Back to Journals » International Journal of Nanomedicine » Volume 11
Real-time colorimetric detection of DNA methylation of the PAX1 gene in cervical scrapings for cervical cancer screening with thiol-labeled PCR primers and gold nanoparticles
Authors Huang J, Liou YL, Kang YN, Tan ZR, Peng MJ, Zhou HH
Received 2 July 2016
Accepted for publication 25 August 2016
Published 12 October 2016 Volume 2016:11 Pages 5335—5347
DOI https://doi.org/10.2147/IJN.S116288
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Linlin Sun
Jin Huang,1,2 Yu-Ligh Liou,1,2 Ya-Nan Kang,3 Zhi-Rong Tan,1,2 Ming-Jing Peng,1,2 Hong-Hao Zhou1,2
1Department of Clinical Pharmacology, Xiangya Hospital, Central South University, 2Institute of Clinical Pharmacology, Hunan Key Laboratory of Pharmacogenetics, Central South University, 3Department of Obstetrics and Gynecology, Xiangya Hospital, Central South University, Changsha, Hunan, People’s Republic of China
Background: DNA methylation can induce carcinogenesis by silencing key tumor suppressor genes. Analysis of aberrant methylation of tumor suppressor genes can be used as a prognostic and predictive biomarker for cancer. In this study, we propose a colorimetric method for the detection of DNA methylation of the paired box gene 1 (PAX1) gene in cervical scrapings obtained from 42 patients who underwent cervical colposcopic biopsy.
Methods: A thiolated methylation-specific polymerase chain reaction (MSP) primer was used to generate MSP products labeled with the thiol group at one end. After bisulfite conversion and MSP amplification, the unmodified gold nanoparticles (AuNPs) were placed in a reaction tube and NaCl was added to induce aggregation of bare AuNPs without generating polymerase chain reaction products. After salt addition, the color of AuNPs remained red in the methylated PAX1 gene samples because of binding to the MSP-amplified products. By contrast, the color of the AuNP colloid solution changed from red to blue in the non-methylated PAX1 gene samples because of aggregation of AuNPs in the absence of the MSP-amplified products. Furthermore, PAX1 methylation was quantitatively detected in cervical scrapings of patients with varied pathological degrees of cervical cancer. Conventional quantitative MSP (qMSP) was also performed for comparison.
Results: The two methods showed a significant correlation of the methylation frequency of the PAX1 gene in cervical scrapings with severity of cervical cancer (n=42, P<0.05). The results of the proposed method showed that the areas under the receiver operating characteristic curve (AUCs) of PAX1 were 0.833, 0.742, and 0.739 for the detection of cervical intraepithelial neoplasms grade 2 and worse lesions (CIN2+), cervical intraepithelial neoplasms grade 3 and worse lesions (CIN3+), and squamous cell carcinoma, respectively. The sensitivity and specificity for detecting CIN2+ lesions were 0.941 and 0.600, respectively, with a cutoff value of 31.27%. The proposed method also showed superior sensitivity over qMSP methods for the detection of CIN2+ and CIN3+ (0.941 vs 0.824 and 1.000 vs 0.800, respectively). Furthermore, the novel method exhibited higher AUC (0.833) for the detection of CIN2+ than qMSP (0.807).
Conclusion: The results of thiol-labeled AuNP method were clearly observed by the naked eyes without requiring any expensive equipment. Therefore, the thiol-labeled AuNP method could be a simple but efficient strategy for cervical cancer screening.
Keywords: colorimetric detection, gold nanoparticles, DNA methylation, cervical cancer screening, UV-vis, high sensitivity, quantitative detection
Introduction
Cervical cancer is the second most common cancer in women worldwide and the major cause of death in developing countries.1,2 Approximately 500,000 new cases of cervical cancer and 200,000 related deaths are reported annually of which more than 80% occur in developing countries.3–5 In the People’s Republic of China, 53,000 deaths associated with and 131,500 new cases of cervical cancer are recorded, which account for 30% of the global statistics. The incidence of cervical cancer has decreased considerably since the introduction of the thinprep cytologic test (TCT) for the detection of cervical lesions.6–9 However, the prognosis of cervical cancer remains poor.
DNA methylation, which is a type of epigenetic silencing in tumor suppressor genes, could be the mechanism underlying carcinogenesis.10–12 DNA methylation has been extensively studied and widely used in classification, early diagnosis, treatment, and prediction of metastasis as well as cancer recurrence.13,14 The aberrant methylation of CpG islands in the promoter region of tumor suppressor genes could impede DNA transcription, a key mechanism of tumor.15 Consequently, DNA methylation could be an effective biomarker for early diagnosis of cancers and prediction of prognosis in cancer patients. Previous studies showed the presence of aberrant DNA hypermethylation of classic tumor suppressor genes in cervical cancer; these genes include the paired box gene 1 (PAX1),16 genes for sex-determining region Y-box 1,17 epidermal growth factor receptor, cyclooxygenase-2, genes for protein tyrosine phosphatase receptor type R,18 and zinc finger protein 582.19 PAX1 shows the highest potential to be a methylation biomarker. PAX1 could be used for the detection of cervical intraepithelial neoplasms (CIN) grade 3 and worse lesions (CIN3+) with a high sensitivity and specificity.20 Moreover, incorporating detection of PAX1 methylation with human papillomavirus (HPV) test of cervical cells could improve the efficacy of cervical cancer screening.21 Hence, the detection of promoter hypermethylation is an effective tool for early diagnosis of cervical cancer and can be valuable for monitoring the tumor behavior and determining tumor responses to targeted therapy.
Researchers have developed novel nanotechnology-based methods, such as DNA methylation detection based on single-base extension reaction and surface-enhanced Raman spectroscopy,22 surface plasmon resonance,23 methylation-specific microarray, and methylation-specific fluorescence resonant energy transfer.24 These techniques utilize the special properties of nanomaterials and thus show superior performance in the detection of methylation.25 However, these methods require professional design and operations, thereby restricting their applications. Hence, a simple but effective method, with high sensitivity, superior specificity, and easy operation must be developed for the detection of DNA methylation; such a technique should not require the use of special equipment and should be based on novel nanotechnologies.
Noble metal nanoparticles, particularly gold nanoparticles (AuNPs),26 are widely used as a biosensor for DNA detection because of their special optical properties.27 DNA-modified AuNPs were used as a signal amplification unit in a sensitivity and selective electrochemical method for the detection of DNA methylation by Jing et al.28 Also, AuNPs could be an effective biomarker for colorimetric detection of DNA.29 The transition from dispersion to aggregation states of AuNPs can induce color changes in the AuNP colloid solution; such changes can be monitored using various optical spectra, including the corresponding peak shift in the ultraviolet-visible spectrophotometry (UV-vis) or surface plasmon absorption spectrum. Color changes can be easily observed by the naked eyes in the AuNP colloid solution with nanomolar concentration. Therefore, these nanoparticles exhibit potential for colorimetric sensing. Colorimetric assays based on AuNPs for the detection of cancer has been reported in a previous study, it displayed many great advantages.30 Chen et al utilized AuNP probes for the DNA methylated detection of E-cadherin, p15, and p16 genes in three human colon cancer cell lines, which showed that this method is simple, rapid, and has a high sensitivity to simultaneously detect methylation.30 However, there were no studies utilizing nanoparticles for the detection of cervical cancer.
In this study, we present a novel method for direct colorimetric detection of PAX1 gene methylation in cervical scrapings by using thiol-labeled primers and bare AuNPs. The proposed method enables real-time detection by the naked eyes without the need for any detection equipment.
Materials and methods
Patients, gDNA isolation, and bisulfite conversion
All participants provided written informed consent to participate in the study. This study was approved by the Institutional Review Board of Department of Clinical Pharmacology, Xiangya Hospital, Central South University (registration number: CTXY-110009) and by Chinese Clinical Trial Registry (registration number: ChiCTR-DOD-14005446). Cervical scrapings were obtained from TCT samples of 42 female patients (aged ≥20 years) who underwent colposcopic cervical biopsy from June to November in 2014 in Xiangya Hospital. Approximately 8–10 mL of TCT sample was centrifuged at 3,000 rcf for 5 minutes. The centrifuged sediment was washed with 200 μL of phosphate-buffered saline. Genomic DNA (gDNA) was extracted from the tissue by using iSat Nucleic Acid Extraction Kit (iStat Biomedical Co., Ltd., New Taipei City, Taiwan) in accordance with the manufacturer’s protocol. gDNA concentration was determined using a BioSpec-Nano spectrophotometer (Shimadzu Company, Kyoto, Japan). Samples with DNA ≥500 ng were used for further assay.
DNA bisulfite conversion was performed with iStat Bisulfite Conversion Kit (iStat Biomedical Co., Ltd.) according to the manufacturer’s protocol. Bisulfite DNA concentration was determined using BioSpec-Nano spectrophotometer (Shimadzu Company).
Quantification of DNA methylation through qMSP
Quantitative methylation-specific polymerase chain reaction (qMSP) by TaqMan-based technologies was performed in Lightcycler LC480 real-time polymerase chain reaction (PCR) system (Hoffman-La Roche Ltd., Basel, Switzerland) to detect DNA methylation.
The methylation status of PAX1 was detected by quantitative PCR (qPCR) kits (iStat Biomedical Co., Ltd.), with the VIC gene as an internal reference. The crossing point (Cp) value for VIC indicates the validity of the test and should be less than 35.
qMSP was performed in a mixture with a total volume of 20 μL, including 2 μL of bisulfite template DNA, 1 μL of 2× custom detection mix, and 10 μL of 2× custom Universal PCR Master Mix. The mixture was subjected to pre-incubation at 95°C for 10 minutes; followed by 50 cycles at 95°C for 10 seconds, annealing and extension at 60°C for 40 seconds, and detection using the LC480. Fluorescence data were collected during annealing or extension to determine Cp. The Cp values from the PAX1 and VIC genes were obtained in each sample. The DNA methylation status was calculated based on the differences between the two Cp values (ΔCp = CpPAX1 − CpVIC). CaSki and C33A cancer cell lines were used as methylation and non-methylation controls,31,32 respectively, to ensure the quality of bisulfite conversion and qPCR.
AuNPs-based colorimetric assay
Synthesis of AuNPs
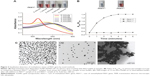
AuNPs (13 nm diameter) were synthesized through citrate reduction of HAuCl4 (Guo Yao Group, Shanghai, People’s Republic of China). Briefly, 50 mL of 0.01 wt% HAuCl4 was brought to a rolling boil with vigorous stirring. Five millilitres of 38.8 mM sodium citrate (Guo Yao Group) was rapidly added to the solution and boiled for 15 minutes. Subsequently, heating was terminated, and the solution was continuously stirred for another 15 minutes. The color of the solution changed from pale yellow to deep red. The resulting solution of the colloidal particles was filtered and characterized through transmission electron microscopy (TEM) (Figure S1A). The synthesized AuNPs were further characterized at 520 nm by using a UV–Vis spectrometer (Figure S1B). The concentration of AuNPs was then calculated.
PCR with thiol-labeled primer and colorimetric assay
Unmethylated (NL) DNA was obtained from peripheral blood of a healthy volunteer. Methylated controls (IVD) were obtained from DNA of NL treated with CpG methyltransferase (Thermo Fisher Scientific, Waltham, MA, USA).
Specific sites of the PAX1 gene were amplified by PCR reaction on Mastercycler Nexus (Eppendorf, Leipzig, Germany) in a 50 μL solution containing 100 ng of the template, 0.4 μM of each primer, 5 μL of 10× EpiTaq PCR buffer, 0.3 mM dNTPs, and 1.25 U EpiTaq DNA polymerase (TaKaRa EpiTaq™ HS for bisulfite-treated DNA; Takara, Nagoya, Japan). The primers used were similar to those used in the standard qMSP technique, except that the forward primer was modified by adding thiol located at the 5′ end for conjugation to AuNPs. PCR was programmed for 4 minutes at 95°C; followed by 16 cycles at 95°C for 30 seconds, 55°C for 30 seconds, and 72°C for 1 minute; and 7 minutes at 72°C. The PCR products were purified with QIA quick PCR Purification Kit (Qiagen NV, Venlo, the Netherlands).
For colorimetric assay, 5 μL of the PCR product was added to 20 μL of the gold colloid solution and mixed with 2 μL of 5 M NaCl.
Quantitative analysis
The defined methylation levels ranged from 100% of the total 100 ng of input DNA by varying the quantities of IVD diluted in NL. The mixtures were used as input template for 16 cycles of the PCR reaction with thiol-labeled PAX1 primers.
UV–vis spectroscopy
UV–vis absorbance spectra were recorded by BioSpec-Nano spectrophotometer (Shimadzu Company) at 520 nm.
TEM characterization
The mixture of AuNPs and PCR products was drop casted on a carbon-coated copper transmission electron microscope grid and dried at room temperature to prepare TEM samples. The mixture was then analyzed using a field-emission transmission electron microscope (JEM-2100, JEOL, Tokyo, Japan).
Statistical analysis
The correlation between clinical characteristics and methylated frequency was determined by chi-square test. The correlation among continuous variables was analyzed by Mann–Whitney U-test. Percentage of methylation rate (PMR) and ΔCp were evaluated to distinguish CIN grade 2 and worse lesions (CIN2+), CIN grade 3 and worse lesions (CIN3+), or squamous cell carcinoma (SCC) by calculating the area under the receiver operating characteristic (ROC) curve (AUC). P<0.05 was considered statistically significant. All statistical analyses were performed with IBM SPSS 19.0 (IBM Corporation, Armonk, NY, USA).
Results
Overall strategy
The overall strategy of the novel method for methylation detection is illustrated in Figure 1. gDNA was extracted and subjected to sodium bisulfite conversion; in this process, unmethylated cytosines were converted into uracil, whereas methylated cytosines were not affected. The target DNA sequence was then amplified using thiol-labeled forward and unlabeled reverse primers through PCR to form amplified nucleic acids with thiol groups at only one end. When mixed with AuNPs, the DNA strands of the PCR product bound to the unmodified AuNPs because of the strong interaction between the thiol groups and surface of AuNPs. This structure could prevent particle interaction after salt addition. By contrast, AuNPs in the absence of the thio-labeled PCR products showed color changes from red to gray because of aggregation upon salt addition. Consequently, the methylated and unmethylated DNAs can be distinguished based on color change upon salt addition. Moreover, the UV–vis spectra differed between aggregated and well-dispersed AuNPs.
Feasibility of methylation detection by thiol-labeled AuNPs in cervical cell lines
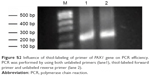
The influence of thiol-labeled primers on PCR efficiency was analyzed in Figure S2. PCR was performed using unlabeled primers (lane 1) as well as thiol-labeled forward primer and unlabeled reverse primer (lane 2) under identical conditions for comparison. PCR efficiency was investigated by gel electrophoresis. Analysis of the gel bands showed PCR efficiency was not reduced when thiol-labeled primers were used.
Control experiments were conducted using the extracted DNA of IVD and NL, which represent unmethylated PAX1 gene (PAX1m−) and methylated PAX1 gene (PAX1m+), respectively. Instantaneous color change from red to purple-gray was observed from 0 to 6 minutes in the AuNP colloid solution containing the PAX1m− specimen. The color change could be due to salt addition. UV–vis spectroscopy showed a narrow peak located at 520 nm at the beginning, and the peak shifted from 520 to 660 nm within 6 minutes. The band became broad, and the optical density value of absorbance decreased as the time increased. This finding indicated the aggregation of AuNPs after salt addition (Figure 2A). By contrast, the color of the AuNP colloid solution with the PAX1m+ gene remained red after salt addition. In addition, no peak shift was observed and the color of the solution remained red.
Changes in the UV–vis spectra were recorded using the ratio of absorbance at 650 and 520 nm to confirm the differences between the methylated and unmethylated samples of the PAX1 gene. Figure 2B shows that the UV–vis spectra for PAX1m− and PAX1m+ samples shifted after 6 minutes. A620/A520 was observed after salt addition every minute. The A650/A520 of AuNPs for the PAX1m− sample was 0.3 at the beginning, increased rapidly, and then slowed down. At 4 minutes, the A650/A520 reached 1.4 and remained stable. This condition showed that AuNPs functionalized with the amplified DNA fragment of the PAX1 gene should be treated with salt for 6 minutes in the following experiment. The AuNP solution underwent rapid color change from red to purple-gray; this change was associated with AuNP aggregation after salt addition. The results observed from the case of PAX1m+ differed from those of the PAX1m−. The A650/A520 of AuNPs for PAX1m+ remained unchanged all the time. The AuNP solution retained its reddish color, indicating that AuNPs remained well dispersed. PAX1m+ case also led to a small degree of AuNP aggregation especially when high salt concentrations were used. However, the color change (from red to purple-gray) in the case of PAX1m+ can be clearly distinguished from PAX1m− even by the naked eye.
TEM analysis was performed to confirm the results of PAX1m+ and PAX1m− cases after salt-induced aggregation of AuNPs (Figure 2C). Bare AuNPs without thiol-labelling were well dispersed (Figure 2C1). AuNPs labeled with the amplified PAX1m+ fragment after salt addition were well dispersed but more sparsely distributed during dilution in the PCR amplification (Figure 2C2). However, the thiol-labeled AuNPs for the PAX1m− sample were aggregated (Figure 2C3) because of the lack of the amplified PCR products.
Quantification of methylation by thiol-labeled AuNPs
The gDNA of PAX1m+ and PAX1m− was mixed in different ratios and analyzed at the PAX1 promoter with methylation-specific primers through UV–vis spectroscopy after 16 cycles. Figure 3A shows that the value of A650/A520 decreased with increasing amount of input methylated DNA in the mixture (with a fixed total DNA concentration). The corresponding relationship between DNA methylated rates with the value of A650/A520 was calculated by curve fitting. The relationship is
Y = 1.45−0.03 * X − 1.22 * X2, | (1) |
where X represents the methylation rate, and Y represents the value of A650/A520 in the UV–vis spectra. A standard curve was created to quantify and compare the methylation status of unknown samples.
Changes in A650/A520 in various percentages of mixed gDNA of PAX1m+ as the time increased within 6 minutes were recorded (Figure 3B). After the addition of NaCl, the A650/A520 in every group started to increase, which indicated the aggregation of AuNPs in the colloid solution. However, mixing of large amounts of methylated DNAs resulted in rapid increase in A650/A520. Finally, the value of A650/A520 remained stable after 4 minutes. The instability of the nanoparticles could be due to poor protection of ssDNA molecules coated on the nanoparticle surface.
The color of the AuNP colloid solutions of every group was recorded (Figure 3C). The color of the AuNP colloid solution remained red in 100% PAX1m+ but turned gray in 0% PAX1m+. The mixing of high amounts of methylated DNA led to a red AuNP colloid solution after 6 minutes.
Quantitative detection of methylation status of PAX1 in cervical scrapings by using thiol-labeled AuNPs
Cervical scrapings were obtained from 42 patients who underwent colposcopic cervical biopsy in Xiangya Hospital to validate the clinical application of the proposed method for methylation detection. Methylation status was detected by the thiol-labeled AuNP method. The value of A620/A520 of the UV–vis spectrum was recorded using the amplified DNA fragment of the PAX1 gene in cervical scraping samples. PMR was calculated by Equation (1). The PMR for the PAX1 gene significantly increased with worsening cervical lesions (Figure 4D). Table 1 lists the PMR for each disease category. The median PMR for PAX1 was 45.14%, 74.64%, 67.81%, and 82.64% in CIN1, CIN2, CIN3, and SCC, respectively. These values are significantly higher than that in normal controls (33.11%). Moreover, the PMR calculated by A620/A520 can significantly distinguish CIN2+ and CIN1- (including CIN1 and normal cervix). In the detection of CIN2+, CIN3+, and SCC lesions, the AUCs of PAX1 methylation detected by UV–vis spectroscopy of AuNPs were 0.833, 0.742, and 0.739, respectively (Figure 4B). The cutoff values of PMR for detection of CIN2+ and CIN3+ were both 31.27%, whereas that of SCC was 75.20% (Table 2). At a cutoff value of 31.27%, the sensitivity and specificity of PAX1 for detection of CIN2+ were 0.941 and 0.600, respectively.
Table 3 shows the distribution of PMR in terms of different clinical and pathological characteristics. The methylation of the PAX1 gene was not associated with patients’ age, cytology results, or HPV.
Comparison of the novel method and conventional qMSP for quantitative detection of PAX1 methylation
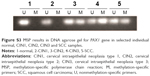
The methylation status of this region of PAX1 gene was verified in a small scale of individual clinical samples by methylation-specific polymerase chain reaction (MSP). The results of agarose gel electrophoresis (Figure S3) showed that methylation of PAX1 gene occurred in CIN2, CIN3, and SCC, while it did not occur in normal and CIN1 samples.
Furthermore, the methylation level ΔCp of the PAX1 gene was detected by qMSP as controls to validate the accuracy of the proposed method. The results are similar to those obtained using AuNP-labeled method. The methylation status of PAX1 increased along with cervical cancer severity (Figure 4C). Table 1 lists the methylation level ΔCp in each disease category. The median ΔCp values for PAX1 were 14.9, 8.9, 4.2, and 3.9 in CIN1, CIN2, CIN3, and SCC, respectively, which are significantly lower than that in the normal controls (15.0).
The AUC values of PAX1 methylation detected by qMSP were 0.807, 0.844, and 0.795 for CIN2+, CIN3+, and SCC lesions, respectively (Figure 4A). The cutoff value for the detection of CIN2+ was 10.57, whereas that of CIN3+ and SCC was 5.65 and 5.60, respectively (Table 2). At a cutoff value of 10.57, the sensitivity and specificity of PAX1 for the detection of CIN2+ were 0.824 and 0.840, respectively.
We adopted 10.57 as the end-point of ΔCp for qMSP and 31.27% the end-point of PMR for AuNP-labeled method because of the same cutoff value for CIN3+ and CIN2+. Methylated frequency was calculated using different clinical and pathological characteristics (Table 3). In accordance with the degrees of cervical cytology results, the samples were divided into atypical squamous cells of undetermined significance (ASCUS), high-grade ASCUS (ASCUS-H), low-grade squamous intraepithelial lesion (LSIL), high-grade squamous intraepithelial lesion (HSIL), cervical SCC, and healthy controls. The methylated percentages detected by AuNP-labeled method in ASCUS, ASCUS-H, LSIL, HSIL, and SCC were 41.67%, 62.50%, 71.43%, 80%, and 100%, respectively. The methylated percentages detected by qMSP in ASCUS, ASCUS-H, LSIL, HSIL, and SCC were 25%, 37.50%, 57.14%, 50%, and 100%, respectively. The methylated percentages detected by AuNP-labeled method in normal, CIN1, CIN2, CIN3, and SCC were 16.67%, 15.38%, 57.14%, 100%, and 100%, respectively. The methylated percentages detected by qMSP in normal, CIN1, CIN2, CIN3, and SCC were 33.33%, 46.15%, 85.71%, 100%, and 100%, respectively. The results of the two methods showed that the percentage of methylation of the PAX1 gene is higher in HSIL or SCC than that in minor cervical lesions, such as ASCUS, ASCUS-H, and LSIL.
The distribution of methylation level ΔCp in different clinical and pathological characteristics is shown in Table 3. The methylation status of the PAX1 gene was not associated with patients’ age, cytology, and HPV. The results of both methods showed that the methylated frequency of the PAX1 gene was significantly associated with pathological features.
Discussion
DNA methylation could be a suitable biomarker for the detection of cervical cancer. The methylation of the PAX1 gene showed clinical potential as a biomarker in cervical screening. The PAX1 gene is a tumor suppressor belonging to the PAX family.33 The PAX gene family is classified into four classes. PAX genes of class 1 and 3 play an essential role in maintaining tissue-specific stem cells and are overexpressed in the development of several cancers. However, members of class 4 genes are less overexpressed in cancers. The PAX1 proteins are transcription factors important in developmental processes, such as self-renewal in embryogenesis. PAX1 participates in some events in carcinogenesis with normal function in the development of the skeleton, thymus, and parathyroid glands. In the development of cervical cancer, the PAX1 gene is silenced by hypermethylation. The methylation of PAX1 showed a great clinical potential as a biomarker in cervical cancer screening. In the present study, we used the thiol-labeled AuNP method to quantitatively analyze the methylation level of the PAX1 gene in cervical scrapings obtained from 42 patients. The results showed that the methylated frequency as well as the PMR of the PAX1 gene increased with worsening cervical cancer. The sensitivity of PAX1 is higher than 90% for the detection of CIN2+, CIN3+, and SCC; the corresponding AUC values are higher than 0.700.
Cervical scraping could be a suitable target for testing for cervical neoplasia. Assessment of DNA methylation in cervical scrapings, instead of tissues, could be effectively used for clinical cervical screening. In previous studies, cervical scraping was used to detect cancer-specific gene methylation.34 Detection of methylation in cervical scrapings combined with HPV test may improve the unsatisfactory sensitivity for CIN3+ detection in cervical cancer.35 This study is the first to apply novel nanotechnology for the detection of methylation in cervical scrapings, with high efficiency. As to the methylated gene in cervical tissues, the negative control groups were normal-appearing adjacent tissues but not in the remote tissues. Moreover, collecting cervical scrapings is more convenient than obtaining cancer tissues from patients.
In addition to the novel method using thiol-labeled PCR primers with AuNPs, we used conventional qMSP to detect methylation status of the PAX1 gene in the same cervical scraping samples. Comparatively, conventional qMSP showed detection results consistent with that of the AuNP-labeled method. The proposed method showed excellent feasibility and is superior to conventional qMSP in several aspects. The proposed method is easier to operate and less expensive than traditional methods, such as qMSP or pyrosequencing. The proposed method is faster, consuming 6 minutes from beginning to finish, and utilizes only 16 PCR cycles, which is lower than the 50 cycles required by qMSP. This condition will greatly reduce the probability of base mismatches during the PCR reaction and greatly improve the accuracy of PCR. Moreover, ROC curves used to evaluate specificity and sensitivity of CIN2+ displayed higher AUC values with AuNP-labeled method (0.833) than those with conventional qMSP method (0.807). The sensitivity for the detection of CIN2+, CIN3+, and SCC by AuNP-labeled method is all higher than that by conventional qMSP method. Specially, the sensitivity for the detection of CIN3+ and SCC by AuNP-labeled method both reached 100%.
The high sensitivity of the novel method is mainly attributed to the excellent optical characteristics of AuNPs. In a previous study,36 AuNPs showed large surface area, good conductivity, and excellent porosity, which led to a high sensitivity in DNA detection by colorimetric assay and UV–vis spectroscopy. In previous studies, the excellent optical performance was characterized through detection of the activity of DNA adenine methylation (Dam) methyltransferase (MTase) and endonuclease Dpn I by using DNA-modified AuNPs coupled with enzyme-linkage reactions. As such, the detection limit of endonuclease is 0.3 U/mL, which is lower than that in previous reports. Detection was performed within 500 seconds, which is faster than that in other methods. Moreover, another research of colorimetric detection method to identify PCR-amplified nucleic acids by thiol-labeled PCR primers and AuNPs was performed for the detection of the disease causing bacterium Chlamydia trachomatis in human urine sample.37 As few as 100 copies of the target template can be detected by observing the color of the AuNP colloid solution because of the advantages of AuNPs. Also, the latest research that applied upconversion nanoparticles to the detection of CDKN2A methylation also showed more sensitivity than that by either qPCR or pyrosequencing.38
The proposed quantitative method is mainly based on the relationship between the ratio of absorbance A620/A520 of AuNPs and methylated ratio of PAX1 gene. The more methylated DNAs were mixed in the template, the more thiol-labeled DNAs were amplified. Then, an increasing resistance to salt-induced aggregation of AuNPs was caused by negative charges on the AuNP surface in the presence of thiol-labeled DNA. The absorbance of 520 nm represents the original state of AuNPs before aggregation, whereas the absorbance of 620 nm represents the salt-induced aggregation of AuNPs. Consequently, the value of A620/A520 could represent the degree of the salt-induced aggregation of AuNPs, which was consistent with the degree of color change of AuNP colloid solution. These results were in good agreement with the previous reports. For instance, in the colorimetric detection of PCR-amplified nucleic acids for the disease causing bacterium C. trachomatis in human urine sample, the results37 also showed that the degree of color change of AuNP colloid solution and ratio of absorbance A522/A700 of AuNPs are linearly dependent on the concentration of thiol-labeled-amplified DNA. The results36 of the detection of Dam MTase by AuNPs coupled with enzyme-linkage reactions showed that with the increase of the inhibitor concentration, the velocity of methylation decreased and the ratio of absorbance A620/A520 increased slowly.
Apart from the direct colorimetric methods of AuNPs and UV–vis spectroscopy, many studies have demonstrated that AuNPs exhibit various optical properties for the detection of DNA in cancer therapy.39–41 AuNPs hold great promises in the detection of cancer because of their good biocompatibility, easy synthesis and functionalization, chemo-physical stability, and optical tunable characteristics. AuNPs, such as gold nanorods or nanoshells,42 have optical properties of light absorbance and scattering in near-infrared (NIR) wavelengths (650–900 nm).43 The therapy with NIR laser and AuNPs was effective in cancer cell lines in vitro.44,45 AuNPs could act as contrast agents by microscopy for the diagnosis for cancer. For instance, two-photon-induced photoluminescence could be used to visualize tumor cells embedded with AuNPs in vivo.46
Conclusion
This study proposes a novel and improved method for colorimetric detection of the methylation of the PAX1 gene by using thiol-labeled MSP primers and unmodified AuNPs. To our knowledge, this study is the first to employ thiol-labeled PCR primers with AuNPs for colorimetric detection of DNA methylation. On the basis of the colorimetric assay and ratios of absorbance A620/A520, the quantitative detection of methylation level of the PAX1 gene was performed using cervical scrapings obtained from 42 patients in various cervical neoplasias. Methylation level at a cutoff value of 31.27% detected by the method proposed in this study could be a prognostic and diagnostic tool for the screening of cervical cancer. However, prospective population-based studies are still necessary for further implementation of this novel method for the detection of methylation for the PAX1 gene. The results were further verified by conventional qMSP method. Both two methods demonstrated that the methylation status and frequencies of PAX1 increased along with disease severity in cervical cancer. Compared with conventional qMSP method, the novel method proposed in this study possessed several significant advantages in the quantitative detection of DNA methylation, including more effective, less time consuming, superior sensitivity, and less probability of base mismatches during the PCR process, greatly improving the accuracy of PCR results. Therefore, the thiol-labeled AuNP method could be a substitute for conventional qMSP in clinical applications. This simple colorimetric method exhibits potential to be a novel and effective method for early cervical cancer screening and other clinical research.
Acknowledgment
This study was supported by the National Natural Science Foundation of China (No 81503563).
Disclosure
The authors report no conflicts of interest in this work.
References
Christopherson WM, Parker JE, Mendez WM, Lundin FE. Cervix cancer death rates and mass cytologic screening. Cancer. 1970;26(4):808–811. | ||
Liu S, Semenciw R, Probert A, Mao Y. Cervical cancer in Canada: changing patterns in incidence and mortality. Int J Gynecol Cancer. 2001;11(1):24–31. | ||
Forouzanfar MH, Foreman KJ, Delossantos AM, et al. Breast and cervical cancer in 187 countries between 1980 and 2010: a systematic analysis. Lancet. 2011;378(9801):1461–1484. | ||
Jemal A, Siegel R, Naishadham D. Cancer statistics, 2012. CA Cancer J Clin. 2012;62(1):10–29. | ||
Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61(4):212–236. | ||
Kun-He WU. Comparison of the liquid-based ThinPrep cytologic screening and biopsy in cervical lesions. Journal of Modern Oncology. 2005;13(2):191–193. | ||
Fei Z, Xiang-yu Z, Rui-ning Y, Laboratory Do, PLA Ho. The value of diagnosing cervical lesions by Thinprep cytologic test(TCT) combined with high-risk type HPV DNA testing. Military Medical Journal of Southeast China. 2016;18(1):28–31. | ||
Zhao YQ. Value of thinprep cytologic test and human papillomavirus test in diagnosis of cervical cancer. Journal of Regional Anatomy & Operative Surgery. 2011;20(6):638–639. | ||
Cao J. The clinical significance of thinprep cytologic test and HPV-PCR quantitative analysis in the diagnosis of cervical cancer and precancerous lesions. China Mod Doct. 2015;36(6):S234–S240. | ||
Teodoridis JM, Strathdee G, Brown R. Epigenetic silencing mediated by CpG island methylation: potential as a therapeutic target and as a biomarker. Drug Resist Updat. 2004;7(4–5):267–278. | ||
Keshet I, Schlesinger Y, Farkash S, et al. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat Genet. 2006;38(2):149–153. | ||
Ellis L, Atadja PW, Johnstone RW. Epigenetics in cancer: targeting chromatin modifications. Mol Cancer Ther. 2009;8(6):1409–1420. | ||
Hildebrandt MA, Gu J, Lin J, et al. Hsa-miR-9 methylation status is associated with cancer development and metastatic recurrence in patients with clear cell renal cell carcinoma. Oncogene. 2010;29(42):5724–5728. | ||
Hayashi M, Wu G, Roh JL, et al. Correlation of gene methylation in surgical margin imprints with locoregional recurrence in head and neck squamous cell carcinoma. Cancer. 2015;121(12):1957–1965. | ||
Chela-Flores J, Migoni RL. CG methylation in DNA transcription. Int J Theor Phys. 1990;29(8):853–862. | ||
Kan YY, Liou YL, Wang HJ, et al. PAX1 methylation as a potential biomarker for cervical cancer screening. Int J Gynecol Cancer. 2014;24(5):928–934. | ||
Lai HC, Ou YC, Chen TC, et al. PAX1/SOX1 DNA methylation and cervical neoplasia detection: a taiwanese gynecologic oncology group (TGOG) study. Cancer Med. 2014;3(4):1062–1074. | ||
Chang CC, Huang RL, Wang HC, Liao YP, Yu MH, Lai HC. High methylation rate of LMX1A, NKX6-1, PAX1, PTPRR, SOX1, and ZNF582 genes in cervical adenocarcinoma. Int J Gynecol Cancer. 2014;24(2):201–209. | ||
Huang RL, Chang CC, Su PH, et al. Methylomic analysis identifies frequent DNA methylation of zinc finger protein 582 (ZNF582) in cervical neoplasms. PLoS One. 2012;7(7):e41060. | ||
Lai HC, Lin YW, Huang RL, et al. Quantitative DNA methylation analysis detects cervical intraepithelial neoplasms type 3 and worse. Cancer. 2010;116(18):4266–4274. | ||
Zhang Y, Liou YLL, Chang CF, Zhang Y, Zhou H. The combination of pax1 methylation gene with oncogenic HPV typing is a new molecular pap smear for cervical cancer detection in China. Asia Pac J Clin Oncol. 2014;10:210–211. | ||
Hu J, Zhang CY. Single base extension reaction-based surface enhanced Raman spectroscopy for DNA methylation assay. Biosens Bioelectron. 2011;31(1):451–457. | ||
Li X, Song T, Guo X. DNA methylation detection with end-to-end nanorod assembly-enhanced surface plasmon resonance. Analyst. 2015;140(18):6230–6233. | ||
Bailey VJ, Keeley BP, Razavi CR, Griffiths E, Carraway HE, Wang TH. DNA methylation detection using MS-qFRET, a quantum dot-based nanoassay. Methods. 2010;52(3):237–241. | ||
Dadmehr M, Hosseini M, Hosseinkhani S, et al. DNA methylation detection by a novel fluorimetric nanobiosensor for early cancer diagnosis. Biosens Bioelectron. 2014;60(30):35–44. | ||
Li H, Rothberg L. Colorimetric detection of DNA sequences based on electrostatic interactions with unmodified gold nanoparticles. Proc Natl Acad Sci U S A. 2004;101(39):14036–14039. | ||
Rashid JIA, Yusof NA, Abdullah J, Hashim U. A novel disposable biosensor based on SINWs/AUNPs modified-screen printed electrode for dengue virus DNA oligomer detection. IEEE Sensors J. 2015;15(8):4420–4427. | ||
Jing X, Cao X, Wang L, Lan T, Li Y, Xie G. DNA-AuNPs based signal amplification for highly sensitive detection of DNA methylation, methyltransferase activity and inhibitor screening. Biosens Bioelectron. 2014;58:40–47. | ||
Zou B, Cao X, Wu H, et al. Sensitive and specific colorimetric DNA detection by invasive reaction coupled with nicking endonuclease-assisted nanoparticles amplification. Biosens Bioelectron. 2015;66:50–54. | ||
Chen K, Zhang M, Chang YN, et al. Utilizing Gold Nanoparticle Probes to Visually Detect DNA Methylation. Nanoscale Res Lett. 2016;11(1):304. | ||
Liou YL, Zhang Y, Liu Y, et al. Comparison of HPV genotyping and methylated ZNF582 as triage for women with equivocal liquid-based cytology results. Clin Epigenetics. 2014;7(1):1–9. | ||
Parashar G, Capalash N. Promoter methylation-independent reactivation of PAX1 by curcumin and resveratrol is mediated by UHRF1. Clin Exp Med. 2016;16(3):471–478. | ||
Paixão-Côrtes VR, Salzano FM, Bortolini MC. Origins and evolvability of the PAX family. Semin Cell Dev Biol. 2015;44:64–74. | ||
Reesink PN, Jeronimo CY, Klip HG, et al. Detecting cervical cancer by quantitative promoter hypermethylation assay on cervical scrapings: a feasibility study. Mol Cancer Res. 2004;2(5):289–295. | ||
Snijders PJ, Hogewoning CJ, Hesselink AT, et al. Determination of viral load thresholds in cervical scrapings to rule out CIN 3 in HPV 16, 18, 31 and 33-positive women with normal cytology. Int J Cancer. 2006;119(5):1102–1107. | ||
Liu T, Zhao J, Zhang D, Li G, Chem A. Novel method to detect DNA methylation using gold nanoparticles coupled with enzyme-linkage reactions. Anal Chem. 2010;82(1):229–233. | ||
Jung YL, Jung C, Parab H, Li T, Park HG. Direct colorimetric diagnosis of pathogen infections by utilizing thiol-labeled PCR primers and unmodified gold nanoparticles. Biosens Bioelectron. 2010;25(8):1941–1946. | ||
Kim S, Hwang SH, Im SG, et al. Upconversion nanoparticle-based förster resonance energy transfer for detecting DNA methylation. Sensors (Basel). 2016;16(8):pii:E1259. | ||
Bucharskaya A, Maslyakova G, Terentyuk G, et al. Towards effective photothermal/photodynamic treatment using plasmonic gold nanoparticles. Int J Mol Sci. 2016;17(8):E1295. | ||
Tian L, Lu L, Qiao Y, Ravi S, Salatan F, Melancon MP. Stimuli-responsive gold nanoparticles for cancer diagnosis and therapy. J Funct Biomater. 2016;7(3):E19. | ||
Ahmad R, Fu J, He N, Li S. Advanced gold nanomaterials for photothermal therapy of cancer. Journal of Nanoscience and Nanotechnology. 2016;16(1):67–80. | ||
Loo C, Lowery A, Halas N, West J, Drezek R. Immunotargeted nanoshells for integrated cancer imaging and therapy. Nano Lett. 2005;5(4):709–711. | ||
Hirsch LR, Stafford RJ, Bankson JA, et al. Nanoshell-mediated near-infrared thermal therapy of tumors under magnetic resonance guidance. Proc Nat Acad Sci U S A. 2003;100(23):13549–13554. | ||
Loo C, Lin A, Hirsch L, et al. Nanoshell-enabled photonics-based imaging and therapy of cancer. Technol Cancer Res Treat. 2004;3(1):33–40. | ||
Lee J, Chatterjee DK, Lee MH, Krishnan S. Gold nanoparticles in breast cancer treatment: promise and potential pitfalls. Cancer Lett. 2014;347(1):46–53. | ||
Park J, Estrada A, Sharp K, et al. Two-photon-induced photoluminescence imaging of tumors using near-infrared excited gold nanoshells. Opt Express. 2008;16(3):1590–1599. |
Supplementary materials
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.