Back to Journals » OncoTargets and Therapy » Volume 12
RASSF1A promoter methylation correlates development, progression, and poor cancer-specific survival of renal cell carcinoma: trial sequential analysis
Authors Zhuang Q, Chen Z, Shen J, Fan M, Xue D, Lu H, Xu R, He X
Received 8 August 2018
Accepted for publication 1 November 2018
Published 20 December 2018 Volume 2019:12 Pages 119—134
DOI https://doi.org/10.2147/OTT.S183142
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Leo Jen-Liang Su
Qianfeng Zhuang,* Zhen Chen,* Jie Shen,* Min Fan, Dong Xue, Hao Lu, Renfang Xu, Xiaozhou He
Department of Urology, The Third Affiliated Hospital of Soochow University, Changzhou 213003, China
*These authors contributed equally to this work
Background: This meta-analysis evaluated the clinicopathologic and prognostic significance of RASSF1A promoter methylation in renal cell carcinoma (RCC).
Materials and methods: The ORs or HRs and their 95% CIs were calculated. Trial sequential analysis was conducted.
Results: Twenty-two articles that included 1,421 patients with RCC and 724 controls were identified. RASSF1A promoter methylation correlated with RCC in tissue, blood, and urine samples. On multivariate analysis, RASSF1A promoter methylation was associated with tumor grade (grade 3–4 vs 1–2: OR=3.59), clinical stage (stage 3–4 vs 1–2: OR=2.15), T classification (pT2–4 vs pT1: OR=2.66), histologic subtypes (papillary vs clear cell: OR=2.91), and cancer-specific survival (HR=1.78), but it was not linked to age, gender, lymph node status, distant metastasis, or overall survival. The Cancer Genome Atlas data also showed that RASSF1A methylation was significantly more likely to be seen in papillary vs clear-cell RCC (OR=23.19).
Conclusion: RASSF1A promoter methylation may be associated with the development and progression of RCC, as well as poor cancer-specific survival. Methylation was more frequent in papillary vs clear-cell RCC. More studies are needed to confirm these findings in blood or urine samples.
Keywords: RAS association domain family protein 1A, methylation, survival, clinical features
Introduction
Renal cell carcinoma (RCC) is the most common malignant tumor affecting the kidneys, accounting for about 90% of kidney carcinomas.1 Approximately 63,990 new RCC cases were diagnosed in the USA in 2017, and these were associated with an estimated 14,400 deaths.2 There are two common histological subtypes of RCC. Clear-cell RCC (ccRCC) is the most common, accounting for 70%–80% of all renal cancer cases. Papillary RCC (pRCC) represents another 10%–20% of cases.3,4 Approximately 25%–30% of patients with RCC present with advanced or metastatic disease, and the 5-year survival rate is poor.5
DNA methylation within the promoter regions is an important mechanism underlying epigenetic modifications, which may cause inactivation of gene expression and play a crucial role in the carcinogenesis, progression, and prognosis of various human cancers.6–8 Previous studies have suggested that promoter methylation of some cancer-related genes is found in RCC, such as HOXB139 and CDKN2A/2B.10 RASSF1A is a key isoform of RASSF1 located on the chromosomal region 3p21.3.11 An important tumor suppressor gene, RASSF1A is involved in cell cycle regulation, microtubule stabilization, cellular adhesion and motility, and cell apoptosis.12–14
RASSF1A promoter methylation has been reported in tissue, blood, and urine samples from patients with RCC.15–17 There are, however, inconsistent results regarding the level of RASSF1A promoter methylation in patients with RCC and controls. For example, Ellinger et al reported that the RASSF1A promoter had a similar methylation rate in RCC and adjacent normal tissue samples.18 In contrast, RASSF1A promoter methylation was more frequent in RCC than in adjacent normal tissue samples in a study by Loginov et al.19 With this background of conflicting results, we conducted a meta-analysis to assess differences in RASSF1A promoter methylation between RCC and control tissue, blood, and urine samples. Moreover, we evaluated the association of RASSF1A promoter methylation with clinicopathologic features and prognosis in patients with RCC.
Materials and methods
Search strategy
A systematic literature search of the PubMed, EMBASE, EBSCO, Wanfang, and CNKI databases was conducted to identify eligible studies published through December 1, 2017, without any language restrictions. The following keywords and scientific search terms were used: (kidney OR renal) AND (cancer OR tumor OR neoplasm OR carcinoma) AND (methylation OR methylated OR hypermethylation OR epigene*) AND (RAS association domain family protein 1A OR RASSF1A OR RASSF1 OR RAS association domain family protein 1). We manually searched the relevant references from all eligible articles to find other potential publications.
Selection criteria
Articles that met the following inclusion criteria were selected for the meta-analysis: 1) patients were confirmed with adult RCC by histopathologic examination; 2) studies reported sufficient data to evaluate differences in RASSF1A promoter methylation between the RCC and control groups; 3) studies had sufficient data to assess the correlation of RASSF1A promoter methylation with clinicopathologic features; and 4) studies provided enough survival data to evaluate the prognostic effect of RASSF1A promoter methylation in RCC. When multiple papers using the same patient population were published, the study with more information was included in the meta-analysis.
Data extraction
The following information was extracted from the included publications: surname of the first author, year of publication, country, ethnic population, cancer stage, mean or median age, sample type, detection method, histologic type, number of cases and controls, survival data with multivariate analysis, and clinicopathologic features such as age (≥50 vs <50 years), gender (male vs female), tumor grade (3–4 vs 1–2), clinical stage (3–4 vs 1–2), T classification (pT2–4 vs pT1), histologic subtypes (pRCC vs ccRCC), lymph node metastasis (yes vs no), and distant metastasis (yes vs no).
The Cancer Genome Atlas (TCGA) dataset
Clinical information for RCC, which included two sets of samples (methylation 450 K dataset: 275 pRCCs and 319 ccRCCs), was downloaded from the TCGA data portal (https://portal.gdc.cancer.gov/repository). The cutoff value of RASSF1A promoter methylation was set by its median value. The association between clinicopathologic characteristics and RASSF1A promoter methylation was analyzed using logistic regression (R; v.3.4.3). Multivariate Cox analysis was used to analyze the impact of RASSF1A promoter methylation on overall survival (R; v.3.4.3).
Statistical analysis
All data analyses were performed using Stata software 12.0 (StataCorp LP, College Station, TX, USA). Differences in RASSF1A promoter methylation between RCC and control samples and the correlation of RASSF1A promoter methylation with the clinicopathologic characteristics of patients with RCC were calculated using pooled ORs and the corresponding 95% CIs. Overall HRs with their 95% CIs were also calculated to determine the prognostic role of RASSF1A promoter methylation, using multivariate analysis if possible. The Cochran’s Q statistic was used to estimate possible heterogeneity among studies.20,21 A random-effects model was applied in the meta-analysis. When substantial heterogeneity was measured (P<0.1), a sensitivity analysis was carried out to determine the influence of an individual study on the pooled OR and heterogeneity by deleting one study at a time.22,23 For results covered by more than nine studies, possible publication bias was detected with Egger’s test.24 We performed trial sequential meta-analyses (TSA) to reduce type I error and to calculate the estimated required sample size information.25,26 For significant results with more than one study, the type I error rate was set at 5% and the type II error rate was considered to be 20% (a statistical test power of 80%). The relative risk reduction was set at 20% in the meta-analysis. If the cumulative Z-curve crossed the trial sequential monitoring boundary or the required information size, the statistical evidence was deemed conclusive. Otherwise, additional studies would be needed for a definitive result.27,28
Results
Study characteristics
Figure 1 summarizes the details of the study selection procedure; 22 publications with a total of 1,421 patients with RCC and 724 controls fulfilled the inclusion criteria and were selected for the meta-analysis.15–19,29–45 Of the included publications, 15 assessed differences in RASSF1A promoter methylation between RCC and control samples using tissue samples and 6 used blood or urine samples. Sixteen studies evaluated the relationships between RASSF1A promoter methylation and the clinicopathologic characteristics of patients with RCC. Two studies reported information on survival in patients with RCC using multivariate analysis. The baseline characteristics of the included publications are presented in Tables 1 and S1.
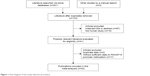  | Figure 1 Flow diagram of the study selection procedure. |
Correlation between RASSF1A promoter methylation and RCC in cancer vs control tissue samples
In 15 studies that included the comparison of 829 patients with RCC and 467 adjacent/normal tissue samples (Figure 2), RASSF1A promoter methylation was notably higher in RCC than in adjacent/normal tissue samples (OR=5.64, 95% CI=1.82–17.51, P=0.003).
Subgroup and sensitivity analyses in cancer vs control tissue samples
We conducted subgroup analyses by ethnicity (Asians and Caucasians) and testing method (methylation-specific polymerase chain reaction [MSP] and non-MSP) (Table 2). In the ethnicity analysis, RASSF1A promoter methylation was associated with RCC in Caucasians (OR=4.90, 95% CI=1.45–16.64, P=0.011), but not in Asians (OR=9.27, 95% CI=0.35–243.63, P=0.182). In the testing method analysis, RASSF1A promoter methylation was associated with RCC in the MSP subgroup (OR=16.32, 95% CI=5.25–50.69, P<0.001), but not in the non-MSP subgroup (OR=1.85, 95% CI=0.27–12.48, P=0.527).
  | Table 2 Subgroup analyses of RASSF1A promoter methylation in cancer vs control tissue samples |
There was evidence of significant heterogeneity in cancer vs control tissue samples, so we performed a sensitivity analysis. We successively removed four studies – Tokinaga et al38 in Japan, Costa et al33 in Portugal, Gonzalgo et al36 in the USA, and Onay et al32 in Turkey – and recalculated the overall OR (OR=19.78, 95% CI=11.09–35.29, P<0.001), resulting in no heterogeneity (P=0.693).
Correlation between RASSF1A promoter methylation and RCC in cancer vs control blood or urine samples
In four studies that included the comparison of 237 patients with RCC with 142 nonmalignant blood samples, RASSF1A promoter methylation was significantly more likely in RCC than in nonmalignant blood samples (OR=11.70, 95% CI=4.82–28.39, P<0.001; Figure 2). In addition, in a comparison of 76 RCCs and 115 nonmalignant urine samples, RASSF1A promoter methylation was significantly higher in RCC than in nonmalignant urine samples (OR=17.54, 95% CI=6.60–46.66, P<0.001; Figure 2).
Correlation of RASSF1A promoter methylation with age and gender in RCC
Seven studies that included 321 patients with RCC demonstrated that RASSF1A promoter methylation was not correlated with age (OR=1.00, 95% CI=0.50–2.04, P=0.99; Figure 3). Eight studies that included 537 patients with RCC showed that RASSF1A promoter methylation was not correlated with gender (OR=0.95, 95% CI=0.51–1.76, P=0.86; Figure 3).
Correlation of RASSF1A promoter methylation with lymph node status and distant metastasis in RCC
In seven studies that included 438 patients with RCC, RASSF1A promoter methylation was not associated with lymph node metastasis (OR=1.72, 95% CI=0.76–3.87, P=0.192; Figure 4). Four studies that included 257 patients with RCC showed that there was no correlation between RASSF1A promoter methylation and distant metastasis (OR=1.66, 95% CI=0.75–3.69, P=0.21; Figure 4).
Correlation of RASSF1A promoter methylation with tumor grade and clinical stage in RCC
In 13 studies that included 686 patients with RCC, a significant relationship was observed between RASSF1A promoter methylation and tumor grade (OR=3.59, 95% CI=1.85–6.95, P<0.001; Figure 5). RASSF1A promoter methylation was also linked to clinical stage in eight studies that included 463 patients with RCC (OR=2.15, 95% CI=1.34–3.45, P=0.001; Figure 5).
Correlation of RASSF1A promoter methylation with T classification and histologic subtypes in RCC
In seven studies that included 306 patients with RCC, a significant correlation was found between RASSF1A promoter methylation and T classification (OR=2.66, 95% CI=1.11–6.39, P=0.029; Figure 6). RASSF1A promoter methylation was also significantly associated with histologic subtypes in eight studies that included 472 patients with RCC (OR=2.91, 95% CI=1.61–5.23, P<0.001; Figure 6).
Prognostic role of RASSF1A promoter methylation using multivariate analysis
Kawai et al reported that RASSF1A promoter methylation was a poor prognostic factor in terms of cancer-specific survival among 179 patients with ccRCC (HR=1.78, 95% CI=1.18–2.78).31 Klacz et al reported that RASSF1A promoter methylation was not associated with overall survival using multivariate analysis among 58 patients with ccRCC.16 More studies with large patient population are needed to further investigate the prognostic role of RASSF1A promoter methylation in RCC.
Publication bias
There was no evidence of publication bias using Egger’s test for the comparison of RCC vs control tissue samples (P=0.782) or in relation to tumor grade (P=0.547; Figure 7).
  | Figure 7 Forest plot of potential publication bias using Egger’s test in RCC vs control tissue samples (P=0.782) and in relation to tumor grade (P=0.547). |
Trial sequential meta-analysis
As shown in Figures 8 and 9, based on the a priori anticipated information size method for significant results, when cancer was compared with control tissue samples, the cumulative Z-curve crossed the trial sequential monitoring boundary and the required information size (Figure 8), suggesting conclusive results. When cancer was compared with control blood or urine samples, the cumulative Z-curve was more than the conventional boundary, but did not cross the trial sequential monitoring boundary (Figure 8), which suggests that more studies are needed to inform these two results. In relation to tumor grade, clinical stage, and histologic subtypes, the cumulative Z-curve was more than the trial sequential monitoring boundary (Figures 8 and 9), which suggests that additional studies are not necessary. In relation to T classification, the cumulative Z-curve crossed the conventional boundary, but did not cross the trial sequential monitoring boundary (Figure 8), suggesting that further studies are essential.
  | Figure 9 Trial sequential analysis assessing the required sample information in relation to tumor histology. |
TCGA dataset
After adjusting for tumor stage (stage 3–4 vs stage 1–2) and tumor histology (pRCC vs ccRCC), RASSF1A promoter methylation was not associated with overall survival using multivariate analysis (HR=0.921, P=0.687) in 567 RCCs.
RASSF1A promoter methylation was not significantly linked to gender (594 patients: OR=1.35, 95% CI=0.95–1.91, P=0.094), but it did correlate with clinical stage (568 patients: P=0.023) and tumor histology (594 patients: pRCC vs ccRCC: OR=23.19, 95% CI=15.07–35.7, P<0.001; Table 3).
Discussion
Tumor suppressor genes are commonly inactivated via promoter methylation within the CpG islands, which may affect several biological processes, including cell proliferation, cell death, cell migration, and cell invasion, and contribute to the initiation and progression of human cancers.46,47 Studies have indicated that methylation of the promoter of the tumor suppressor gene RASSF1A reduces its expression, which may play an important role in RCC carcinogenesis.40,43 However, potential differences in methylation between RCC and control tissue samples have remained unclear owing to conflicting evidence from previous studies. Two studies showed that RASSF1A promoter methylation correlated negatively with RCC.33,38 Four studies reported no association between RASSF1A promoter methylation and RCC.18,32,34,36 Also, nine other studies showed that RASSF1A promoter methylation correlated positively with RCC.19,37,39–45 The present meta-analysis including all eligible publications with large patient populations demonstrated that RASSF1A promoter methylation was notably higher in RCC than in adjacent or normal tissue samples; TSA revealed that the result was conclusive. This suggests that RASSF1A promoter methylation is significantly associated with RCC carcinogenesis.
When RCC was compared with control tissue samples, a subgroup analysis of ethnicity showed that RASSF1A promoter methylation was associated with RCC in Caucasians, but not in Asians, which suggests that only the Caucasian population is susceptible to RASSF1A promoter methylation. A subgroup analysis of detection method demonstrated that RASSF1A promoter methylation correlated with RCC in the MSP subgroup, but not in the non-MSP subgroup, which indicates that the MSP method may be sensitive to the detection of RASSF1A promoter methylation. We performed a sensitivity analysis because substantial heterogeneity was measured in the comparison of cancer and control tissue samples. When four studies32,33,36,38 were successively removed and the pooled OR was recalculated, remaining significant, there was no evidence of heterogeneity (P=0.693). It is possible that the main cause of heterogeneity in this meta-analysis was contamination of adjacent normal tissue samples by cancer cells in these four studies. In addition, Egger’s test showed no publication bias. The relevant analyses supported the stability and credibility of our results.
RASSF1A promoter methylation was associated with RCC in blood and urine samples (cancer vs nonmalignant controls), which suggested that RASSF1A promoter methylation may become a promising noninvasive biomarker for the detection of RCC in the future. According to the results of TSA, additional prospective clinical studies with large sample sizes are required to further investigate whether RASSF1A promoter methylation could be used as a biomarker for the diagnosis of RCC based on blood or urine samples.
Finally, we evaluated whether RASSF1A promoter methylation was linked to clinicopathologic characteristics and prognosis in patients with RCC. RASSF1A promoter methylation did not correlate with age, gender, lymph node status, or distant metastasis. Significant relationships were observed between RASSF1A promoter methylation and tumor grade, clinical stage, and T classification, with methylation notably higher in high-grade vs low-grade tumors, advanced vs early-stage patients, and high (pT2–4) vs low (pT1) T classification. These analyses suggest that RASSF1A promoter methylation may be closely associated with RCC progression. TSA showed that additional studies are essential to inform the analyses of T classification, but that the analyses of tumor grade, clinical stage, and tumor histology were robust. Additionally, TCGA data showed that RASSF1A promoter methylation remained significantly associated with pRCC vs ccRCC (OR=23.19, P<0.001), suggesting that it may play a more important role in the pathogenesis of pRCC. On multivariate analysis, RASSF1A promoter methylation was associated with poorer cancer-specific survival among 179 patients with ccRCC patients,31 but did not correlate with overall survival among 58 patients with ccRCC patients.16 Further analysis using TCGA data showed that no correlation was found between RASSF1A promoter methylation and overall survival on multivariate analysis (HR=0.921, P=0.687) in 567 RCCs. More studies using multivariate analysis will be crucial to confirm the prognostic impact of RASSF1A promoter methylation on cancer-specific survival.
The current results compare favorably with the previous meta-analyses by Yu et al48 and Huang et al.49 Yu et al only analyzed whether RASSF1A promoter methylation was correlated with RCC in cancer vs nontumor controls,48 and RASSF1A promoter methylation did not correlate with RCC in tissue samples.48 Our result involving a greater number of eligible studies with a larger population (15 studies with 1,296 tissue samples) showed that RASSF1A promoter methylation was significantly associated with RCC in tissue samples. In addition, Yu et al48 did not report whether RASSF1A promoter methylation was linked to clinical features (eg, gender, tumor grade, clinical stage, T classification, histologic subtypes, lymph node metastasis, and distant metastasis) and did not include an analysis of overall survival. Huang et al only analyzed whether RASSF1A promoter methylation was linked to tumor stage (five studies with 252 cases) and grade (four studies with 190 cases), showing that RASSF1A promoter methylation had a borderline significant correlation with tumor stage (P=0.051) and a significant association with tumor grade (P=0.001).49 Our meta-analysis involving more patients suggested that RASSF1A promoter methylation was significantly linked to tumor grade (13 studies with 686 patients with RCC, P<0.001) and clinical stage (8 studies with 463 patients with RCC, P=0.001). Additionally, Huang et al49 did not analyze whether RASSF1A promoter methylation was associated with prognosis (cancer-specific survival or overall survival) or other clinical features such as gender, T stage, and lymph node status.
The current meta-analysis had several limitations. First, the size of the population with blood or urine samples was small. Second, the populations of the included studies mainly consisted of Asians and Caucasians, with limited numbers of other ethnic subgroups, such as Africans. Third, only two studies reported the prognostic role of RASSF1A promoter methylation using multivariate analysis in RCC.
Conclusion
The present findings show that RASSF1A promoter methylation correlates with RCC in tissue, blood, and urine samples. RASSF1A promoter methylation is not linked to age, gender, lymph node status, distant metastasis, or overall survival, but it is associated with tumor grade, clinical stage, T classification, histologic subtypes, and cancer-specific survival on multivariate analysis. Based on TSA, additional studies with large sample sizes are needed to validate these results in cancer vs control blood and urine samples and to confirm the findings regarding T classification and prognosis.
Data sharing statement
All relevant data are included in the paper.
Ethics approval
Although this study was not primary research involving human samples, our study was a secondary analysis regarding human subject data published in the public domain. For this type of study, formal consent is not required.
Acknowledgment
This study was supported by the National Science Foundation of Jiangsu Province (Grant No BK20150251), the Youth Medical Talent Project of Jiangsu Province (QNRC2016292), and the China Postdoctoral Science Foundation (Grant No 63, 2018 M632371).
Disclosure
The authors report no conflicts of interest in this work.
References
Chow WH, Dong LM, Devesa SS. Epidemiology and risk factors for kidney cancer. Nat Rev Urol. 2010;7(5):245–257. | ||
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. | ||
Courthod G, Tucci M, Di Maio M, Scagliotti GV. Papillary renal cell carcinoma: a review of the current therapeutic landscape. Crit Rev Oncol Hematol. 2015;96(1):100–112. | ||
Lopez-Beltran A, Scarpelli M, Montironi R, Kirkali Z. 2004 WHO classification of the renal tumors of the adults. Eur Urol. 2006;49(5):798–805. | ||
Jonasch E, Gao J, Rathmell WK. Renal cell carcinoma. BMJ. 2014;349:g4797. | ||
Ye M, Huang T, Ni C, Yang P, Chen S. Diagnostic capacity of RASSF1A promoter methylation as a biomarker in tissue, brushing, and blood samples of nasopharyngeal carcinoma. EBioMedicine. 2017;18:32–40. | ||
Zhao R, Choi BY, Lee MH, Bode AM, Dong Z. Implications of genetic and epigenetic alterations of CDKN2A (p16(INK4a)) in cancer. EBioMedicine. 2016;8:30–39. | ||
Carrió E, Suelves M. DNA methylation dynamics in muscle development and disease. Front Aging Neurosci. 2015;7:19. | ||
Okuda H, Toyota M, Ishida W, et al. Epigenetic inactivation of the candidate tumor suppressor gene HOXB13 in human renal cell carcinoma. Oncogene. 2006;25(12):1733–1742. | ||
Girgis AH, Iakovlev VV, Beheshti B, et al. Multilevel whole-genome analysis reveals candidate biomarkers in clear cell renal cell carcinoma. Cancer Res. 2012;72(20):5273–5284. | ||
Burbee DG, Forgacs E, Zöchbauer-Müller S, et al. Epigenetic inactivation of RASSF1A in lung and breast cancers and malignant phenotype suppression. J Natl Cancer Inst. 2001;93(9):691–699. | ||
Vichalkovski A, Gresko E, Cornils H, Hergovich A, Schmitz D, Hemmings BA. NDR kinase is activated by RASSF1A/MST1 in response to Fas receptor stimulation and promotes apoptosis. Curr Biol. 2008;18(23):1889–1895. | ||
Donninger H, Vos MD, Clark GJ. The RASSF1A tumor suppressor. J Cell Sci. 2007;120(Pt 18):3163–3172. | ||
Rong R, Jiang LY, Sheikh MS, Huang Y. Mitotic kinase Aurora-A phosphorylates RASSF1A and modulates RASSF1A-mediated microtubule interaction and M-phase cell cycle regulation. Oncogene. 2007;26(55):7700–7708. | ||
Skrypkina I, Tsyba L, Onyshchenko K, et al. Concentration and methylation of cell-free DNA from blood plasma as diagnostic markers of renal cancer. Dis Markers. 2016;2016:1–10. | ||
Klacz J, Wierzbicki PM, Wronska A, et al. Decreased expression of RASSF1A tumor suppressor gene is associated with worse prognosis in clear cell renal cell carcinoma. Int J Oncol. 2016;48(1):55–66. | ||
Hoque MO, Begum S, Topaloglu O, et al. Quantitative detection of promoter hypermethylation of multiple genes in the tumor, urine, and serum DNA of patients with renal cancer. Cancer Res. 2004;64(15):5511–5517. | ||
Ellinger J, Holl D, Nuhn P, et al. DNA hypermethylation in papillary renal cell carcinoma. BJU Int. 2011;107(4):664–669. | ||
Loginov VI, Khodyrev DS, Pronina IV, et al. Methylation of promoter region of RASSF1A gene and frequencies of allelic imbalances in chromosome 3 critical regions are correlated with progression of clear cell renal cell carcinoma. Mol Biol (Mosk). 2009;43(3):429–438. | ||
Zintzaras E, Ioannidis JP. HEGESMA: genome search meta-analysis and heterogeneity testing. Bioinformatics. 2005;21(18):3672–3673. | ||
Han S, Zong S, Shi Q, et al. Is Ep-CAM expression a diagnostic and prognostic biomarker for colorectal cancer? A systematic meta-analysis. EBioMedicine. 2017;20:61–69. | ||
Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. | ||
Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med. 1997;127(9):820–826. | ||
Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. | ||
Brok J, Thorlund K, Gluud C, Wetterslev J. Trial sequential analysis reveals insufficient information size and potentially false positive results in many meta-analyses. J Clin Epidemiol. 2008;61(8):763–769. | ||
Kulinskaya E, Wood J. Trial sequential methods for meta-analysis. Res Synth Methods. 2014;5(3):212–220. | ||
Holst LB, Petersen MW, Haase N, Perner A, Wetterslev J. Restrictive versus liberal transfusion strategy for red blood cell transfusion: systematic review of randomised trials with meta-analysis and trial sequential analysis. BMJ. 2015;350:h1354. | ||
Wetterslev J, Thorlund K, Brok J, Gluud C. Estimating required information size by quantifying diversity in random-effects model meta-analyses. BMC Med Res Methodol. 2009;9:86. | ||
Hauser S, Zahalka T, Fechner G, Müller SC, Ellinger J. Serum DNA hypermethylation in patients with kidney cancer: results of a prospective study. Anticancer Res. 2013;33(10):4651–4656. | ||
de Martino M, Klatte T, Haitel A, Marberger M. Serum cell-free DNA in renal cell carcinoma: a diagnostic and prognostic marker. Cancer. 2012;118(1):82–90. | ||
Kawai Y, Sakano S, Suehiro Y, et al. Methylation level of the RASSF1A promoter is an independent prognostic factor for clear-cell renal cell carcinoma. Ann Oncol. 2010;21(8):1612–1617. | ||
Onay H, Pehlivan S, Koyuncuoglu M, Kirkali Z, Ozkinay F. Multigene methylation analysis of conventional renal cell carcinoma. Urol Int. 2009;83(1):107–112. | ||
Costa VL, Henrique R, Ribeiro FR, et al. Quantitative promoter methylation analysis of multiple cancer-related genes in renal cell tumors. BMC Cancer. 2007;7:133. | ||
Peters I, Rehmet K, Wilke N, et al. RASSF1A promoter methylation and expression analysis in normal and neoplastic kidney indicates a role in early tumorigenesis. Mol Cancer. 2007;6:49. | ||
Hori Y, Oda Y, Kiyoshima K, et al. Oxidative stress and DNA hypermethylation status in renal cell carcinoma arising in patients on dialysis. J Pathol. 2007;212(2):218–226. | ||
Gonzalgo ML, Yegnasubramanian S, Yan G, et al. Molecular profiling and classification of sporadic renal cell carcinoma by quantitative methylation analysis. Clin Cancer Res. 2004;10(21):7276–7283. | ||
Loginov VI, Maliukova AV, Seregin I, et al. Methylation of the promoter region of the RASSF1A gene, a candidate tumor suppressor, in primary epithelial tumors. Mol Biol (Mosk). 2004;38(4):654–667. | ||
Tokinaga K, Okuda H, Nomura A, Ashida S, Furihata M, Shuin T. Hypermethylation of the RASSF1A tumor suppressor gene in Japanese clear cell renal cell carcinoma. Oncol Rep. 2004;12(4):805–810. | ||
Dulaimi E, Ibanez de Caceres I, Uzzo RG, et al. Promoter hypermethylation profile of kidney cancer. Clin Cancer Res. 2004;10(12 Pt 1):3972–3979. | ||
Yano T, Ito F, Kobayashi K, et al. Hypermethylation of the CpG island of connexin 32, a candidate tumor suppressor gene in renal cell carcinomas from hemodialysis patients. Cancer Lett. 2004;208(2):137–142. | ||
Battagli C, Uzzo RG, Dulaimi E, et al. Promoter hypermethylation of tumor suppressor genes in urine from kidney cancer patients. Cancer Res. 2003;63(24):8695–8699. | ||
Yoon JH, Dammann R, Pfeifer GP. Hypermethylation of the CpG island of the RASSF1A gene in ovarian and renal cell carcinomas. Int J Cancer. 2001;94(2):212–217. | ||
Morrissey C, Martinez A, Zatyka M, et al. Epigenetic inactivation of the RASSF1A 3p21.3 tumor suppressor gene in both clear cell and papillary renal cell carcinoma. Cancer Res. 2001;61(19):7277–7281. | ||
Duan JM, Zhi LI, Min Z, Zhang JY, Bo P, Huang JH. Abnormal methylation of RASSF1A and BLU genes in renal carcinoma. Acad J Sec Mil Med Univ. 2007;28(10):1068–1071. | ||
Yuan JH, Zhou JM, Huang HY. Methylation of CpG region in promoter region of RASSF1A in renal cell carcinoma. J Trop Med. 2008;8(6):539–540. Chinese. | ||
Shi J, Fu H, Jia Z, He K, Fu L, Wang W. High expression of CPT1A predicts adverse outcomes: a potential therapeutic target for acute myeloid leukemia. EBioMedicine. 2016;14:55–64. | ||
Maziveyi M, Alahari SK. Breast cancer tumor suppressors: a special emphasis on novel protein nischarin. Cancer Res. 2015;75(20):4252–4259. | ||
Yu GS, Lai CY, Xu Y, Bu CF, Su ZX. Aberrant methylation of RASSF1A gene contribute to the risk of renal cell carcinoma: a meta-analysis. Asian Pac J Cancer Prev. 2015;16(11):4665–4669. | ||
Huang YQ, Guan H, Liu CH, et al. Association between RASSF1A promoter methylation and renal cell cancer susceptibility: a meta-analysis. Genet Mol Res. 2016;15(2). |
Supplementary material
References
Skrypkina I, Tsyba L, Onyshchenko K, et al. Concentration and methylation of cell-free DNA from blood plasma as diagnostic markers of renal cancer. Dis Markers. 2016;2016:1–10. | ||
Klacz J, Wierzbicki PM, Wronska A, et al. Decreased expression of RASSF1A tumor suppressor gene is associated with worse prognosis in clear cell renal cell carcinoma. Int J Oncol. 2016;48(1):55–66. | ||
Ellinger J, Holl D, Nuhn P, et al. DNA hypermethylation in papillary renal cell carcinoma. BJU Int. 2011;107(4):664–669. | ||
Loginov VI, Khodyrev DS, Pronina IV, et al. Methylation of promoter region of RASSF1A gene and frequencies of allelic imbalances in chromosome 3 critical regions are correlated with progression of clear cell renal cell carcinoma. Mol Biol (Mosk). 2009;43(3):429–438. | ||
Kawai Y, Sakano S, Suehiro Y, et al. Methylation level of the RASSF1A promoter is an independent prognostic factor for clear-cell renal cell carcinoma. Ann Oncol. 2010;21(8):1612–1617. | ||
Onay H, Pehlivan S, Koyuncuoglu M, Kirkali Z, Ozkinay F. Multigene methylation analysis of conventional renal cell carcinoma. Urol Int. 2009;83(1):107–112. | ||
Costa VL, Henrique R, Ribeiro FR, et al. Quantitative promoter methylation analysis of multiple cancer-related genes in renal cell tumors. BMC Cancer. 2007;7:133. | ||
Hori Y, Oda Y, Kiyoshima K, et al. Oxidative stress and DNA hypermethylation status in renal cell carcinoma arising in patients on dialysis. J Pathol. 2007;212(2):218–226. | ||
Gonzalgo ML, Yegnasubramanian S, Yan G, et al. Molecular profiling and classification of sporadic renal cell carcinoma by quantitative methylation analysis. Clin Cancer Res. 2004;10(21):7276–7283. | ||
Loginov VI, Maliukova AV, Seregin I, et al. Methylation of the promoter region of the RASSF1A gene, a candidate tumor suppressor, in primary epithelial tumors. Mol Biol. 2004;38(4):549–560. | ||
Tokinaga K, Okuda H, Nomura A, Ashida S, Furihata M, Shuin T. Hypermethylation of the RASSF1A tumor suppressor gene in Japanese clear cell renal cell carcinoma. Oncol Rep. 2004;12(4):805–810. | ||
Dulaimi E, Ibanez de Caceres I, Uzzo RG, et al. Promoter hypermethylation profile of kidney cancer. Clin Cancer Res. 2004;10(12 Pt 1):3972–3979. | ||
Battagli C, Uzzo RG, Dulaimi E, et al. Promoter hypermethylation of tumor suppressor genes in urine from kidney cancer patients. Cancer Res. 2003;63(24):8695–8699. | ||
Yoon JH, Dammann R, Pfeifer GP. Hypermethylation of the CpG island of the RASSF1A gene in ovarian and renal cell carcinomas. Int J Cancer. 2001;94(2):212–217. | ||
Morrissey C, Martinez A, Zatyka M, et al. Epigenetic inactivation of the RASSF1A 3p21.3 tumor suppressor gene in both clear cell and papillary renal cell carcinoma. Cancer Res. 2001;61(19):7277–7281. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.









