Back to Journals » OncoTargets and Therapy » Volume 13
Rap2c as a Novel Biomarker for Predicting Poor Prognosis in Glioma
Authors Wang X, Wang C, Xi L , Yu Z
Received 30 January 2020
Accepted for publication 26 March 2020
Published 14 April 2020 Volume 2020:13 Pages 3073—3083
DOI https://doi.org/10.2147/OTT.S247731
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Takuya Aoki
Xiucun Wang,1,2,* Cheng Wang,2,* Lin Xi,2 Zhengquan Yu1
1Department of Neurosurgery, The First Affiliated Hospital of Soochow University, Suzhou, Jiangsu 215006, People’s Republic of China; 2Department of Neurosurgery, The Affiliated Hospital of Xuzhou Medical University, Xuzhou, Jiangsu 221002, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Zhengquan Yu Email [email protected]
Objective: Rap2c is a member of the Ras superfamily that has been implicated in various types of cancers. However, its role in glioma remains elusive. This study aimed to elucidate the role of Rap2c in glioma and its specific molecular mechanism.
Methods: We determined the expression of Rap2c in glioma tissues by Western blotting and immunohistochemistry (IHC) assays. The proliferation and apoptosis of cells were explored using CCK-8 and flow cytometry assay, whereas the migration and invasion of glioma cells were determined using transwell assay. The potential mechanism of Rap2c in the migration of glioma cell lines was investigated through Western blotting analysis and transwell assay. BALB/c nude mice were used to establish tumor models to test the effect of Rap2c on cancer metastasis in vivo.
Results: Our data showed that the protein expression of Rap2c was significantly up-regulated in glioma tissues compared with normal brain tissues, and Rap2c overexpression negatively correlated with 5-year overall survival rate. However, there was no correlation between Rap2c expression and clinicopathological parameters of glioma patients. Overexpression of Rap2c promoted the migration and invasion abilities of glioma cells but had no significant effect on the proliferation of glioma cells. Western blotting analysis revealed that Rap2c overexpression increased the phosphorylation level of extracellular signal-related kinase1/2 (ERK1/2), and this effect was abolished with U0126, a selective MEK inhibitor. Furthermore, overexpression of Rap2c induced lung metastasis of glioma cells in xenograft models.
Conclusion: These findings indicate that high Rap2c expression predicts poor prognosis in glioma. Rap2c-mediated ERK1/2 phosphorylation initiates EMT cascade and promotes migration and invasion of glioma cells. Thus, targeting Rap2c and ERK signaling pathway could be a novel treatment modality for glioma.
Keywords: Rap2c, migration, invasion, MAPK, ERK, glioma
Introduction
Gliomas comprise the most common type of primary malignant brain tumor, and except for pilocytic astrocytoma and subependymal giant cell astrocytoma, nearly all are characterized by high recurrence rates, high mortality rates, and short survival times.1 Of note, the 5-year survival rate of glioma patients postdiagnosis is only 5.5% whereas the median survival is approximate 14.5–16.6 months. These statistics indicate poor prognosis of this cancer despite availability of multiple treatments including surgery, radiotherapy and chemotherapy.2 Several molecular pathways and therapeutic targets have been proposed for glioma, but the clinical translation of such targets has been challenging.3,4 This points to the need to unravel the molecular mechanisms that drive glioma progression and metastasis and expose novel therapeutic targets for glioma.
Rap2c, a fifth member of the Rap superfamily of small GTP-binding proteins, was first identified by Paganini et al.5 Similar to other members of Ras family, Rap proteins work as molecular switches of multiple signal transduction cascades and play a pivotal role in cell/matrix, cell/cell adhesion and cytoskeletal rearrangement.6,7 Compelling evidence showed that Rap2a and Rap2b were involved in tumor progression.8,9 Hence, there has been renewed interest to study the novel oncogenic roles of Rap2 family.10 Zhang et al indicated that the expression levels of Rap2b was predominantly upregulated in many types of tumors, and supported the oncogenic status.11 Subsequent studies demonstrated that Rap2a was significantly increased in various human tumors and involved in cancer cell migration and invasion.8 However, little is known about the role of Rap2c in cancers, although Rap2c gene has more than 90% sequence homology with Rap2b and Rap2a.5
Different intracellular signaling pathways regulate the proliferation, differentiation and motility of cancer cells, among which the role of Ras/Raf/MAP kinase-ERK kinase (MEK)/extracellular signal-regulated kinase (ERK) (MAPK) pathway in the pathogenesis of cancer has long been established.12 Ras protein binds to the plasma membrane in its active form and recruits Raf. Then, Raf phosphorylates and activates MEK, which in turn phosphorylates ERK. Therefore, MAPK/ERK pathway is a convergent signaling node receiving input from numerous stimuli, including DNA damage pathways and internal metabolic stress. Epithelial-mesenchymal transition (EMT) is a process in which epithelial cells acquire a mesenchymal phenotype with increased migration and invasion abilities, and MAPK/ERK signaling pathway is involved in this process.13,14 Recent study shows that Rap2c promotes proliferation and inhibits apoptosis of breast cancer cells via MAPK signaling pathway.15 Considering that Rap2c is a member of the Ras superfamily of GTPases, we speculate that there is a relationship between Rap2c and MAPK/ERK signaling pathway which influence the migration of glioma cells.
In the current study, we compared the expression of Rap2c in glioma tissues and cell lines, and explored whether its expression correlated with clinicopathological characteristics of glioma patients. We found that high expression of Rap2c was significantly associated with glioma occurrence. In addition, activation of Rap2c altered the expression of EMT markers and promoted invasion and metastasis of glioma cells through phosphorylating ERK. This study revealed that the novel Rap2c-ERK signaling pathway promoted migration and metastasis of glioma cells.
Materials and Methods
Patients and Specimens
All experiments involving human participants were approved by the Ethics Committee of the Affiliated Hospital of Xuzhou Medical University. All patients provided written informed consent to participate in the study. Tissue microarray (TMA) cohorts were utilized in this study. TMA, including 15 normal brain tissues and 180 glioma tissues, was obtained from Shanghai Xinchao Biotechnology (Shanghai, China). The array dot diameter was 1.5 mm and each dot represented a tissue spot from one individual specimen that was pathologically confirmed. According to the WHO criteria for pathologic grading of tumors, 105 cases of malignant tumors were Grade I-II whereas 75 cases of malignant tumors were Grade III–IV.
Immunohistochemistry
The streptavidin-peroxidase (Sp) method was employed to perform IHC of tumor tissues using the Sp Kit (Zhongshan Biotech,China). Paraffin-embedded slides were dewaxed at 60°C for 2h followed by 15 min washes with dimethylbenzene. Next, the slide was rehydrated with graded ethanol and distilled water. For antigen retrieval, the slides were heated at 95°C for 30 min in 0.01M citrate buffer (pH=6.0). Endogenous peroxidase activity was inhibited by incubation with 3% hydrogen peroxide for 20 min. After blocking for 30 min with 10% normal goat serum, the sections were incubated with polyclonal rabbit anti-Rap2c antibody (1:100 dilution; Abcam) overnight at 4°C. Next, the slides were incubated with a biotinylated secondary antibody (Zhongshan Biotech, China) at room temperature for 30 min. The slides were then treated with avidin-peroxidase reagent and 3, 3ʹ-diaminobenzidine substrate (DAB, Zhongshan Biotech, China). Subsequently, slides were counterstained with hematoxylin, dehydrated, and sealed with cover slips. Negative controls were obtained by phosphate buffered saline (PBS) with non-immune serum. Results of Rap2c staining were analyzed by two pathologists independently who were blinded to each other.Any discrepancies in the score results were resolved by consensus. Intensity scores of Rap2c staining were assigned as follows: 0 (negative), 1 (weak), 2 (moderate), and 3 (strong). The values of percentage of positive malignant cells were assigned as follows: 1 for 0–25%, 2 for 26–50%, 3 for 51–75% and 4 for 76–100%. The immunoreactive score (IRS) of each section was calculated by the products of the staining intensity and the percentage of tumor cells. Staining patterns were divided into two classes: negative (IRS:0–6), and positive (IRS: 8–12) based on IRS.
Cell Lines, Plasmids and Transfection
Human glioma cell lines (U87, U118 and U251) were purchased from Biochemistry and Cell Biology department, Chinese Academy of Sciences (Shanghai, China) and cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Hyclone,USA) supplemented with 10% fetal bovine serum (Invitrogen, USA). Normal human astrocyte cells were obtained from Sciencell Research Laboratories (Carlsbad,USA) and cultured in RPMI 1640 medium (Gibco,USA). These cells lines were incubated in a humidified incubator at 37°C with 5% CO2. The plasmid of pcDNA3.1-Rap2c was constructed by subcloning Rap2c cDNA into the eukaryotic expression vector pcDNA3.1 through two restriction enzyme sites EcoRI and Hind III, and confirmed by DNA sequencing. Subsequently, the pcDNA3.1 and pcDNA3.1-Rap2c were transiently transfected into U87 and U118 cells using Lipofectamine 2000 transfection reagent (Invitrogen, USA) following the manufacturer’s protocol. Stable cell lines were established by the infection with Rap2c-lentivirus (LV-Rap2c; GenePharma, China) and control-lentivirus (LV-Ctrl; GenePharma, China) and were selected with puromycin (Vicmed,China) for 4 weeks. Rap2c siRNA (5ʹ-GAAGCAAGAUCAGUGUUGUTT-3ʹ, 5’-ACAACACUGAUCUUGCUUCTT-3ʹ) was purchased from GenePharma (Shanghai, China). Transfection of Rap2c siRNA was carried out using siLentFect Lipid Reagent (Bio-Rad, USA) according to the manufacturer’s instructions. A non-specific siRNA (5ʹ-UUCUCCGAACGUGUCACGUTT-3ʹ, 5ʹ-ACGUGACACGUUCGGAGAATT-3ʹ was transfected as a control.
Cell Proliferation Assay
After transfection, the cells were seeded into 96-well culture plates at a density of 5×103/well. Cell proliferation was evaluated using Cell Counting Kit-8 (CCK8; BeyotimeBiotech, China) at the indicated time points. Briefly, 10 μL CCK-8 solutions and 100 μL serum-free culture medium were added to each well and incubated at 37°C for 2 h. The absorbance of the plate was read at 450 nm and the optical density values were used to calculate the degree of cell proliferation. All assays were repeated three times.
Apoptosis by Flow Cytometry with Annexin V-FITC/PI
The cells were plated in six-well plates and transfected with Rap2c expression plasmid for 24 h, or siRNA for 48 h. The rate of apoptosis was measured with Annexin V/FITC staining (KeyGen Biotech, China) using a fluorescence-activated cell sorting machine (FACSCaliburTM, USA). All procedures were carried out following the manufacturer’s instructions.
Western Blotting
Pre-treated cells were harvested and washed three times with PBS. They were digested with lysis buffer (Beyotime Biotech, China) and centrifuged to obtain total cellular protein. The concentration of proteins was determined using bicinchoninic acid (BCA) kit (Beyotime Biotech, China). Next, 80 μg proteins were separated by 12.5% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and electro-transferred to polyvinylidene fluoride (PVDF) membrane. The membrane was blocked for 2 h in 5% bovine serum albumin (BSA; Beyotime Biotech, China), and incubated overnight at 4 °C with the following primary antibodies: Rap2c (anti-rabbit; 1:1000; Abcam), MEK1/2 (anti-mouse; 1:1000; CST), p-MEK1/2 (anti-rabbit; 1:1000; CST), E-cadherin (anti-mouse; 1:1000; Proteintech), Vimentin (anti-mouse; 1:1000; Proteintech),ERK1/2 (anti-rabbit; 1:1000; CST), p-ERK1/2 (anti-rabbit; 1:1000; CST), β-actin (anti-mouse; 1:5000; Proteintech). Subsequently, the membranes were washed with PBS, and probed with corresponding HRP secondary antibodies for 2 h at room temperature. Finally, the protein bands were detected semiquantitatively with TanonTM High-sig ECL Western Blotting Substrate (Tanon, China).
Wound Healing Assay
Briefly, cells were seeded into 6-well plates in culture medium and reached about 90% confluence. A wound was created by dragging a 200 μL pipette tip along the center of the plate. The confluent monolayer was washed three times with PBS to remove debris and observed every 24 h using a light microscope (Nikon, Japan) at ×100 magnification. The rate of wound healing was determined by comparing the wound width at 24 h to the wound width at 0 h.
Cell Migration and Invasion Assay
Transwell chambers (BD Bioscience, USA) containing 8 μm pores were coated with or without Matrigel (BD Biosciences, USA) for migration and invasion assays, respectively. Briefly, 1×104 U87 cells orU118 cells was suspended in serum-free medium and put in the upper chamber. After incubation for 24 h at 37°C, cells in the upper chamber were carefully removed and migrated cells were fixed in methanol, and stained with crystal violet. The migration and invasion of glioma cells were calculated and photographed under a light microscopy at ×200 magnification. All assays were repeated three times.
Xenograft Mouse Metastatic Model
Animal experiments were performed in strict accordance with the protocols approved by the Institutional Animal Care and Use Committee of Xuzhou Medical University. Twenty female BALB/c nude mice were purchased from Huafukang Biotechnology (Beijing, China) and randomly divided into experimental groups. Stable U87 cells (LV-Ctrl and LV-Rap2c) were diluted to 2×106/100μL PBS and injected into mice via the lateral tail vein. After 5 weeks, all mice were euthanized and lungs were then excised and photographed. The number of metastatic tumors per lung and its weight were counted and recorded. Finally, the lung tissues were harvested and prepared for H&E and Western blotting.
Statistical Analysis
All analyses were performed using SPSS 16.0 software (SPSS, USA). Quantitative data were presented as means ± SD (standard deviation). Means of different groups were compared using Student’s t-tests or one-way analysis of the variance (ANOVA). The association between Rap2c staining and the clinicopathologic parameters of the glioma patients was determined by χ2 test. Survival analysis was estimated by the Kaplan-Meier method and the Log rank test. All experiments were performed at least three times unless otherwise indicated. P value<0.05 was considered statistically significant.
Results
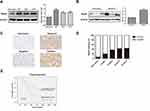
Rap2c Is Upregulated in Glioma
Western blotting results revealed that Rap2c protein level was significantly higher in diverse glioma cell lines (U87, U118 and U251) than in astrocyte cells (Figure 1A). Given the high expression of Rap2c in U87 and U118 cells, these cells were used in subsequent experiments. Rap2c was also highly expressed in glioma tissues (Figure 1B). To further explore the correlation between Rap2c expression and glioma progression, we conducted IHC staining using 15 normal brain tissues and 180 malignant tumor tissues (Grade I–IV). We found that Rap2c staining was more intense in malignant glioma tissues, while its staining was weaker in normal brain tissue (Figure 1C). Positive Rap2c staining was recorded in 37.2% (67 of 180 cases) in glioma tissues. Of the non-cancerous normal tissues from 15 patients, positive Rap2c expression was observed in 6.7% (1 of 15 cases) (Figure 1D). These results suggested that Rap2c expression was up-regulated in both glioma cells and tissues. We further performed Kaplan-Meier survival analysis and Log rank test based on Rap2c expression. The result showed that high Rap2c expression negatively correlated with 5-year overall survival rate (Figure 1E, P<0.05), suggesting that Rap2c predicts poor outcomes in glioma patients.
Correlation of Rap2c Expression with Clinicopathological Parameters
The clinical relationship between Rap2c expression and clinicopathological parameters in glioma was further analyzed to explore the importance of Rap2c expression. However, we did not find significant correlations between Rap2c expression and WHO grade (P=0.112) or histologic type (P=0.359). In addition, Rap2c expression was not significantly correlated with other clinicopathologic variables, such as patient age (P=0.836) and gender (P=0.591) (Table 1).
 |
Table 1 The Correlation Between Rap2c Expression and Clinicopathological Characteristics Based on IHC Analysis |
Rap2c Has No Effect on the Proliferation of Glioma Cells
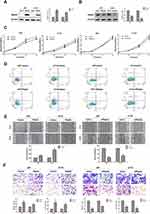
We further determined whether Rap2c regulates the growth of glioma cells in vitro. Initially, pcDNA3.1 control and pcDNA3.1-Rap2c plasmids were transiently transfected into both U87 and U118 cells. After 24 h transfection, Rap2c protein was significantly increased (Figure 2A). In contrast, siRNA transfection led to a marked reduction in Rap2c expression in glioma cells (Figure 2B). Cell viability was determined by CCK-8 and Annexin V-FITC/PI assays. The result showed that Rap2c had no significant effect on cell proliferation ability (Figure 2C). Expectedly, cell apoptosis was not different between Rap2c and control groups (Figure 2D).
Effect of Rap2c on Invasion and Migration of Glioma Cells
Given that there are significant correlations between increased Rap2c expression and aggressive features in glioma, we inferred that Rap2c enhanced tumor progression. It is noteworthy that malignant gliomas are aggressive and lethal neoplasms featured by the rapid growth and persistent infiltration.3 In this study, wound healing assay revealed that Rap2c overexpression accelerated wound healing, as evidenced by the higher rate of wound healing in Rap2c overexpressing cells compared to cells transfected with the control vector. Similarly, the wound filling was obviously retarded in glioma cells with siRap2c (Figure 2E). In addition, transwell assay showed that overexpression of Rap2c markedly strengthened the migration and invasion abilities of glioma cells compared with negative control vectors. Knockdown of Rap2c effectively reduced the number of invaded cells in both cell lines (Figure 2F).
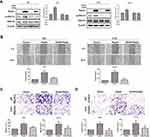
Rap2c-Mediated ERK Phosphorylation Alters Expression of EMT Markers in Glioma Cells
Based on the results from cell migration and invasion assays, we explored the mechanism by which Rap2c influenced these cellular processes. The transdifferentiation of epithelial cells into motile mesenchymal cells, a process known as EMT, contributes to fibrosis formation and cancer progression. Western blotting results showed that high Rap2c protein expression decreased the expression of E-cadherin, and enhanced the protein level of vimentin (Figure 3A). These results were consistent with those of transwell assays shown in Figure 2E-F. Since ERK phosphorylation results in the activation of multiple substrates that are responsible for cell movement and adhesion, we wondered whether Rap2c communicates with MAPK/ERK signaling pathway. As shown in Figure 3B, Rap2c overexpression resulted in the up-regulation of MEK and ERK phosphorylation (Figure 3B). These findings suggested that MEK/ERK might be a downstream target of Rap2c that mediates Rap2c-induced migration and invasion of glioma cells.
Rap2c Promotes Migration and Invasion of Glioma Cells via Rap2c-ERK Signaling Pathway
To further determine whether the effects of Rap2c on migration of glioma cells were mediated by the MAPK/ERK signaling pathway, cells were treated with U0126, a selective MEK1/2 inhibitor, and then the migration and invasion abilities of cells were detected. As expected, selective inhibition of MEK significantly blocked Rap2c-mediated increase in ERK phosphorylation (Figure 4A). Moreover, U0126 pretreatment canceled the ability of Rap2c to enhance the migration and invasion of glioma cells (Figure 4B-D). Taken together, these results indicated that ERK works as a downstream signaling molecule for Rap2c to promote EMT in glioma cells in vitro.
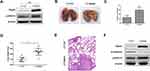
Rap2c Promotes Glioma Metastasis via Regulating ERK Phosphorylation in Xenograft Model
To further determine whether Rap2c can promote the metastasis of glioma cells in vivo, we developed metastasis models of murine xenograft. U87 cells were transfected with lentivirus encoding Rap2c and then intravenously injected into 6-week-old BALB/c nude mice via tail. Western blotting was performed to detect the protein levels of ERK and Rap2c in stable U87 cells before intravenous injection (Figure 5A). After five weeks of feeding, all mice were sacrificed and numbers of metastatic nodules on the lung surfaces were counted. The number of lung metastasis in LV-Rap2c group was markedly higher compared with LV-Ctrl group (Figure 5B-C). Additionally, the lung weights in LV-Rap2c group were higher than that in LV-Ctrl group (Figure 5D). H&E staining confirmed that the nidi in mice lungs were metastatic tumors (Figure 5E). Consistently, western blotting showed that treatment with Rap2c increased ERK phosphorylation and decreased E-cadherin protein in lungs (Figure 5F). Taken together, these results indicated that Rap2c overexpression enhanced the metastatic capability of glioma cells through up-regulating phosphorylated ERK in vivo.
Discussion
Gliomas are the most common malignant brain tumors. The available treatment options are not effective enough, leading to poor prognosis. Ras family of small GTPases regulates various cellular processes including cell adhesion, cell cycle control, differentiation, cytoskeletal organization and metabolic turnover.16–18 Its subfamily Rap2 includes three members: Rap2a, Rap2b and Rap2c.19 So far, Rap2b and Rap2a have been reported to influence the development of tumor.8,9 Wu et al demonstrated that Ras-related protein Rap2c promotes the migration and metastasis of osteosarcoma cells via regulating Akt signaling pathway.20 Subsequent studies confirmed that miR-188-5p inhibits the proliferation of breast cancer cells by targeting Rap2c.15 However, the effects of Rap2c on tumorigenesis and metastasis of glioma have never been reported by others.
In this study, we assessed whether Rap2c participates in the pathogenesis of malignant brain tumors. Our results showed that, similar to Rap2a and Rap2b, Rap2c was highly expressed in glioma cells than in normal astrocyte cells. Strong Rap2c staining was detected in glioma tissues, whereas absent or low expression was showed in most normal brain tissues and low-grade glioma. We further evaluated the association of Rap2c expression with the clinicopathological features and prognostic survival in glioma. Our results showed that high levels of Rap2c strongly reduced the overall survival of patients with glioma. These indicated that Rap2c plays a pivotal role in tumorigenesis and progression of glioma, which is consistent with previous studies to support cancer-promoting characters of Rap2c.20 However, there was no difference in Rap2c expression between high-grade gliomas, and Rap2c expression was not significantly associated with age, gender, WHO grade and histologic type. This may be due to the limited number of glioma samples or the limitation of Rap2c to distinguish high-grade malignancies.
MAPK pathway has four signaling families, the MAPK/ERK family, c-Jun N-terminal kinases (JNK), p38 and Big MAP kinase-1 (BMK-1).2 Generically, these kinases are named MAPK kinase-kinase (MAPKKK), MAPK kinase (MAPKK) and MAPK from upstream to downstream, whereas MAPKK consists of MEK1 and MEK2. Finally, further downstream are ERK1/2, which are the final effectors of the MAPK pathway.21 Numerous evidences showed that MAPK/ERK pathway was associated with cell proliferation, differentiation, migration and EMT of glioma.22,23 Furthermore, Zhu et al found that miR-188-5p promotes apoptosis and inhibits the proliferation of breast cancer cells via the MAPK signaling pathway by targeting Rap2c.15 We, therefore, speculate that Rap2c, as a member of Ras family, could recruit Raf to the cell membrane to activate MEK, which in turn phosphorylates MAPK/ERK, and co-regulates the progression of glioma. Our results showed that overexpression of Rap2c promoted migration and invasion abilities of tumor cells but did not affect the proliferation of glioma cells. Animal model of glioma metastasis also confirmed that Rap2c promoted lung metastasis in vivo. Moreover, mesenchymal marker, vimentin was significantly increased in U87 and U118 cells with Rap2c overexpression. Conversely, epithelial marker, E-cadherin was reduced by Rap2c upregulation. Mechanistically, Rap2c increased phosphorylation of MEK1/2 and ERK 1/2.
Considering that Rap2c overexpression increased the level of ERK phosphorylation, we used U0126, a selective blocker of its upstream kinase MEK, to observe the changes in migration and invasion of glioma cells by Rap2c. Our data showed that ERK1/2 phosphorylation induced by Rap2c was blocked by U0126, whereas the total levels remain unchanged. Furthermore, migration and invasion abilities of glioma cells induced by Rap2c can be suppressed by inhibitor U0126. These results suggested that ERK signaling pathway positively mediated Rap2c-induced cell migration and invasion. Consistent with this result, metastasis models of Rap2c overexpression showed that higher level of ERK phosphorylation and lower level of E-cadherin. This indicates that Rap2c mediated MAPK/ERK signaling pathway and facilitated cell migration by downregulating E-cadherin. Nevertheless, further experiments are needed to establish the precise molecular mechanism of Rap2c-induced invasion and target Rap2c for anti-tumoral effect in vivo.
Conclusion
In summary, our results provide evidence for the emerging connections among Rap2c, EMT markers and glioma metastasis mediated by ERK. Rap2c is highly expressed in glioma tissues and cell lines. High expression of Rap2c is significantly linked to the progression of glioma and it could act as a prognostic factor of glioma. Rap2c induces EMT and enhances metastasis of glioma cells by phosphorylating ERK. Therefore, targeting Rap2c-ERK may be a potential therapeutic strategy for glioma.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Ethics and Consent Statement
Written informed consent was obtained from each patient and the study was approved by the Ethics Committee of the Affiliated Hospital of Xuzhou Medical University.
Funding
This research was not supported by any funding agency in the public, commercial, or not-for-profit sectors.
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Ostrom QT, Gittleman H, Stetson L, Virk S, Barnholtz-Sloan JS. Epidemiology of intracranial gliomas. Prog Neurol Surg. 2018;30:1–11. doi:10.1159/000464374
2. Sun Y, Liu WZ, Liu T, Feng X, Yang N, Zhou HF. Signaling pathway of MAPK/ERK in cell proliferation, differentiation, migration, senescence and apoptosis. J Recept Signal Transduct Res. 2015;35(6):600–604. doi:10.3109/10799893.2015.1030412
3. Furnari FB, Fenton T, Bachoo RM, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21(21):2683–2710. doi:10.1101/gad.1596707
4. Krakstad C, Chekenya M. Survival signalling and apoptosis resistance in glioblastomas: opportunities for targeted therapeutics. Mol Cancer. 2010;9:135. doi:10.1186/1476-4598-9-135
5. Paganini S, Guidetti GF, Catricala S, et al. Identification and biochemical characterization of Rap2C, a new member of the Rap family of small GTP-binding proteins. Biochimie. 2006;88(3–4):285–295. doi:10.1016/j.biochi.2005.08.007
6. Volk L, Chiu SL, Sharma K, Huganir RL. Glutamate synapses in human cognitive disorders. Annu Rev Neurosci. 2015;38:127–149. doi:10.1146/annurev-neuro-071714-033821
7. Wittchen ES, van Buul JD, Burridge K, Worthylake RA. Trading spaces: rap, Rac, and Rho as architects of transendothelial migration. Curr Opin Hematol. 2005;12(1):14–21. doi:10.1097/01.moh.0000147892.83713.a7
8. Wu JX, Zhang DG, Zheng JN, Pei DS. Rap2a is a novel target gene of p53 and regulates cancer cell migration and invasion. Cell Signal. 2015;27(6):1198–1207. doi:10.1016/j.cellsig.2015.02.026
9. Di J, Huang H, Qu D, et al. Rap2B promotes proliferation, migration, and invasion of human breast cancer through calcium-related ERK1/2 signaling pathway. Sci Rep. 2015;5:12363. doi:10.1038/srep12363
10. Fu G, Liu Y, Yuan J, et al. [Identification and functional analysis of a novel candidate oncogene RAP2B in lung cancer]. Zhongguo Fei Ai Za Zhi. 2009;12(4):273–276. doi:10.3779/j.issn.1009-3419.2009.04.03
11. Zhang X, He Y, Lee KH, et al. Rap2b, a novel p53 target, regulates p53-mediated pro-survival function. Cell Cycle. 2013;12(8):1279–1291. doi:10.4161/cc.24364
12. De Luca A, Maiello MR, D’Alessio A, Pergameno M, Normanno N. The RAS/RAF/MEK/ERK and the PI3K/AKT signalling pathways: role in cancer pathogenesis and implications for therapeutic approaches. Expert Opin Ther Targets. 2012;16 Suppl 2:S17–S27. doi:10.1517/14728222.2011.639361
13. Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2(6):442–454. doi:10.1038/nrc822
14. Chen Y, Deng G, Fu Y, et al. FOXC2 promotes oxaliplatin resistance by inducing epithelial-mesenchymal transition via MAPK/ERK signaling in colorectal cancer. Onco Targets Ther. 2020;13:1625–1635. doi:10.2147/OTT.S241367
15. Zhu X, Qiu J, Zhang T, et al. MicroRNA-188-5p promotes apoptosis and inhibits cell proliferation of breast cancer cells via the MAPK signaling pathway by targeting Rap2c. J Cell Physiol. 2020;235(3):2389–2402. doi:10.1002/jcp.29144
16. Itoh M, Nelson CM, Myers CA, Bissell MJ. Rap1 integrates tissue polarity, lumen formation, and tumorigenic potential in human breast epithelial cells. Cancer Res. 2007;67(10):4759–4766. doi:10.1158/0008-5472.CAN-06-4246
17. Lin KB, Tan P, Freeman SA, Lam M, McNagny KM, Gold MR. The Rap GTPases regulate the migration, invasiveness and in vivo dissemination of B-cell lymphomas. Oncogene. 2010;29(4):608–615. doi:10.1038/onc.2009.345
18. Park HO, Chant J, Herskowitz I. BUD2 encodes a GTPase-activating protein for Bud1/Rsr1 necessary for proper bud-site selection in yeast. Nature. 1993;365(6443):269–274. doi:10.1038/365269a0
19. Goitre L, Trapani E, Trabalzini L, Retta SF. The Ras superfamily of small GTPases: the unlocked secrets. Methods Mol Biol. 2014;1120:1–18. doi:10.1007/978-1-62703-791-4_1
20. Wu J, Du W, Wang X, et al. Ras-related protein Rap2c promotes the migration and invasion of human osteosarcoma cells. Oncol Lett. 2018;15(4):5352–5358. doi:10.3892/ol.2018.7987
21. Robinson MJ, Cobb MH. Mitogen-activated protein kinase pathways. Curr Opin Cell Biol. 1997;9(2):180–186. doi:10.1016/s0955-0674(97)80061-0
22. Tian Y, Guan Y, Jia Y, Meng Q, Yang J. Chloride intracellular channel 1 regulates prostate cancer cell proliferation and migration through the MAPK/ERK pathway. Cancer Biother Radiopharm. 2014;29(8):339–344. doi:10.1089/cbr.2014.1666
23. Pan HC, Jiang Q, Yu Y, Mei JP, Cui YK, Zhao WJ. Quercetin promotes cell apoptosis and inhibits the expression of MMP-9 and fibronectin via the AKT and ERK signalling pathways in human glioma cells. Neurochem Int. 2015;80:60–71. doi:10.1016/j.neuint.2014.12.001
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.