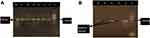Back to Journals » Infection and Drug Resistance » Volume 12
Rapid diagnosis of neonatal sepsis by PCR for detection of 16S rRNA gene, while blood culture and PCR results were similar in E.coli-predominant EOS cases
Authors EL-Amir MI , El-Feky MA, Abo Elwafa DA , Abd-Elmawgood EA
Received 13 May 2019
Accepted for publication 9 August 2019
Published 30 August 2019 Volume 2019:12 Pages 2703—2710
DOI https://doi.org/10.2147/IDR.S213958
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Joachim Wink
Mostafa I EL-Amir,1 Mohamed Ali El-Feky,1,2 Doaa A Abo Elwafa,1 Eman Ahmed Abd-Elmawgood3
1Department of Medical Microbiology and Immunology, Faculty of Medicine, South Valley University, Qena, Egypt; 2Department of Medical Microbiology and Immunology, Faculty of Medicine, Assiut University, Assiut, Egypt; 3Department of Pediatrics, Faculty of Medicine, South Valley University, Qena, Egypt
Correspondence: Mostafa I EL-Amir
Lecturer of Medical Microbiology and Immunology, Faculty of Medicine, South Valley University, Qena 83511, Egypt
Tel +20 100 772 5553
Email [email protected]
Purpose: To determine the bacteriological pattern and antibiotic susceptibility of bacterial isolates causing neonatal sepsis in Qena University Hospitals and compare polymerase chain reaction (PCR) and blood culture results in a trial for rapid diagnosis.
Patients and methods: Blood samples from 75 clinically suspected cases of neonatal sepsis were subjected to identification of bacteria and determination of their antibiotic sensitivity through blood culture, and rapid detection of 16S rRNA and the uidA gene (to confirm the presence of E. coli) by PCR from extracted bacterial DNA.
Results: Most patients were preterm (64%) and low birth weight (LBW) (68%). In total, 42.7% presented with early onset sepsis (EOS). LBW was significantly associated with EOS (P-value=0.03). Although the blood culture and PCR results were similar in EOS, the PCR results were significantly higher than those of blood culture in detecting bacteria (85.3% vs 68%, respectively, P-value=0.001). Blood culture showed 100% specificity. The most common pathogen was E. coli (86.2%) in EOS and Staphylococcus spp. (45.5%) in late-onset sepsis (LOS) (P-value=0.001 and 0.02, respectively). The most effective antibiotics against Gram-negative bacteria were ofloxacin, ciprofloxacin, imipenem, and amikacin, while vancomycin, oxacillin, and imipenem were the most effective antibiotics against Gram-positive bacteria.
Conclusion: EOS was mainly caused by E. coli, while LOS was mainly caused by Staphylococcus spp. The 16S rRNA PCR showed higher sensitivity with rapid and accurate diagnosis. Blood culture is the most suitable method for antimicrobial sensitivity testing.
Keywords: neonatal sepsis, 16S rRNA, blood culture, EOS, LOS
Introduction
Neonatal sepsis is a clinical syndrome characterized by systemic manifestations due to bacteremia in the first month of life.1 The disease characterized by high morbidity and mortality rates in developing and developed countries, especially in premature infants.2 Despite the presence of quality care in developed countries for cases of neonatal sepsis, approximately 40% of the patients die or suffer from major permanent disability, especially neurological complications.3
Neonatal sepsis can be differentiated into two types according to the age of onset. Early onset sepsis (EOS), which is mainly due to maternal origin, infection occurring during the first 3 days of life, and late-onset sepsis (LOS), which manifests 3 days after delivery, is mainly due to acquiring the pathogen from prolonged hospitalization, especially in preterm newborns.4,5
The disease is characterized by non-specific manifestations, leading to difficult diagnosis by routine clinical practices. Blood culture is considered the gold standard for the diagnosis of neonatal sepsis, but it takes a long time to obtain a positive result and the inability to isolate causative microorganisms for neonatal sepsis by blood culture does not exclude sepsis.6
Spread of antimicrobial resistance (AMR) in bacteria by different mechanisms occurs mainly due to inappropriate use of antimicrobial agent. Deficiency in diagnostic procedure leads to irresponsive prescription, especially in developing countries. Global surveillance report published by the World Health Organization (WHO) showed that about 50% of the Escherechia coli and Staphylococcus aureus were resistant to third-generation cephalosporin and methicillin, respectively.7 Although antibiotics stewardship programs were established in different developed countries, in developing countries, implementation of such strategies is defective due to resources deficiency.8
Accurate and reliable tests are needed for the rapid diagnosis of neonatal sepsis to differentiate between truly infected and non-infected newborns to minimize the prolonged and inappropriate use of empirical antibiotics and decrease the emergence of resistant bacterial strains.9 Several viruses are implicated as a cause of neonatal sepsis as herpes simplex virus (HSV), enterovirus, and parechovirus. Diagnosis of viral sepsis depends on exclusion.10 Early diagnosis plays an important role in effective treatment by appropriate antiviral drug and limits inappropriate use of antibiotics. Viral cause of neonatal sepsis must be suspected in blood culture and 16S rRNA gene negative cases.11
The 16S rRNA gene is present in all bacteria but not in other organisms. The identification of a conserved gene region by polymerase chain reaction (PCR) can be used for the rapid diagnosis of bacterial infection. The 16S rRNA can be used for bacterial identification at the genus and species levels in approximately 90% of the cases.12
The aim of this study was to determine the most common types of bacteria causing neonatal sepsis and its antibiotic sensitivity pattern. Additionally, we aimed to determine the most common risk factors associated with disease pathogenesis and evaluate the diagnosis of neonatal sepsis by amplification of the 16S rRNA conserved gene in bacteria by PCR in comparison with blood culture results.
Patients and methods
This prospective study was conducted over a period of one year from July 2017 to June 2018 in the Neonatal Intensive Care Unit (NICU) at Qena University Hospitals. Seventy-five patients with clinically suspected neonatal sepsis were enrolled in the study. Inclusion criteria included all neonates admitted in our NICU showing clinical signs suggestive of neonatal sepsis (at least two clinical manifestations) which included: core temperature greater than 38.5°C or less than 36°C and/or temperature instability, cardiac manifestation as (bradycardia, tachycardia, poor perfusion, or hypotension), respiratory manifestation as (tachypnea, apnea, cyanosis, or respiratory distress), gastrointestinal manifestation as (feeding difficulty or abdominal distension), non-specific manifestation as (lethargy, hypotonia, or irritability).
Full histories were taken, including age, sex, birth weight, premature rupture of membranes (PROM), mode of delivery, maternal fever, antepartum hemorrhage, and gestational age. If sepsis manifestation appears at the first 72 hrs after birth, the case considered as EOS, while LOS manifestations occur after 72 hrs of age. Clinical and laboratory data were also included (respiratory distress, poor feeding, temperature, blood pressure, and CRP). Infected neonates were enrolled in the study after written informed consent was obtained from their parents. The study was approved by the Ethics Committee, Qena Faculty of Medicine, South Valley University. The study was conducted in accordance with the principles of the Declaration of Helsinki.
Three milliliters of venous blood were obtained from all neonates under complete aseptic conditions. The presence of bacteria in blood samples was diagnosed in different ways: the first by blood culture and the other method by molecular diagnosis through bacterial DNA extraction from blood samples after 12 hrs of enhancement by trypticase soy broth (TSB) (Hi-Media Laboratories-India), as previously described.13
The top of each blood culture bottle (Bio-Lab, UK) was wiped using a fresh ethanol swab, and then 2 mL of patient blood were drawn inside the blood culture bottle. Each bottle was clearly labeled with the patient ID, date, and time of collection. The media were incubated at 37°C and inspected visually every morning for turbidity. Subcultures were performed 24 hrs after inoculation and then every day for up to 7 days. If there was no bacterial growth after 7 days of incubation, blood culture was reported as negative.14
Two milliliters were withdrawn from the fresh blood culture and then inoculated in 4 mL TSB for 12 hrs of incubation at 37°C to support bacterial growth. After bacterial enhancement, DNA was extracted as described previously.13 Samples were then preserved at −20°C until molecular analysis for the detection of the universal bacterial gene (16S rRNA gene) was performed.
Several drops were withdrawn from the incubated blood culture by sterile syringe and subcultured onto blood agar and chocolate agar. All of the plates were incubated at 37°C for 24–48 hrs. Bacterial colonies were identified by Gram staining and several biochemical tests, such as Simmon’s citrate agar, triple sugar iron, and oxidase and catalase tests.15
Antibiograms of various isolates were determined by the Kirby–Bauer method on Mueller–Hinton agar medium (Hi-Media Laboratories, India) according to the Clinical and Laboratory Standards Institute (CLSI).16 Antibiotic discs used were ampicillin (10 µg), amoxicillin clavulanic acid (30 µg), oxacillin (1 µg), vancomycin (30 µg), ceftriaxone (30 µg), cefoxitin (30 µg), ciprofloxacin (5 µg), ofloxacin (5 µg), amikacin (30 µg), gentamycin (10 µg), azithromycin (30 µg), piperacillin (100 µg), and imipenem (10 µg).
Amplification of the bacterial 16S rRNA gene by PCR was performed on DNA extracts under the following PCR conditions: initial denaturation at 95°C for 3 mins, followed by 35 cycles of 94°C for 1 min, 53°C for 2 mins, 72°C for 2 mins, and a final extension at 72°C for 10 mins. The expected PCR product was 380 base pairs and separated by electrophoresis on a 1.5% agarose gel using ethidium bromide and visualized by UV transillumination, as shown in Figure 1. E. coli strains were confirmed by PCR by the presence of the uidA gene as previously described.17 The PCR product was 212 base pairs, as shown in Figure 2.
 |
Figure 2 E. coli isolates were confirmed by the presence of the uidA gene, which shows a 212 bp band in gel electrophoresis. |
Statistical analysis was performed using SPSS software version 22. Categorical variables were presented as proportions and percentages. Numerical variables were presented as the mean and standard deviation when normally distributed, and median and interquartile ranges when not normally distributed. Chi-square tests or Fisher’s exact tests were performed when appropriate. The odds ratio was calculated. P-value of 0.05 or less was considered significant.
Results
Seventy-five patients with clinically suspected neonatal sepsis were enrolled in the study. A total of 40/75 (53.3%) were males and 35/75 (46.7%) were females, with a mean birth weight of 2.12±0.6 kg and mean gestational age of 33.7±3 weeks. Most of the cases were preterm 48/75 (64%) and low birth weight (LBW) 51/75 (68%). In total, 49/75 (65.3%) of neonates were delivered by spontaneous vaginal delivery, and 21/75 (28%) were exposed to PROM. History of maternal risk factors, such as maternal fever, meconium aspiration syndrome, and preeclampsia, was less common among our patients than in PROM (5.3%, 13.3%, and 1.3%, respectively).
As shown in Table 1, 32/75 (42.7%) of neonates were diagnosed with EOS based on clinical data, and 43/75 (57.3%) neonates were diagnosed with LOS. LBW neonates mostly presented with EOS 26/32 (81.2%) with a significant P-value (P=0.03; OR=3.12), while only 25/43 (58.1%) of LOS neonates were LBW. Although 23 of 32 neonates (71.9%) that presented with EOS were preterm, this result showed a nonsignificant P-value (P=0.22) when compared with LOS. For EOS cases, 11/32 (34.3%) were associated with PROM, while it was less common among LOS cases 10/43 (23.2%), with a nonsignificant P-value (P-value=0.2.; OR=1.7).
 |
Table 1 Different risk factors associated with early onset (EOS) and late-onset sepsis (EOS) |
Blood culture result revealed that 51/75 (68%) of neonates were positive for bacterial growth. Ten cases of blood culture positive samples showed mixed infection with two organisms (10/51, 19.5%). As high as 20.5% of EOS and 18.5% of LOS were presented with mixed bacterial infection with two organisms.
The microorganisms detected by blood culture (73.8%) were Gram-negative organisms, 24.6% were Gram-positive organisms, and 1.6% were Candida spp. The total number of isolated microorganisms was sixty-one. E. coli was the most frequently isolated bacteria 34/61 (55.5%), followed by coagulase-negative staphylococci (CoNS) 8/61 (13.1%), Staphylococcus aureus 7/61 (11.5%), Klebsiella spp. 6/51 (9.8%), Pseudomonas aeruginosa 3/61 (4.9%), Proteus 2/61 (3.2%), and Candida spp. 1/61 (1.6%), as shown in Table 2.
 |
Table 2 Bacteriological profile of blood culture-positive cases in EOS and LOS cases |
Most of the patients with EOS 25/29 (86.2%) were infected with E. coli in comparison with LOS, with 9/22 (40.9%), with a significant P-value (P=0.001). S. aureusand CoNS were isolated from 10/22 (45.5%) LOS patients and found in 5/29 (17%) EOS patients with significant P-values (P=0.02). Pseudomonas was isolated from LOS cases only (0% in EOS vs 13.6% in LOS), with a P-value=0.04, as shown in Table 2.
In Table 3, the least amount of antibiotic resistance among Gram-negative bacilli was observed in ofloxacin (15.5%), ciprofloxacin (20%), and imipenem (35.7%), while 40% were resistant to amikacin. The antibiotic resistance for E.coli isolated from EOS cases showed the same sensitivity pattern as Gram-negative bacilli (Ofloxacin 16%, ciprofloxacin 20%, and imipenem 32%, while 36% were resistant to amikacin).
 |
Table 3 Antimicrobial-resistant patterns of bacterial strains identified by blood culture |
In contrast, all Gram-positive bacteria were sensitive to vancomycin, while 26.6% were resistant to imipenem, and 33.3% were resistant to oxacillin and amikacin. All S. aureus isolated from LOS cases were sensitive to vancomycin, while 20% were resistant to imipenem and oxacillin.
As shown in Table 4, 24/75 (32%) of the cases showed negative blood culture. Most of the negative cases were LOS (21/24). In EOS (90.6%), the cases had positive blood culture results, while in LOS, only 51.2% of the cases were positive. The PCR and blood culture results were the same in EOS cases (90.6% were positive and 9.4% were negative). While a difference was found in LOS cases, 48.8% of the cases showed no bacterial growth by blood culture, while 18.6% of the cases were negative by PCR (16S rRNA not identified in the sample).
 |
Table 4 Pattern of positive and negative cases of EOS and LOS according to blood culture and PCR results |
As shown in Table 5, 51/75 (68%) of the cases were positive by blood culture. A total of 64/75 (85.3%) of the cases were truly positive for bacterial infection by detection of 16S rRNA by PCR, while 11/75 (14.7%) of the cases were negative for bacterial infection by PCR and had no need for antibiotic treatment at all. No cases were positive by blood culture and negative by PCR. No false-positive results for blood culture were detected, but false-negative results were 13/75 (17.3%). Compared with PCR, the diagnosis of neonatal sepsis by blood culture revealed 79.6% sensitivity, 100% specificity, 100% positive predictive value (PPV), 45.8% negative predicted value (NPV), and 82.6% overall accuracy.
 |
Table 5 Comparison between PCR and blood culture results |
Discussion
Rapid identification of the bacterial cause of neonatal sepsis was performed by identifying the 16S rRNA gene from the enriched blood sample. In 85.3% of the cases, bacteria were identified by PCR with a significantly higher result than those in blood culture (68%) as described previously.18 No false-positive cases were detected by blood culture (100% specificity). Blood culture was less accurate in the detection of positive cases than 16S rRNA gene (79.6%). It is serious for clinician to consider viral causes of neonatal sepsis in blood culture negative and 16S rRNA gene negative cases, which limits inappropriate use of antibiotics.
In our study, the percentage of males was higher than that of females (53.3% vs 46.7%), which is similar to other studies from Arab Gulf countries and Taiwan. The percentages of males in those studies were 54.9% and 52.8%, respectively.19,20 None of the previous studies found a significant difference between males and females in neonatal sepsis, which is similar to our study.
In our study, 68% of the cases were LBW and 64% were preterm. LBW and preterm are considered the most predisposing factors for infections in neonates as described previously.21 As high as 65.3% of the patients were delivered by normal vaginal delivery, while Stoll et al22 found that 47% of the sepsis patients were delivered by normal vaginal delivery. However, other studies have determined that in normal vaginal delivery, the newborn may be exposed to infected secretions in the birth canal or normal maternal bacterial flora, which can effectively produce sepsis.23 According to delivery circumstances, about one-third of our patients were exposed to PROM, which is similar to other studies that concluded that the risk of neonatal sepsis increases with the duration of membrane rupture.24
LBW was significantly higher in EOS among our patients (81.2% of the EOS cases were LBW), which is similar to Wójkowska-Mach et al,25 who found that one of the factors that significantly increased the risk of EOS was LBW.
In the current study, 68% of the cases had microbiologically confirmed sepsis by blood culture. A similar study from Egypt found that 40.7% of the suspected neonatal sepsis cases had confirmed positive blood culture results.26 Our high results may be because blood samples drawn from neonates for blood culture were at least 1.5 mL. Some studies recommended that 1 mL is the least amount of blood that should be withdrawn for blood culture, doubling the likelihood of positivity compared with 0.5 mL blood.27
In the current study, E. coli was the most frequent microorganism isolated from EOS cases (86.2%), which is similar to other studies that found that there is a change in pathogens causing EOS from group B streptococci infections, which have shown a significant decline in recent years due to intrapartum antibiotic prophylaxis to a significant increase in E. coli infections that commonly colonize maternal enteric canal.28 Unlike several studies from Egypt, which found that the most common Gram-negative bacteria causing EOS was Klebsiella, we confirmed our result by detecting the uidA gene encoding the b-glucuronidase enzyme, which is frequently used to identify E. coli.29–31
Candida spp. were isolated from 1.6% of the cases of EOS, which is similar to a previous study by Stoll and Hansen,28 who found that Candida causes 2.4% of the infections in EOS. Another study4 found that Candida spp. were isolated more frequently from LOS than EOS due to nosocomial infection as delayed removal of the central catheter, while in our study, Candida was isolated only from EOS cases, which may be due to infection through the birth canal, as two-thirds of our cases were delivered normally through the vagina.
S. aureus and CoNS were isolated from 45.5% of the LOS cases, which is similar to other studies, such as Kung et al,20 who found that Staphylococcus spp. cause 45.2% of the cases of LOS. In addition, Stoll et al and Resende et al32,33 found similar percentages to those in previous studies (56% and 57.9%, respectively), while Hammoud et al19 found lower percentages than those in previous studies (39%). Increasing the care and extensive intervention for premature newborns in the NICU, in addition to increasing nurses’ workload, elevates the possibility of infection with CoNS. Another study also correlated health care workers’ training level and infection rates in the NICU.34 In our study, Pseudomonas was recognized in LOS with a significant P-value than in EOS, which is similar to another study,35 which found that Pseudomonas is significantly higher in LOS than EOS because Pseudomonas is well known as a hospital-acquired pathogen, and neonates presented with LOS were hospitalized for a long time.
Gram-negative bacteria isolated from blood culture showed antibiotic susceptibility to ofloxacin, ciprofloxacin, imipenem, and amikacin, and marked resistance to ampicillin, amoxicillin clavulanic acid, ceftriaxone, piperacillin, cefoxitin, and azithromycin. According to Gram-positive bacteria, they showed marked susceptibility to vancomycin followed by imipenem, then oxacillin, and amikacin. On the other hand, Gram-positive bacteria showed marked resistance against ampicillin, amoxicillin clavulanic acid, ofloxacin, ceftriaxone, and ciprofloxacin. E.coli which was the commonest organism isolated from EOS cases showed susceptibility to ofloxacin, ciprofloxacin, imipenem followed by amikacin, while S. aureus which was the commonest cause for LOS showed susceptibility to vancomycin followed by oxacillin and imipenem. Our results are similar to those of another study from Egypt, which found that imipenem and quinolones are the best antibiotics against Gram-negative bacteria, while 100% of the Gram-positive bacteria were susceptible to vancomycin.26 Although some studies support the use of quinolones and ciprofloxacin to life-threating neonatal infections,36,37 others considered that there is no sufficient data to allow safe and effective use of these antibiotics in neonates.38 The emergence of resistant strains due to inappropriate uses of antibiotics before hospitalization or hospital-acquired infection has been described previously.39
When comparing the results of blood culture with those of 16S rRNA, we found no difference between the two previous tests in EOS. No cases were identified with true infection and negative blood culture in EOS (confirmed by the absence of the 16S rRNA gene in these cases), as previously described by Stoll et al,22 who concluded that frequent use of intrapartum antibiotics increases the incidence of infection with more virulent and rapidly multiplying Gram-negative bacteria.
Conclusion
EOS was mainly caused by E. coli, imipenem was the best empirical antibiotic for this case followed by amikacin, while LOS was mainly caused by Staphylococcus spp. and vancomycin was the best empirical antibiotic for this case followed by oxacillin and imipenem. The 16S rRNA PCR showed higher sensitivity with rapid and accurate differentiation between bacteria and non-bacterial causes of neonatal sepsis. Blood culture is the most suitable method for antimicrobial sensitivity testing.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Edwards MS, Baker CJ. Sepsis in the newborn. In: Gershon AA, Hotez PJ, Katz SL, editors. Krugman’s Infectious Diseases of Children.
2. Schlapbach LJ, Aebischer M, Adams M, et al. Impact of sepsis on neurodevelopmental outcome in a Swiss National Cohort of extremely premature infants. Pediatrics. 2011;128(2):e348–357.
3. Brocklehurst P, Farrell B, King A, et al. Treatment of neonatal sepsis with intravenous immune globulin. N Engl J Med. 2011;365(13):1201–1211.
4. Hornik CP, Fort P, Clark RH, et al. Early and late onset sepsis in very-low-birth-weight infants from a large group of neonatal intensive care units. Early Hum Dev. 2012;88 Suppl 2:S69–S74.
5. Edwards MS, Gonik B. Preventing the broad spectrum of perinatal morbidity and mortality through group B streptococcal vaccination. Vaccine. 2013;31:D66–D71.
6. Wynn JL, Polin RA. Progress in the management of neonatal sepsis: the importance of a consensus definition. Pediatr Res. 2018;83:13–15.
7. World Health Organization. Antimicrobial Resistance Global Report on Surveillance. Geneva: World Health Organization; 2014.
8. Tiong JJ, Loo JS, Mai CW. Global antimicrobial stewardship: a closer look at the formidable implementation challenges. Front Microbiol. 2016;7:1860.
9. Prashant A, Vishwanath P, Kulkarni P, et al. Comparative assessment of cytokines and other inflammatory markers for the early diagnosis of neonatal sepsis – a case control study. PLoS One. 2013;8(7):e68426.
10. Verboon-Maciolek MA, Krediet TG, Gerards LJ, de Vries LS, Groenendaal F, van Loon AM. Severe neonatal parechovirus infection and similarity with enterovirus infection. Pediatr Infect Dis J. 2008;27:241–245.
11. Gupta N, Richter R, Robert S, Kong M. Viral sepsis in children. Front Pediatr. 2018;6:252.
12. Mignard S, Flandrois JP. 16S rRNA sequencing in routine bacterial identification: a 30-month experiment. J Microbiol Methods. 2006;67(3):574–581.
13. Dutta S, Narang A, Chakraborty A, Ray P. Diagnosis of neonatal sepsis using universal primer polymerase chain reaction before and after starting antibiotic drug therapy. Arch Pediatr Adolesc Med. 2009;163(1):6–11.
14. Kirn TJ, Weinstein MP. Update on blood cultures: how to obtain, process, report, and interpret. Clin Microbiol Infect. 2013;19(6):513–520.
15. Murray B, Pfaller T. Manual of Clinical Microbiology.
16. Clinical Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing.
17. Lindsey RL, Garcia-Toledo L, Fasulo D, Gladney LM, Strockbine N. Multiplex polymerase chain reaction for identification of Escherichia coli, Escherichia albertii and Escherichia fergusonii. J Microbiol Methods. 2017;140:1–4. doi:10.1016/j.mimet.2017.06.005
18. Tong MQ, Shang SQ, Wu YD, Zhao ZY. Rapid diagnosis of neonatal sepsis by 16SrRNA genes PCR amplification and genechip hybridization. Zhonghua Er Ke Za Zhi. 2004;42(9):663–667.
19. Hammoud MS, Al-Taiar A, Al-Abdi SY, et al. Late-onset neonatal sepsis in Arab states in the Gulf region: two-year prospective study. Int J Infect Dis. 2017;55:125–130. doi:10.1016/j.ijid.2017.01.006
20. Kung YH, Hsieh YF, Weng YH, et al. Risk factors of late-onset neonatal sepsis in Taiwan: a matched case-control study. J Microbiol Immunol Infect. 2016;49(3):430–435. doi:10.1016/j.jmii.2013.10.001
21. Khinchi YR, Kumar A, Yadhav S. Profile of neonatal sepsis. J Coll Med Sci. 2010;6(2):1–6.
22. Stoll BJ, Hansen NI, Sánchez PJ, et al. Early onset neonatal sepsis: the burden of group B Streptococcal and E. coli disease continues. Pediatrics. 2011;127(5):817–826. doi:10.1542/peds.2010-2217
23. Chan GJ, Lee AC, Baqui AH, Tan J, Black RE. Risk of early-onset neonatal infection with maternal infection or colonization: a global systematic review and meta-analysis. PLoS Med. 2013;10(8):e1001502. doi:10.1371/journal.pmed.1001502
24. Herbst A, Källén K. Time between membrane rupture and delivery and septicemia in term neonates. Obstet Gynecol. 2007;110(3):612–618. doi:10.1097/01.AOG.0000277632.36186.84
25. Wójkowska-Mach J, Borszewska-Kornacka M, Domańska J, et al. Early-onset infections of very-low-birth-weight infants in Polish neonatal intensive care units. Pediatr Infect Dis J. 2012;31(7):691–695. doi:10.1097/INF.0b013e3182567b74
26. Shehab El-Din EM, El-Sokkary MM, Bassiouny MR, Hassan R. Epidemiology of neonatal sepsis and implicated pathogens: a study from Egypt. Biomed Res Int. 2015;2015:509484.
27. Polin RA; Committee on Fetus and Newborn. Management of neonates with suspected or proven early-onset bacterial sepsis. Pediatrics. 2012;129(5):1006–1015. doi:10.1542/peds.2012-0541
28. Stoll BJ, Hansen N. Infections in VLBW infants: studies from the NICHD Neonatal Research Network. Semin Perinatol. 2003;27(4):293–301. doi:10.1016/S0146-0005(03)00046-6
29. Seliem WA, Sultan AM. Etiology of early onset neonatal sepsis in neonatal intensive care unit - Mansoura, Egypt. J Neonatal Perinatal Med. 2018;11(3):323–330. doi:10.3233/NPM-17128
30. Fahmey SS. Early-onset sepsis in a neonatal intensive care unit in Beni Suef, Egypt: bacterialisolates and antibiotic resistance pattern. Korean J Pediatr. 2013;56(8):332–337. doi:10.3345/kjp.2013.56.8.332
31. Kishk RM, Mandour MF, Farghaly RM, Ibrahim A, Nemr NA. Pattern of blood stream infections within Neonatal Intensive Care Unit, Suez Canal UniversityHospital, Ismailia, Egypt. Int J Microbiol. 2014;2014:276873. doi:10.1155/2014/276873
32. Stoll BJ, Hansen N, Fanaroff AA, et al. Late-onset sepsis in very low birth weight neonates: the experience of the NICHD Neonatal Research Network. Pediatrics. 2002;110:285–291. doi:10.1542/peds.110.2.285
33. Resende DS, Peppe AL, dos Reis H, Abdallah VO, Ribas RM, Gontijo Filho PP. Late onset sepsis in newborn babies: epidemiology and effect ofa bundle to prevent central lineassociated bloodstream infections in the neonatal intensive care unit. Braz J Infect Dis. 2015;19(1):52–57. doi:10.1016/j.bjid.2014.09.006
34. Gray J, Omar N. Nosocomial infections in neonatal intensive care units in developed and developing countries: how can we narrow the gap? J Hosp Infect. 2013;83(3):193–195. doi:10.1016/j.jhin.2012.12.006
35. Vergnano S, Menson E, Kennea N, et al. Neonatal infections in England: the NeonIN surveillance network. Arch Dis Child Fetal Neonatal Ed. 2011;96(1):F9–F14. doi:10.1136/adc.2009.178798
36. Gürpinar AN, Balkan E, Kiliç N, Kiriştioğlu I, Doğruyol H. The effects of a fluoroquinolone on the growth and development of infants. J Int Med Res. 1997;25(5):302–306. doi:10.1177/030006059702500508
37. Drossou-Agakidou V, Roilides E, Papakyriakidou-Koliouska P, et al. Use of ciprofloxacin in neonatal sepsis: lack of adverse effects up to one year. Pediatr Infect Dis J. 2004;23(4):346–349.
38. Kaguelidou F, Turner MA, Choonara I, Jacqz-Aigrain E. Ciprofloxacin use in neonates: a systematic review of the literature. Pediatr Infect Dis J. 2011;30:e29–e37. doi:10.1097/INF.0b013e3181fe353d
39. Shrestha RK, Rai SK, Khanal LK, Mandal PK. Bacteriological study of neonatal sepsis and antibiotic susceptibility pattern in Kathmandu, Nepal. Nepal Med Coll J. 2013;15:71–73.
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.