Back to Journals » Infection and Drug Resistance » Volume 15
Rapid Detection of Carbapenem-Resistant Klebsiella pneumoniae Using Machine Learning and MALDI-TOF MS Platform
Authors Wang J , Xia C , Wu Y, Tian X, Zhang K, Wang Z
Received 20 March 2022
Accepted for publication 2 July 2022
Published 12 July 2022 Volume 2022:15 Pages 3703—3710
DOI https://doi.org/10.2147/IDR.S367209
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Jinyu Wang,* Cuiping Xia,* Yue Wu, Xin Tian, Ke Zhang, Zhongxin Wang
Department of Clinical Laboratory, The First Affiliated Hospital of Anhui Medical University, Hefei, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Zhongxin Wang, Department of Clinical Laboratory, The First Affiliated Hospital of Anhui Medical University, Hefei, People’s Republic of China, Tel +8613866709500, Fax +5516-5908076, Email [email protected]
Background: Rapid detection of carbapenem-resistant Klebsiella pneumoniae (CRKP) is essential for specific antimicrobial therapy. Machine learning techniques combined with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) can be used as a rapid, reliable, sensitive, and low-cost species identification method.
Methods: Clinically collected K. pneumoniae were subjected to MALDI-TOF MS analysis. A random forest (RF) algorithm and non-linear support vector machine (SVM) were used to construct the RF, SVM, and dimension reduction (SVM-K) models, and their performance was assessed for accuracy, sensitivity, specificity, and area under the subject worker curve (AUC).
Results: The RF, SVM and SVM-K models showed good classification performance with 0.88, 0.88, and 0.91 accuracy, 0.82, 0.85, and 0.89 sensitivity, 0.93, 0.92, and 0.94 specificity with an AUC of 0.9013, 0.9298, and 0.9356, respectively. For the SVM-K model, the optimal dimension reduction was 105 to 153, and the average accuracy was > 0.9. The top 10 peak features of significance according to the RF algorithm with 6515 Da appeared in 56.8% of CRKP isolates and 5.3% of CSKP isolates, which indicated the best classification performance.
Conclusion: The three RF, SVM, and SVM-K models showed excellent classification performance differentiating the CRKP from CSKP; the SVM-K model was the best. Data analysis with machine learning combined with MALDI-TOF MS can be employed as a rapid and inexpensive alternative to existing detection methods.
Keywords: Klebsiella pneumoniae, RF, SVM, SVM-K, MALDI-TOF MS
Introduction
Klebsiella pneumoniae is commonly encountered opportunistic gram-negative bacterium that causes higher morbidity and mortality.1 With the widespread use of broad-spectrum antimicrobials such as lactamides and aminoglycosides, bacteria are prone to become multi-drug resistant by producing β-lactamases and cephalosporinases. Worldwide, treatment of carbapenem resistance K. pneumoniae (CRKP) has become a serious challenge. The resistance mechanism of CRKP involves the absence of membrane porins OmpK35 and OmpK36 and the production of broad-spectrum lactamases (ESBLs) or carbapenases.2 The emergence of CRKP greatly limits the selection of antimicrobial therapy, often resulting in poor outcomes.3
Meanwhile, antimicrobial drug susceptibility tests are time-consuming and expensive. A longer bacterial resistance testing cycle further aggravates the problem of carbapenem resistance. Therefore, developing rapid and reliable detection methods for the pathogenic bacteria resistant to antibiotics is crucial for the accurate and timely treatment. In the past few decades, matrix-assisted laser-resolved ionization time-of-flight mass spectrometry (MALDI-TOF MS), with resistance testing potential, has been widely used for rapid species identification of clinical microbes. It is faster, precise, and cost-effective than conventional microbial identification tests.4
Previous studies used direct analysis of characteristic hydrolysis peaks to rapidly detect drug resistance and classification in the MALDI-TOF MS mass spectrometry data acquired from enzyme-mediated antimicrobial hydrolysis.5 However, the method is complicated and requires an additional 3–4 h time. Meanwhile, for species identification, MALDI-TOF MS only depends on a few features, such as m/z and peak height, making it a fast and effective method. Though, the mass spectrum data information yet remains largely unutilized. Related studies discovered that extraneous variables could be removed to change the expression of MS data using machine learning to fully utilize the obscured information in mass spectrum data. By using intelligent data analysis, machine learning can maximize the mining of the information encoded in these mass spectra, exceeding most other methods.
Several studies used machine learning algorithms to make full use of MALDI-TOF MS data for species identification and simplified antimicrobial resistance assays.6 With self-supervised learning and continuously refining processes, machine learning can deeply mine the non-linear correlations in the data. It can swiftly evaluate drug resistance by detecting differences in strain mass spectrometry data. For example, Mather et al7 used the SVM (support vector machine) algorithm to select some characteristic bacterial peaks and build a classification model to successfully differentiate vancomycin-intermediate S. aureus (VISA) from vancomycin-sensitive S. aureus (VSSA). Therefore, this study aimed to construct RF, SVM, and SVM-K (RBF; radial basis function kernel) models to distinguish CRKP from CSKP (carbapenem sensitive K. pneumoniae).
Materials and Methods
Bacterial Strains
CRKP, n= 95 and CSKP, n=76, a total of 171 isolates were randomly collected from the First Affiliated Hospital of Anhui Medical University from January 2020 to December 2021. All strains were obtained from Hefei City, China. All strains were inoculated on Colombian blood agar plates, and individual colonies were isolated after 18–24 h of incubation at 35 °C. Carbapenem resistance tests were performed using the disk diffusion method and VITEK-2 compact system (BioMerieux, France). The isolates used in this study were resistant to imipenem, meropenem, or ertapenem. All species had a “carbapenemase” phenotype expressed through the Advanced Expert System (AES) of the VITEK 2 system. Resistant (R) and sensitive (S) isolates were interpreted following the Clinical Laboratory Standards Association of Standards (CLSI).8
Analysis of MALDI-TOF MS
Well-growing individual colonies were selected and evenly applied to the MALDI-TOF MS target plate with 1 µL of CHCA (-cyanogen-4-hydroxycinnamic acid) and dried at room temperature. E. coli ATCC8739 of the VITEK-MS system was used as the quality control strain. Spectrums were obtained in the RUO mode using the MALDI-TOF MS instrument (BioMerieux, France) with a laser frequency of 75 Hz, 100 shots, and a 2000 to 20,000 Da mass-charge ratio. The collected spectra were analyzed through the SARAMIS software. The most original data were processed without baseline correction and denoising.
Feature Selection
The peak data of CRKP and CSKP strains were exported to Microsoft Excel from the SARAMIS software. Feature peaks were selected for all recorded spectral peak data, considering the polarisation formed during instrument acquisition, as reported by Pena et al.9 To unify the small polarisation generated by the spectrum and facilitate subsequent data processing and model construction, similar peaks (tolerance ± 3Da) were grouped to balance their differences. These corrected peaks were sorted for importance by the RF algorithm. A set of peaks obtained simultaneously during data processing is known as “housekeeping peaks” for their presence in almost all the spectrum of K. pneumoniae. The peaks present in at least 70% of the MS data were deemed the housekeeping peaks for the classification of CRKP and CSKP.
Development of Predictive Models
Figure 1 shows the detailed flow of the model construction, and we constructed the corresponding classification model through the data collected by MALDI-TOF MS combined with the random forest algorithm and the support vector machine algorithm.
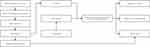 |
Figure 1 Flow chart showing the construction of RF, SVM, and SVM-K models. |
Large probabilistic optimal solutions for important parameters like the number of decision trees and maximum depth were used in the RF model to determine the SVM model’s cost parameter (C). The random forest and SVM models were built through scikit_learn (https://scikit-learn.org/stable/index.html), providing pre-packaged machine learning SVM tools in a Python environment. The SVM model used a radial basis function kernel to optimize data processing performance.
Not all of the peak features used by the model had classification power. Therefore, a dimensionality reduction of the data was applied. According to the peak feature importance ranking, K features were selected to input the SVM model. Firstly, K selected all peak features to construct the dimensionality reduction model. Subsequently, each model construction removed the last ranked features and continued until all features were removed. The dimensionality reduction model was run 100 times/cycle to obtain the average accuracy as the output. Figure 2 shows the effect of data dimensionality reduction on the average accuracy of the model. Finally, the SVM-K model was constructed based on the feature dimension reduction range with the highest average accuracy. This new model was compared with the previous model based on the original data to explore the impact of data dimensionality reduction on the model classification performance. All the three classification models (RF, SVM, and SVM-K) underwent 10-fold cross-validation
 |
Figure 2 Top 10 peaks as per importance and intergroup proportion. |
Model Evaluation
The models were evaluated for accuracy, specificity, sensitivity, and area under the subject working characteristic curve (AUC). Meanwhile, the effect of dimension reduction on the model performance was investigated based on the change in average accuracy.
Data Availability
The raw data supporting the conclusions of this manuscript are available upon request from the corresponding author.
Results
MALDI-TOF MS and Relevant Features
CRKP and CSKP were identified using MALDI-TOF MS (BioMerieux, France); 171 K. pneumoniae strains were successfully identified. Based on SARAMIS software and feature selection, 211 peak features were obtained, including 47 housekeeping peaks. Figure 3 shows the top 10 importance peak features and their proportion among different groups according to the RF algorithm. The SVM-K dimension reduction model was constructed based on the K optimal feature selection. The final optimal K dimension reduction ranged between 105 and 153.
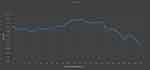 |
Figure 3 A plot of accuracy fluctuations in model construction by continuously removing the lowest-ranked features. For the range 105–153, the model accuracy was >0.9. |
Performance of the Models
Figure 4 shows the evaluation performance of the 3 classification models, including accuracy, sensitivity, and specificity. The optimized RF, SVM, and SVM-K models showed good classification performance. Among them, the SVM-K model with the best results for accuracy (0.91), sensitivity (0.89), and specificity (0.94) exhibited overall best classification performance. The accuracy of RF and SVM models was 0.88. The sensitivity of the SVM model was slightly higher (0.85), while the specificity (0.92) was slightly lower than the RF model. The relative performance of the model was difficult to distinguish direct from the boxplots. Therefore, a more intuitive comparison of the models’ performances was made using the area under the subject worker curve (AUC). Figure 5 shows AUC plots of the 3 classification models. The SVM-K model revealed the best classification performance (AUC=0.9356); the SVM model (AUC=0.9298) slightly outperformed the RF model (AUC=0.9013).
 |
Figure 5 AUC plots of the 3 classification models. |
Discussion
CRKP is a serious threat to public health. With the limited and poor efficacy of antimicrobials, the mortality rate of CRKP patients is much higher than that of CSKP infections.10–12 Also, CRKP-infected patients need longer hospital stays and face a higher economic burden.13 Therefore, rapid drug resistance testing methods are urgently needed.
Traditional drug resistance tests require a drug sensitivity test after bacterial culture isolation, which is time-consuming and costly. In this study, the total mass spectrometric data of CRKP and CSKP were directly classified by MALDI-TOF MS using the RF and SVM algorithms. Strains were cultured for 18–24 h following the regular microbiology laboratory practice. This method can detect carbapenem resistance with a daily laboratory strain identification system saving time without the need for additional drug susceptibility tests. Other rapid resistance detection methods, including the disappearance and appearance of characteristic antimicrobials hydrolysis peaks based on MALDI-TOF MS data, also demonstrated good results.14 However, that approach requires additional culture time after bacterial isolation.
Furthermore, the method cannot detect the mechanisms of carbapenem resistance beyond carbapenemases, such as changes in pore exchange and outflow pump system. Complex resistance mechanisms lead to the K. pneumoniae resistant phenotype. However, there may not be a beta-lactamase or carbapenemase within the bacteria. Therefore, the hydrolysis of antimicrobials does not necessarily occur during the co-culture of CRKP, and no corresponding change in related antimicrobials hydrolysis peak was found in the MALDI-TOF MS data increasing false-negative rate. Most recent studies investigated the relevant resistance genes before K. pneumoniae were co-cultured with related antimicrobials to address this problem. This increases the accuracy of the experimental results but is unrealistic for routine laboratory testing. In contrast, the present study classified data based on drug resistance phenotype and later performed direct analysis using machine learning algorithms. This approach does not consider the specific resistance mechanisms of K. pneumoniae and is more suitable for routine laboratory tests.
As shown in Figure 2, the top 10 peaks of importance and their share in both CRKP and CSKP. Among these, the peaks of the highest importance at 6515 Da appeared in 56.8% of the CRKP isolates and 5.3% of the CSKP isolates. However, previous studies indicated that CRKP and CSKP peaks at 7705.009 Da were significantly different, appearing at 80.4% in CRKP isolates and 2.2% in CSKP isolates.16 The 7705.009 Da peaks in our experiment appeared in 95.8% of CRKP isolates and 69.7% of CSKP isolates; the difference was not significantly higher. This could be due to the different preliminary data processing methods of the two studies. They processed the data directly via the mass-up (http://sing.ei.uvigo.es/mass-up) software, which may fail to correct the peak misalignment. The MALDI-TOF mass spectrometer inevitably causes polarisation during K. pneumoniae data acquisition, leading to subtle alterations in mass spectrometry data.
For instance, in this study, the peak polarisation at 6515 Da ranged between 6512 and 6518 Da. The tolerance peaks (± 3 Da) were grouped to balance these differences, and the features were selected manually for stable and accurate results.9 Further studies with more strains are required to determine whether relevant characteristic peaks such as 6515 Da can be a potential biomarker. The model constructed in this study based on MALDI-TOF MS data can be equally applied to other classification problems to detect other resistance.
Forty-seven peaks were common in almost all spectra, called the K. pneumoniae housekeeping peaks. These housekeeping peaks are not helpful for the model’s classification performance and will drag them down. The dimension reduction processing of the total data significantly improved the model’s classification performance. Previous studies applied visual examination methods, PCA, and Lasso regression for dimension reduction of MALDI-TOF MS data.9,15 We used the feature elimination dimension reduction mode for the models’ performance analysis. The dimensionality reduction model SVM-K runs once per cycle to remove the last importance-ranked feature until all features get removed. Figure 3 shows the change in accuracy obtained during constant dimensionality reduction. For the dimensionality reduction range 105–153, the SVM-K model achieved an accuracy of >0.90, indicating the best classification performance. Notably, dimension reduction >153 times reduced the model accuracy. This indicated that too much lowering of data dimension reduction is not always better as some hidden important information may also be lost, which could be unfavorable for classification model building by data mining.
In several previously reported studies, the model was passed cross-validation (5-fold, 10-fold) to avoid overfitting.15,16 In the present study, 10-fold cross-validation was used to improve the model stability. The final results of accuracy, sensitivity, and specificity of the models were an average of 100 times run on a 10-fold cross-validation basis, which greatly improved the data stability.
All three classification models constructed in this study showed good classification performance with an average accuracy of >0.88. The SVM-K model after dimension reduction showed the best performance with an average accuracy of 0.91. Although the model classification performance is excellent, its universality needs further study. Due to limited capital, time, complex data acquisition, and analysis, we could not conduct external data validation to assess the universality of our model. Many other studies could not verify external data.9,15,16 Since most data analysis in these studies was performed by the marketed (company provided) software, such as file analysis and ClinProTools, re-analysis and external validation of data becomes quite difficult.17,18
Our CRKP and CRSP identification methods are simple, rapid, economical, and suitable for conventional laboratory diagnosis. In addition, K. pneumoniae made 100 shots on the instrument, and the data stability was very reliable. These advantages make it possible to incorporate this approach into clinical practice without changing the current protocol. However, since we only evaluated the classification performance of CRKP and CSKP, other bacteria and antimicrobial agents need to be tested for the universality of the method.
Conclusion
This study demonstrates that machine learning algorithms combined with the MALDI-TOF MS platform can rapidly distinguish between CRKP and CSKP isolates. This method is quick, accurate, and provides valuable information. The carbapenem resistance can be directly predicted based on the MALDI-TOF MS data.
Ethical Approval
There is no ethical concern in this study, and The Medical Ethics Committee approved this experiment at the First Affiliated Hospital of Anhui Medical University. Written informed consent was obtained from patients under the Declaration of Helsinki.
Acknowledgments
This work was financially supported by the Provincial Natural Science Research project of universities in Anhui Province (grant number: KJ2015A337).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Wu X, Shi Q, Shen S, Huang C, Wu H. Clinical and bacterial characteristics of Klebsiella pneumoniae affecting 30-day mortality in patients with bloodstream infection. Front Cell Infect Microbiol. 2021;11:688989. doi:10.3389/fcimb.2021.688989
2. Karampatakis T, Antachopoulos C, Iosifidis E, et al. Molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae in Greece. Future Microbiol. 2016;11:809–823. doi:10.2217/fmb-2016-0042
3. Ventola CL. The antimicrobials resistance crisis: part 1: causes and threats. Pharm Ther. 2015;40:277–283.
4. Haigh JD, Green IM, Ball D, et al. Rapid identification of bacteria from bioMérieux BacT/ALERT blood culture bottles by MALDI-TOF MS. Br J Biomed Sci. 2013;70:149–155. doi:10.1080/09674845.2013.11669949
5. Sakarikou C, Ciotti M, Dolfa C, et al. Rapid detection of carbapenemase-producing Klebsiella pneumoniae strains derived from blood cultures by Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry (MALDI-TOF MS). BMC Microbiol. 2017;17(1):54. doi:10.1186/s12866-017-0952-3
6. Fangous M-S, Mougari F, Gouriou S, et al. Classification algorithm for subspecies identification within the Mycobacterium abscessus species, based on matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2014;52:3362–3369. doi:10.1128/JCM.00788-14
7. Mather CA, Werth BJ, Sivagnanam S, et al. Rapid detection of vancomycin-intermediate Staphylococcus aureus by Matrix-assisted laser desorption ionization-time of flight massspectrometry. J Clin Microbiol. 2016;54(4):883–890. doi:10.1128/JCM.02428-15
8. CLSI. Performance Standards for Antimicrobial Susceptibility Testing.
9. Pena I, Pena-Vina E, Rodriguez-Avial I, et al. Comparison of performance of MALDI-TOF MS and MLST for biotyping carbapenemase-producing Klebsiella pneumoniae sequence types ST11 and ST101 isolates [published online ahead of print, 2020 Dec 15]. Enferm Infecc Microbiol Clin. 2020;40(4):172–178.
10. Hussein K, Raz-Pasteur A, Finkelstein R. Impact of carbapenem resistance on the outcome of patients’ hospital-acquired bacteraemia caused by Klebsiellapneumoniae. J Hosp Infect. 2013;83(83):307–313. doi:10.1016/j.jhin.2012.10.012
11. Li C, Wen X, Ren N, et al. Point-prevalence of healthcare-associated infection in China in 2010: a large multicenter epidemiological survey. Infect Control Hosp Epidemiol. 2014;35(11):1436–1437. doi:10.1086/678433
12. Meng X, Liu S, Duan J, et al. Risk factors and medical costs for healthcare-associated carbapenem-resistant Escherichia coli infection among hospitalized patients in a Chinese teaching hospital. BMC Infect Dis. 2017;17(1):82. doi:10.1186/s12879-016-2176-9
13. Arato V, Raso MM, Gasperini G, et al. Prophylaxis and treatment against Klebsiella pneumoniae: current insights on this emerging anti-microbial resistant global threat. Int J Mol Sci. 2021;22(8):4042. doi:10.3390/ijms22084042
14. Wang G, Song G, Xu Y. A rapid antimicrobial susceptibility test for Klebsiella pneumoniae using a broth micro-dilution combined with MALDI TOF MS. Infect Drug Resist. 2021;14:1823–1831. doi:10.2147/IDR.S305280
15. Liu X, Su T, Hsu YS, et al. Rapid identification and discrimination of methicillin-resistant Staphylococcus aureus strains via matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 2021;35(2):e8972. doi:10.1002/rcm.8972
16. Huang TS, Lee SS, Lee CC, et al. Detection of carbapenem-resistant Klebsiella pneumoniae on the basis of matrix-assisted laser desorption ionization time-of-flight mass spectrometry by using supervised machine learning approach. PLoS One. 2020;15(2):e0228459. doi:10.1371/journal.pone.0228459
17. Esener N, Green MJ, Emes RD, et al. Discrimination of contagious and environmental strains of Streptococcus uberis in dairy herds by means ofmass spectrometry and machine-learning. Sci Rep. 2018;8:17517. doi:10.1038/s41598-018-35867-6
18. Pérez-Sancho M, Vela AI, Horcajo P, et al. Rapid differentiation of Staphylococcus aureus subspecies based on MALDI-TOF MS profiles. J Vet Diagn Invest. 2018;30:813–820. doi:10.1177/1040638718805537
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

