. As shown in Figure 7B, RHss4 transfected 55.92% HEK293T cells at N/P 40:1, and the mean fluorescence intensity was 56.71. In comparison, the positive percentage and mean value of BPEI was 30.2% and 44.08%, respectively, at the optimal N/P ratio of 10:1 in HEK293T cells. The transfection efficacy of RHss, Rss, R8, and BPEI with luciferase gene in HEK293T and HeLa cells was also studied. As shown in Figure 7C, the three RHss polypeptides achieved comparable luciferase gene expression with BPEI in both cells and significantly higher efficacy as compared with Rss and R8.
The transgene expression activity of LRHss4 was measured to investigate the effect of configuration of arginine on transfection efficiency. Compared with RHss4, EGFP expression in the LRHss4 group was significantly lower than that in RHss4 group, and the luciferase expression decreased by sixfold at N/P ratio of 40:1 (Figure S3, Supplementary materials). To check the contribution of the buffering ability of histidine peptides to the transfection efficiency of the polypeptides, chloroquine or bafilomycin A1 was incubated with the polyplexes during transfection in HEK293T cells. As shown in Figure 9, the transfection efficiency of Rss was enhanced markedly and by 10.3-fold in the presence of 100 μM chloroquine, while the luciferase expression in RHss4 group increased by 1.8-fold only and was negligible in the BPEI group. In addition, the transfection of polyplexes was conducted in the presence of bafilomycin A1. In contrast to chloroquine, bafilomycin A1 decreased the transfection efficiency of RHss4, and BPEI and luciferase expression reduced by 20.2-fold and 9.5-fold, respectively. As expected, bafilomycin A1 showed no significant effect on the transfection efficiency of Rss.
To check the effect of disulfide cleavage on transfection efficiency of the polypeptide vectors, BSO, a glutathione (GSH) inhibitor, was incubated with RHss4/DNA polyplexes during transfection in HEK293T cells. As shown in Figure S4 (Supplementary materials), BSO significantly reduced EGFP expression in a dose-dependent manner. When the concentration of BSO was 1,000 μM, the luciferase expression reduced by 28-fold from 1.59×109 to 5.66×107.
Cytotoxicity assay
The cytotoxicity of cationic vectors was detected by CCK-8 and LDH assays in HEK293T (Figure 10) and HeLa cells (Figure S5, Supplementary materials). The results showed that the R8 monomer and RHss4 polypeptides exhibited high cell viability (>80%) after 48-hour incubation at concentrations up to 200 μg/mL and showed low toxicity as compared with Rss polypeptides. In comparison, BPEI showed significant toxicity at concentrations up to 10 μg/mL. Then, early-stage cytotoxicity from membrane disruption during the transfection was detected by LDH release assay, and the result thus obtained was consistent with that of CCK-8 assay. As shown in Figure 10B, LDH release of BPEI obviously increased with the concentration increasing, while the LDH release ratio of R8 peptide, RHss4, and Rss polypeptides remained low when the concentration was <100 μg/mL. However, considerable LDH release was observed in Rss and RHss4 groups at the concentration of 200 μg/mL.
The therapeutic effect of TRAIL gene mediated by polypeptides in vitro
First, cytotoxicity of the polyplexes was detected by CCK-8 assay. As shown in Figure 11A, cell growth was inhibited in RHss4/pTRAIL and Rss/pTRAIL groups, while no significant change in cell morphology was observed in RHss4/pEGFP and Rss/pEGFP groups. The CCK-8 data confirmed the microscopic observation. As illustrated in Figure 11B, the viability of cells treated with RHss4/pEGFP, Rss/pEGFP, and naked pTRAIL was as high as 94.5%, 93.4%, and 92.7%, respectively, indicating that the vectors had no toxic effect at this concentration due to the ineffectiveness of the naked DNA in gene transfection. Cell viability was the lowest in the RHss4/pTRAIL transfection group as compared with the Rss/pTRAIL group (35.2% vs 64.8%).
To further explore the anticancer mechanism of polyplexes, cell apoptosis and cell cycle in HeLa cells were evaluated after 48-hour treatment. As shown in Figure 11C and E, negligible cell apoptosis and necrosis were observed in the control and naked pTRAIL groups, while the percentage of late-apoptotic and necrotic cells increased significantly after RHss4/pTRAIL and Rss/pTRAIL treatment, with the early-apoptotic rate remaining unchanged significantly. In addition, compared with the Rss/pTRAIL group, the late-apoptotic rate and total injury ratio were increased significantly in the RHss4/pTRAIL polyplex group.
The results of the cell cycle assay (Figure 11D and F) showed that most cells were distributed in the G0/G1 phase in the control and naked pTRAIL groups. After RHss4/pTRAIL and Rss/pTRAIL treatment, the percentage of cells in the S phase was increased significantly, and the percentage of cells distributing in the G0/S phase was decreased correspondingly. There was insignificant change in the proportion of cells in the G2/M phase, indicating that the TRAIL protein inhibited the growth of HeLa cells and blocked them in the S phase. In the RHss4/pTRAIL group, the percentage of cells distributing in the S phase was increased as compared with that in the Rss/pTRAIL group.
Effectiveness of gene delivery in vivo
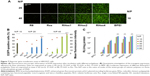
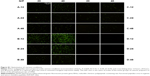
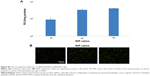
It was found in our in vitro experiment that RHss polypeptides had excellent transgene expression ability, and therefore we further investigated their transfection efficiency in vivo via luciferase reporter gene and TRAIL therapeutic gene. As shown in Figure 12A, the luciferase expression level in RHss4/pGL3 polyplex group was significantly higher than that in the Rss group after repeated intratumoral injections. Data from the homogenate also confirmed the observation from the charge-coupled device image. The luciferase expression level in the RHss4 group was 6.12-fold that in the Rss group (Figure 12B).
After confirming the in vivo gene delivery ability, we further studied the therapeutic effect of RHss4/pTRAIL and Rss/pTRAIL polyplexes in the HeLa mouse xenograft model by measuring the volume and weight of the tumors, to assess the antitumor effect in different groups. As shown in Figure 12C, both PBS and TRAIL exhibited inconspicuous antitumor effect (P>0.05). The inhibitory activity of Rss/pTRAIL polyplexes was significantly stronger than that of the control group (P<0.05). Notably, the therapeutic efficacy in RHss4/pTRAIL group was significantly higher than that in the Rss group (P<0.01). After 3-week treatment, the tumors were excised, measured, and weighed. It was found that both the tumor size and weight in the RHss4/pTRAIL group were significantly smaller than those in the Rss/pTRAIL group (P<0.01) and control group (P<0.01) (Figure 12D and E).
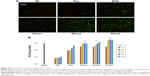
HE-stained tumor sections were observed microscopically to assess tissue damage. As shown in Figure 12F, the nuclei were dense and the tumor tissues were intact in the control group, indicating no therapeutic effect, while in both Rss/TRAIL and RHss4/TRAIL groups, part of the tumor tissue became sparse and the nuclei were fractured as shown in the white circle.
Discussion
Reducible cationic polymers based on disulfide bonds have received much interest recently due to their rapid intracellular gene release and low cytotoxicity.20,21 In this study, we synthesized a series of reducible chimeric polypeptides known as RHss that contained octa-D-arginine and tetra-L-histidine peptides with various ratios. After physical characterization and transfection efficiency evaluation, we screened out a polypeptide known as RHss4 that had the highest transgene expression ability and used it to condense the TRAIL gene for further investigation.
Compared with DMSO, which is widely used in the study of reducible polymers,16,20,22 the oxidative reaction of thiol groups using low-concentration H2O2 in the present study was relatively mild without producing by-products. The reaction conditions were optimized for the best transgene expression ability. The polypeptides were confirmed by 1H-NMR and GPC. Compared with the original peptides, MW of RHss was increased significantly, suggesting that low-concentration H2O2 could mediate the cross-linking reaction by cross-linking the thiol-containing monomers into high-MW products and that polypeptides are sensitive to the reductive environment and could degrade to monomers over a period of 1 hour.
RHss polypeptides can form polyplexes at appropriate N/P ratios with DNA. Compared with R8 peptides, the higher ability of Rss to condense DNA is due to the increased MW caused by disulfide bonds. With the amount of H4 peptides increasing, the condensing ability of RHss reduced gradually, indicating that polypeptide-inserted H4 moieties dispersed the charge density of the arginine peptides and loosened the compact structure of polyplexes, thus facilitating the transgene expression ability.23 DTT-treated polyplexes were incubated in NaCl solution to mimic the intracellular environment. It was found that DTT promoted gene release from the polyplexes in NaCl solution. The mild affinity to DNA of the resultant short R8 peptides was directly related to the lower MW of the polymers.24 In the nonreducing condition, the polyplexes retained the condensing ability and effectively resisted DNase degradation. The particle size and zeta potential of the polyplexes are essential for cellular uptake and gene transfection.25 RHss4 condensed pDNA into nanosized polyplexes (~100 nm) with positive zeta potential (+32 mV), which are suitable for gene transfection. In addition, the polyplexes exhibited an increasing trend in size and PDI value after DTT treatment because polyplexes formed by low-MW cationic polymers tend to aggregate rapidly.12,18
Efficient cellular uptake is the first step for gene transfection. Many studies have demonstrated that the cellular uptake ability of a vector is positively correlated with transfection efficiency.26,27 Therefore, it is a feasible way to improve the vector by promoting the cellular uptake efficiency. Polypeptides with increased MW exhibited a significantly higher ability of cellular uptake than R8 peptides. In addition to MW, the configuration of arginine also affected the cross-membrane transport significantly. Many studies demonstrated28,29 that nondegradable D-arginine peptides exhibited a significantly higher transduction ability than L-arginine peptides. There was no significant difference in cellular uptake between Rss and RHss4, indicating that H4 moieties had little effect on cellular uptake of polycations; rather, they affected the intracellular process after endocytosis. CLSM imaging showed that the polyplexes accumulated in the endosome, presenting as punctate fluorescence spots.30 Numerically more red punctate spots were observed in the cytoplasm in the Rss group, indicating insufficient endosomal escape of the polyplexes, while fewer punctate spots and more uniform distribution were observed in the cytoplasm in RHss4 group, indicating sufficient endosomal escape, which may be attributable to the “proton sponge” effect of the histidine moiety.
The result of our study on transfection efficiency showed that RHss4 had significantly higher transgene expression ability due to high cellular uptake, efficient endosomal escape, and rapid gene release, compared with the other groups. The transfection efficiency in the LRHss4 groups was significantly lower than that in the RHss4 group, suggesting that the efficient cellular uptake had significant impact on gene transfection. However, the transgene expression efficiency of Rss remained low regardless of its high cellular uptake ability, suggesting that histidine played a critical role in the intracellular delivery process. We therefore used chloroquine and bafilomycin A1 to detect whether histidine moieties could promote endosomal escape of the polyplexes. It was found that chloroquine as an endosomolytic agent could significantly increase the transgene expression activity of Rss/DNA polyplexes that lacked the proton-buffering group. However, the enhancement was inconspicuous in the RHss4 and BPEI groups, suggesting that endosomal escape was efficient. Because histidine peptides and other buffering moieties can only be protonated in the acid environment, inhibition of acidification could decrease the endosomolytic ability of histidine peptides. Bafilomycin A1 could inhibit vacuolar type H+-ATPase and then hamper the acidification of the endosome.31 Bafilomycin A1 decreased the transgene expression activity in the BPEI and RHss4 groups, and this effect was not observed in the Rss group. These results confirm that compared with Rss, the enhanced transgene expression activity of RHss4 was mainly attributable to the ability of the buffering group to promote endosomal escape. In addition, the effect of disulfide bond cleavage on the transgene expression activity of polyplexes was investigated. Compared with the extracellular space, the concentration of GSH was at a level (2–20 mM) high enough to generate relatively high redox potential to destabilize disulfide bond and degrade polyplexes.32 As shown in our study, the GSH inhibitor BSO limited the fracture of the disulfide bond and then decreased the transgene expression efficiency of the polyplexes.
The CCK-8 assay and LDH release study were carried out to assess cytotoxicity of the polypeptides. It was found that the cross-linked polypeptides had minimal cytotoxicity as compared with BPEI, a golden standard for nonviral gene vectors. Although BPEI has a high transfection rate, it is highly cytotoxic due to high MW, nondegradation, and high cationic charge density.22,33 The low cytotoxicity of R8 could be attributed to the small MW and degradable feature. After being cross-linked into the polypeptides, RHss4 and Rss showed high cell viability in spite of increased MW and positive charge density. Many studies have demonstrated that reducible degradation of polypeptides is crucial for cytotoxic attenuation. When the concentration of the polypeptides was increased to 200 μg/mL, cell viability in the RHss4 group was increased significantly as compared with that in the Rss group, indicating that the inserted H4 peptides could reduce cationic charge density, which is consistent with the result of gel electrophoresis.
Knowing that TRAIL is a tumor-necrosis-factor superfamily member that can specifically induce tumor cell apoptosis via death receptors 4 and 5 (DR4/DR5),34 we used pTRAIL as a model agent to evaluate the anticancer effect of RHss4/pTRAIL polyplexes in the present study. Our cytotoxicity assay showed that RHss4/pTRAIL had a more potent antitumor effect than Rss/pTRAIL due to more efficient transfection, while no significant therapeutic effect was observed in the RHss4/pEGFP group, indicating that the cytotoxicity of RHss4/pTRAIL was attributable to TRAIL expression rather than to the vector itself. In addition, the subsequent apoptosis-inducing and S phase arrest is the primary antitumor mechanism of the TRAIL gene in HeLa cells. The in vivo therapeutic study using HE staining further demonstrated the superior antitumor effect of RHss4/pTRAIL.
Conclusion
We have developed a reducible chimeric polypeptide, RHss, consisting of octa-D-arginine and tetra-L-histidine peptides for efficient DNA delivery. This RHss showed efficient cellular uptake, high transfection efficiency, and low cytotoxicity in vitro. Remarkable HeLa cell apoptosis and tumor growth inhibition were observed in the mouse model when the RHss4 polypeptide was used to transfer the apoptotic TRAIL gene. Our results suggest that this chimeric polypeptide may prove to be a potential tool for cancer gene therapy.
Acknowledgment
The study was supported by the National Natural Science Foundation of China (numbers 81172514, 81101658, 81372762, 81272819, and 81302212) and National Major Scientific and Technological Special Project for “Significant New Drugs Development” (number 2013ZX09J13109-04B).
Disclosure
The authors report no conflicts of interest in this work.
References
Fortier C, Durocher Y, De Crescenzo G. Surface modification of nonviral nanocarriers for enhanced gene delivery. Nanomedicine. 2014;9(1):135–151. | ||
Kim HH, Lee WS, Yang JM, Shin S. Basic peptide system for efficient delivery of foreign genes. Biochim Biophys Acta. 2003;1640(2–3):129–136. | ||
Ward CM, Read ML, Seymour LW. Systemic circulation of poly(L-lysine)/DNA vectors is influenced by polycation molecular weight and type of DNA: differential circulation in mice and rats and the implications for human gene therapy. Blood. 2001;97(8):2221–2229. | ||
Ou M, Xu R, Kim SH, Bull DA, Kim SW. A family of bioreducible poly(disulfide amine)s for gene delivery. Biomaterials. 2009;30(29):5804–5814. | ||
Yin Q, Shen J, Chen L, Zhang Z, Gu W, Li Y. Overcoming multidrug resistance by co-delivery of Mdr-1 and survivin-targeting RNA with reduction-responsible cationic poly(beta-amino esters). Biomaterials. 2012;33(27):6495–6506. | ||
Lee YS, Kim SW. Bioreducible polymers for therapeutic gene delivery. J Control Release. 2014;190C:424–439. | ||
Son S, Singha K, Kim WJ. Bioreducible BPEI-SS-PEG-cNGR polymer as a tumor targeted nonviral gene carrier. Biomaterials. 2010;31(24):6344–6354. | ||
Khalil IA, Kogure K, Akita H, Harashima H. Uptake pathways and subsequent intracellular trafficking in nonviral gene delivery. Pharmacol Rev. 2006;58(1):32–45. | ||
Li Y, Wen G, Wang D, et al. A complementary strategy for enhancement of nanoparticle intracellular uptake. Pharm Res. 2014;31(8):2054–2064. | ||
Cai X, Zhu H, Dong H, Li Y, Su J, Shi D. Suppression of VEGF by reversible-PEGylated histidylated polylysine in cancer therapy. Adv Healthc Mater. 2014;3(11):1818–1827. | ||
Nakagawa O, Ming X, Carver K, Juliano R. Conjugation with receptor-targeted histidine-rich peptides enhances the pharmacological effectiveness of antisense oligonucleotides. Bioconjug Chem. 2014;25(1):165–170. | ||
Read ML, Singh S, Ahmed Z, et al. A versatile reducible polycation-based system for efficient delivery of a broad range of nucleic acids. Nucleic Acids Res. 2005;33(9):e86. | ||
Kiselev A, Egorova A, Laukkanen A, Baranov V, Urtti A. Characterization of reducible peptide oligomers as carriers for gene delivery. Int J Pharm. 2013;441(1–2):736–747. | ||
Amand HL, Fant K, Norden B, Esbjorner EK. Stimulated endocytosis in penetratin uptake: effect of arginine and lysine. Biochem Biophys Res Commun. 2008;371(4):621–625. | ||
Su Y, Doherty T, Waring AJ, Ruchala P, Hong M. Roles of arginine and lysine residues in the translocation of a cell-penetrating peptide from (13)C, (31)P, and (19)F solid-state NMR. Biochemistry. 2009;48(21): 4587–4595. | ||
Won YW, Kim HA, Lee M, Kim YH. Reducible poly(oligo-D-arginine) for enhanced gene expression in mouse lung by intratracheal injection. Mol Ther. 2010;18(4):734–742. | ||
Won YW, Kim KM, An SS, Lee M, Ha Y, Kim YH. Suicide gene therapy using reducible poly (oligo-D-arginine) for the treatment of spinal cord tumors. Biomaterials. 2011;32(36):9766–9775. | ||
Won YW, Yoon SM, Lee KM, Kim YH. Poly(oligo-D-arginine) with internal disulfide linkages as a cytoplasm-sensitive carrier for siRNA delivery. Mol Ther. 2011;19(2):372–380. | ||
Huang J, Zhang H, Yu Y, et al. Biodegradable self-assembled nanoparticles of poly (d, l-lactide-co-glycolide)/hyaluronic acid block copolymers for target delivery of docetaxel to breast cancer. Biomaterials. 2014;35(1):550–566. | ||
Kang HC, Kang HJ, Bae YH. A reducible polycationic gene vector derived from thiolated low molecular weight branched polyethyleneimine linked by 2-iminothiolane. Biomaterials. 2011;32(4):1193–1203. | ||
Liu H, Wang H, Yang W, Cheng Y. Disulfide cross-linked low generation dendrimers with high gene transfection efficacy, low cytotoxicity, and low cost. J Am Chem Soc. 2012;134(42):17680–17687. | ||
Peng Q, Zhong Z, Zhuo R. Disulfide cross-linked polyethylenimines (PEI) prepared via thiolation of low molecular weight PEI as highly efficient gene vectors. Bioconjug Chem. 2008;19(2):499–506. | ||
Yoshihara C, Shew CY, Ito T, Koyama Y. Loosening of DNA/polycation complexes by synthetic polyampholyte to improve the transcription efficiency: effect of charge balance in the polyampholyte. Biophys J. 2010;98(7):1257–1266. | ||
Manickam DS, Oupicky D. Multiblock reducible copolypeptides containing histidine-rich and nuclear localization sequences for gene delivery. Bioconjug Chem. 2006;17(6):1395–1403. | ||
Zhang Q, Chen S, Zhuo RX, Zhang XZ, Cheng SX. Self-assembled terplexes for targeted gene delivery with improved transfection. Bioconjug Chem. 2010;21(11):2086–2092. | ||
Wang F, Wang Y, Wang H, Shao N, Chen Y, Cheng Y. Synergistic effect of amino acids modified on dendrimer surface in gene delivery. Biomaterials. 2014;35(33):9187–9198. | ||
Kim T-I, Bai CZ, Nam K, Park J-S. Comparison between arginine conjugated PAMAM dendrimers with structural diversity for gene delivery systems. J Control Release. 2009;136(2):132–139. | ||
Verdurmen WP, Bovee-Geurts PH, Wadhwani P, et al. Preferential uptake of L- versus D-amino acid cell-penetrating peptides in a cell type-dependent manner. Chem Biol. 2011;18(8):1000–1010. | ||
Ma Y, Gong C, Ma Y, et al. Direct cytosolic delivery of cargoes in vivo by a chimera consisting of D- and L-arginine residues. J Control Release. 2012;162(2):286–294. | ||
Bonner DK, Leung C, Chen-Liang J, Chingozha L, Langer R, Hammond PT. Intracellular trafficking of polyamidoamine-poly(ethylene glycol) block copolymers in DNA delivery. Bioconjug Chem. 2011;22(8):1519–1525. | ||
Yoshimori T, Yamamoto A, Moriyama Y, Futai M, Tashiro Y. Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J Biol Chem. 1991;266(26):17707–17712. | ||
Read ML, Bremner KH, Oupicky D, Green NK, Searle PF, Seymour LW. Vectors based on reducible polycations facilitate intracellular release of nucleic acids. J Gene Med. 2003;5(3):232–245. | ||
Breunig M, Lungwitz U, Liebl R, Goepferich A. Breaking up the correlation between efficacy and toxicity for nonviral gene delivery. Proc Natl Acad Sci U S A. 2007;104(36):14454–14459. | ||
Pan G, Ni J, Wei YF, Yu G, Gentz R, Dixit VM. An antagonist decoy receptor and a death domain-containing receptor for TRAIL. Science. 1997;277(5327):815–818. |
Supplementary materials
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.