Back to Journals » Cancer Management and Research » Volume 10
Quantifying the prognostic significance of microRNA-17/17-5P in cancers: a meta-analysis based on published studies
Authors Duan F , Yang Y , Liu W, Zhao J, Song X, Li L , Li F
Received 22 January 2018
Accepted for publication 29 March 2018
Published 17 July 2018 Volume 2018:10 Pages 2055—2069
DOI https://doi.org/10.2147/CMAR.S163415
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Harikrishna Nakshatri
Fujiao Duan,1 Yang Yang,2 Weigang Liu,3 Jie Zhao,4 Xiaoqin Song,4 Lifeng Li,4 Fuqin Li2
1Medical Research Office, Affiliated Cancer Hospital of Zhengzhou University, Zhengzhou 450008, Henan, China; 2Department of Nosocomial Infection Management, First Affiliated Hospital of Zhengzhou University, Zhengzhou, 450052, China; 3Medical Record Statistics Office, Affiliated Hospital of Hebei University of Engineering, Handan, Hebei 056002, China; 4Center of Telemedicine, First Affiliated Hospital of Zhengzhou University, Zhengzhou, 450008, China
Objective: The aim of this study was to investigate the prognostic value of mircoRNA-17 and mircoRNA-17-5P (miR-17/17-5P) in patients with cancer.
Materials and methods: We conducted a comprehensive search on published literature following the guidelines of the meta-analysis of observational studies in epidemiology group for design, implementation, and reporting. The methodological qualities for included studies were assessed using the quality in prognosis studies. The pooled hazard ratios (HRs) with 95% CIs for overall survival (OS) and progression-free survival/recurrence-free survival/disease-free survival (PFS/RFS/DFS) were calculated to appraise the associations between miR-17/17-5P expression and cancer prognosis.
Results: A total of 21 studies involving 2099 subjects were analyzed in evidence synthesis. The results showed that high expression of miR-17 was associated with poor OS (HR=2.14; 95% CI: 1.69–2.71, P<0.001) in patients with cancer, especially in Caucasian (HR=2.23; 95% CI: 1.58–3.14, P<0.001) and digestive tract cancer (HR=1.29, 95% CI: 1.03–1.63, P=0.03), and miR-17 expression was significantly correlated with PFS/RFS in cancer patients (HR=1.69, 95% CI: 1.29–2.22, P<0.001). miR-17-5P overexpression was significantly linked with poor OS in cancer patients (HR=1.66; 95% CI: 1.31–2.09, P=0.00), especially in Asian (HR=1.81; 95% CI: 1.37–2.40, P<0.001), digestive tract cancer (HR=1.80; 95% CI: 1.29–2.50, P<0.001), and serum sample (HR=1.76; 95% CI: 1.29–2.41, P<0.001). miR-17-5P expression was significantly associated with DFS in cancer patients (HR=1.58, 95% CI: 1.07–2.35, P=0.02).
Conclusion: High expression of miR-17 and miR-17-5P are significantly associated with poor survival in patients with cancer. This indicated that miR-17/17-5P may be a novel prognostic indicator in cancer.
Keywords: miR-17/17-5P, prognosis, cancer, meta-analysis
Introduction
Cancer is one of the major disease burdens worldwide.1 In 2016, cancer was the second leading cause of death in the United States.2 Cancer has been the leading cause of death with about one-fourth of all deaths in China.3 The burden of disease attributable to the occurrence of cancers was rising.
MicroRNA (miRNA) is a kind of small, non-protein-coding RNA (containing about 22 nucleotides) molecules involving in RNA silencing and post-transcriptional regulation of gene expression through mRNA degradation and translational repression,4,5 which regulate gene expression by binding to the 3’-untranslated region (UTR) of target mRNAs.6
More and more studies had proved that aberrantly expressed miRNAs were involved in different types of cancers, which acted as tumor oncogenes or suppressors and played critical roles in many aspects of cancer carcinogenesis, including cell proliferation, differentiation, angiogenesis, and metastasis.7–10 miRNAs are regarded as biomarkers for cancer prognosis and prediction of treatment response beacuse of the high stability of miRNAs in circulation and formalin-fixed, paraffin-embedded (FFPE) tissue.11–13
MicroRNA-17 (miR-17) family is the major common studied oncogenic miRNA (onco-miRNA) groups.14 miR-17 and miR-17-5P (miR-17/17-5P) belong to miR-17 family, which has been shown to play a key role in the pathogenesis of diverse cancers.15 miR-17 overexpression significantly improve the ability of migration and motility of melanoma cells by suppression of translation of ETV1.16 The miR-17-5p was overexpressed in ovarian cancer cells. It activated AKT by lowering phosphatase and tensin homolog (PTEN) in ovarian cancer cells.17
High levels of the oncogenic miRNA (onco-miR) guide strand were called miR-17-5p. In triple-negative breast cancer, the overexpressed miR-17-5p can inhibit ribosomal translation of cancer suppressor gene mRNAs, such as PTEN or programmed cell death 4 (PDCD4).18 However, expression of miR-17/17-5P in the circulation or tissue have not been predicted or experimentally confirmed, and the overall survival (OS) and progression-free survival/recurrence-free survival/disease-free survival (PFS/RFS/DFS) of the patterns and expression levels of miR-17/17-5P have not been well established.
In this study, a systematic evaluation with the prognostic data of miR-17/17-5P from published studies was first performed. We focused on correlations between the expression level of miR-17/17-5P and prognostic indicator, which can guide clinical decisions.
Materials and methods
The present study was executed in accordance with the guidelines of the Meta-analysis of Observational Studies in Epidemiology (MOOSE)19 group and criteria of Preferred Reporting Items for Systematic Reviews and Meta-analysis (PRISMA).20 The protocol of this meta-analysis has not been published or registered to any databases.
Search strategy
Studies related to miR-17/17-5P and cancers were retrieved from multiple databases: PubMed, EMBASE, Web of Science, Google Scholar, Wanfang (China), and the Chinese National Knowledge Infrastructure (CNKI) database through July 2017. The search items were combinations of “microRNA-17” or “miR-17”, “microRNA-17-5P” or “miR-17-5P” and “neoplasms” or “cancer”. We additionally manually retrieved bibliography of the selected articles to identify further potentially relevant studies that may be missed by online information retrieval.
Study selection and exclusion criteria
Studies were eligible for inclusion if they were: 1) cohort studies that investigated the relationship between miR-17/17-5P and patients with cancer prognosis; 2) provided available data to extrapolate the hazard ratio for survival and corresponding 95% CI; 3) the expression of miR-17/17-5P was measured in cancer tissue or serum; and 4) available in Chinese or English.
Exclusion criteria were: 1) reviews, experimental cell research, non-human research, letters; 2) neither Chinese nor English studies; 3) experiments not conducted on cancer patients; and 4) insufficient data to calculate the HRs and their 95% CIs, or the Kaplan–Meier curve unable to calculate HRs and 95% CI parameters. If a study had overlapping data with other published literature, we selected the study with a larger sample size or the latest published article. Two independent reviewers (FJD and WGL) screened all initially identified articles according to the eligibility for inclusion criteria.
Data extraction
Two reviewers (YY and JZ) independently identified eligibility studies and performed data extraction and quality assessment. Discrepancies were resolved by consensus and by referencing the original reports.
The following data/necessary information was extracted from the eligible studies if they were available: first author, year of publication, original country, histopathological classification and stage, sample size and type, detection method, cutoff value and follow-up, HRs of miR-17/17-5P for OS and/or PFS/RFS/DFS and their 95% CIs. If the study only provided survival data in a Kaplan–Meier curve, the HR and 95% CI were digitized and extracted using the methods designed by Parmar et al21 and Tierney et al.22
Quality assessment
The quality of included studies was assessed according to the Newcastle–Ottawa Scale (NOS).23 The method rates observational studies on a nine-point scale, and a study with a score ≥6 was considered a high-quality study.
The Quality In Prognosis Studies (QUIPS) for specific biases of prognosis was appraised based on the method of Hayden et al.24 Estimation of the potential bias of the items included study participation, study attrition, prognostic factor measurement, outcome measurement, study confounding, statistical analysis, and reporting.
The quality evaluation was processed independently by two authors (FQL and WGL), and in case of any inconsistency, the final decision was reached with consensus.
Data synthesis and statistical analysis
Data were analyzed by using RRevMan (Version 5.3 for Windows, Cochrane Collaboration, Oxford, UK) and STATA (Version 13.1MP, StataCorp, College Station, TX, USA). HRs and corresponding 95% CIs were utilized to assess the relationship strength between miR-17/17-5P expression and prognosis. Inter-study heterogeneity was evaluated using the Cochran’s Q test and Higgin’s I2. Pheterogeneity<0.1 and I2> 50% represents high heterogeneity. The random-effects model (DerSimonian and Laird method)25 was used in a pooled analysis. Afterward, meta-regression was utilized to explore sources of heterogeneity.26 Otherwise, a fixed-effects model (Mantel–Haenszel method)27 was used. The subgroup analyses were performed by ethnicity (Asian, Caucasian) and cancer subtypes (pathological type), if a cancer type was studied in less than 2 individual studies, it was classified as the “other cancers” group. Assessing the impact of included studies on the pooled results, one-way sensitivity analyses were performed to compensate for statistical heterogeneity.
Begg’s test (rank correlation test)28 and Egger’s test (weighted linear regression test)29 are used to assess the extent of publication bias. A two-tailed value of P<0.05 was considered statistically significant.
Results
Study identification
The search process returned 1127 relevant citations and the titles were screened based on the search strategy (Figure 1). According to the exclusion criteria, the abstracts of 197 studies were reviewed. The titles and abstracts were screened by 2 of the authors. Full texts of potentially eligible articles were retrieved for further identification, 19 eligible articles were screened out. The reference lists of eligible articles were screened for potential publications. Finally, a total of 19 articles (21 studies)30–48 including 11 articles (13 studies)30–40 for miR-17 and 841-48 for miR-17-5P were considered in a pooled analysis. One of the articles39 performed three cohort studies in different populations.
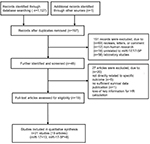  | Figure 1 Flow chart of literature search and study selection. Abbreviations: HR, hazard ratio; miR, microRNA. |
Baseline characteristics of included studies
The main characteristics of eligible studies are presented in Table 1. The studies were published from 2008 to 2017 and included 2099 patients from China, Brazil, America, Norway, Japan, Spain, and Italy. The patients were classified as Asian or Caucasian based on ethnic background. The types of cancer included breast cancer, osteosarcoma, lymphoma, esophageal squamous cell carcinoma (ESCC), oral cancer, glioma, myeloma, colon cancer, non-small cell lung cancer (NSCLC), gastric cancer, colorectal cancer, hepatocellular carcinoma (HCC), endometrial serous adenocarcinoma (ESC) and malignant mesothelioma (MM). The method of miR-17/17-5P detection was quantitative real-time polymerase chain reaction (qRT-PCR) in 21 studies. miR-17/17-5P expression levels were measured in tissue or serum. The cutoff values of miR-17/17-5P were mostly normal or median.
Qualitative assessment
The results based on QUIPS are summarized in Table 2. The estimated items include the participation, attrition, measurement of prognostic factor, confounding measurement and account, outcome measurement, and analysis and reporting 6 bias domains and the risk of bias shown in Figures 2 and 3. According to the NOS (Table S1), 84% (16/19) of these articles were high-quality (quality score ≥ 6).
Test of heterogeneity
The results of heterogeneity tests are shown in Table 3. There was no significant heterogeneity among studies for the miR-17(I2 =23%, P=0.21) and miR-17-5P (I2 =0%, P=0.53) in association between miR-17/17-5P expression and the risk of tumorigenesis. Therefore, the fixed effects were applied to calculate the pooled HR for miR-17/17-5P.
Meta-analysis findings
We applied a fixed-effects model to evaluate the pooled HR value (95% CI).
As for the miR-17, HRs of OS were provided by 11 studies, and a significant relation was observed between high miR-17 expression levels and poor OS (HR=2.14; 95% CI: 1.69–2.71, P<0.001). HRs of disease progression were provided by 3 studies and the miR-17 expression level was significantly correlated with PRS/RFS in cancers (HR=1.69; 95% CI: 1.29–2.22, P<0.001) (Figure 2, Table 3). Subgroup analysis was conducted by ethnicity and miR-17 expression was significantly associated with OS in Asian (HR=2.06; 95% CI: 1.49–2.87, P<0.001) and Caucasian (HR=2.23; 95% CI: 1.58–3.14, P<0.001) (Table 3). Additionally, subgroup analysis was carried out according to cancer subtype and the results indicated that a high expression level of miR-17 significantly predicted poor OS in digestive tract cancer (DTC) (HR=2.10; 95% CI: 1.54–2.87, P<0.001) and other cancer types (HR=2.20; 95% CI: 1.53–3.16, P<0.001) (Table 3).
As for the miR-17-5P, HRs of OS were provided by 8 studies, and a significant association was observed between high miR-17-5P expression level and poor OS (HR=1.66; 95% CI: 1.31–2.09, P<0.001) (Figure 3, Table 3). HRs for disease progression were provided by 2 studies, and miR-17-5P expression was significantly correlated with DFS in cancers (HR=1.58; 95% CI: 1.07–2.35, P=0.02) (Figure 3, Table 3). Subgroups analyses were carried out by ethnicity and miR-17-5P expression level was significantly correlated with OS in Asian (HR=1.81; 95% CI: 1.37–2.40, P<0.001), but not in Caucasian (HR=1.36; 95% CI: 1.89–2.07, P=0.15). Subgroups analyses were also carried out based on cancer subtypes and miR-17-5P expression was significantly correlated with OS in DTC (HR=1.80; 95% CI: 1.29–2.50, P<0.001) and other cancer types (HR=1.53; 95% CI: 1.10–2.13, P<0.001). Furthermore, subgroups analyses were performed according to sample types and the result indicated that a high expression level of miR-17 significantly predicted poor OS in tissue sample (HR=1.53; 95% CI: 1.08–2.18, P=0.02) and serum sample (HR=1.76; 95% CI: 1.29–2.41, P<0.001) (Table 3).
Sensitivity analyses
Sensitivity analysis was performed to assess the contribution of each study to the pooled estimate by omitting individual data sets and recalculating the pooled HR estimates for the remaining studies. The stable pooled HR was found not to be dominantly influenced by any individual study (Figure 4).
  | Figure 4 (A) Sensitivity analysis for OS of miR-17. (B) Sensitivity analysis for OS of miR-17-5P. Abbreviations: OS, overall survival; miR, microRNA. |
Publication bias
Begg’s and Egger’s tests were used to assess the publication bias. No significant publication bias was found in the included studies (Table 4). Meanwhile, the shape of the funnel plots indicated no obvious publication bias (Figures S1 and S2).
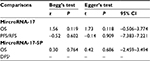  | Table 4 Publication bias of miR-17/17-5P for Begg’s28 test and Egger’s29 test Note: aInsufficient observations. Abbreviations: OS, overall survival; PFS, progression-free survival; RFS, recurrence-free survival; DFS, disease-free survival; miR, microRNA. |
Discussion
Emerging studies have demonstrated that miRNAs can act as oncogenes and tumor suppressor genes and play a crucial role in several processes, including cell proliferation, apoptosis, differentiation and metastasis.49,50 Therefore, exploring the profiles of miRNAs and corresponding target genes involved in tumorigenesis may promote the understanding of underlying mechanisms of tumor formation and provide valuable insights for the treatment and early diagnosis of such diseases.51,52 Results from published studies have continually indicated that the miRNAs which are present in the circulation can be promising diagnostic and prognostic biomarkers for tumors.53,54
In this study, increased expression of miR-17 and miR-17-5P were found to be good predictors of poor survival in patients with a variety of cancers. The combined HRs of OS were 2.14 (95% CI: 1.69–2.71) for miR-17 and 1.69 (95% CI: 1.31–2.09) for miR-17-5P, indicating that elevated miR-17/17-5P levels are closely linked with the prognosis of patients with different types of malignant tumors. This is particularly true for miR-17 in the Caucasian, whose pooled HR of OS was 2.23 (95% CI: 1.58–3.14), and this result further demonstrated the predictive value of miR-17. Subgroup analyses also indicated a closer relationship between rising miR-17 levels and poor survival in the Caucasian subgroup. Among 19 studies reporting on OS in 12 cancer types, 10 were DTC. Due to the limited number of included studies for each cancer type, we carried out a stratified analysis of DTC. The result also revealed that elevated miR-17/17-5P yielded worse OS in DTC (HR=2.10; 95% CI: 1.54–2.87 for miR-17; HR=1.80; 95% CI: 1.29–2.50 for miR-17-5P). Meanwhile, further research is needed to explore whether pathological types of specific cancers affect the prognostic role of miR-17/17-5P.
Because the eligible studies used various indices to assess cancer progression, such as DFS, PRS, and RFS, we combined these prognostic indicators to evaluate the prognostic value of miR-17/17-5P. The results indicated an intimate association between high miR-17 expression levels and PFS/RFS (pooled HR=1.69; 95% CI: 1.29–2.22) and high miR-17-5P expression levels and DFS (pooled HR=1.58; 95% CI: 1.07–2.35). Therefore, high miR-17/17-5P expression levels may be a promising negative prognostic factor in DTC. In addition, subgroup analyses were performed according to sample types. However, further study is needed to confirm the role of miR-17/17-5P in predicting the prognosis of different types of cancer.
Evidence has suggested a controversial role of the miR-17 in tumorigenesis, tumor suppressive,55,56 or oncogenic,57,58 which suggested a tumor/tissue-specific role of miR-17. The miR-17-5p functionally modulates sensitivity to radiation in vitro in esophageal adenocarcinoma cells and alters expression of predicted miR-17-5p target genes, such as C6orf120.59 In vivo, miR-17-5p is decreased significantly, while target gene expression is increased significantly in pretreatment tumor biopsies from patients who have a poor response to neoadjuvant chemoradiation therapy.60 It indicates that miR-17-5p can be regarded as a predictive marker of response to neoadjuvant chemoradiation therapy and novel therapeutic target to enhance efficacy of esophageal adenocarcinoma in chemoradiation therapy.61 Meanwhile, compared to normal tissue, miR-17/17-5P have been found at higher levels in colorectal cancerous tissue, although with varied expression of each individual component.38,62 A similar result was observed by Wang et al,45 who demonstrated that the circulating miR-17-5p expression level might be a molecular marker for gastric cancer.
As a comprehensive review of prognostic studies, one limitation is the heterogeneous nature of the identified studies. The reviewed studies varied in study quality, study populations, and statistical analyses. The influencing factors of heterogeneity included in analysis model, definitions of measurement, pathologic classification, laboratory techniques, and treatments received.63 In some situations, inadequate data from some types of cancer and subgroup analyses in HRs may be the origin of heterogeneity.64 The miRNAs can induce widely variable effects depending on tumor biology, microenvironment, and a multitude of other interconnected factors. If the number of studies is very small, it may be impossible to estimate the between-studies variance (τ2) with any precision.65
To our knowledge, there is no evidence-based meta-analysis to evaluate the prognostic values of miR-17/17-5P. Therefore, we pooled the available evidence from all relevant studies to assess the prognostic values of miR-17/17-5P. Although our meta-analysis is robust, several limitations deserved focus as follows. First, not all the included studies provide multivariate adjusted HRs and corresponding 95CI%. In this case, the data was extracted from Kaplan–Meier survival curves, which can result in several tiny errors on calculated HRs with the 95% CIs. Second, although we find no evidence of publication bias, included studies were mostly in English, which may generate publication bias. Third, the cutoff values (median, mean, etc.) were applied to evaluate the different miR-17/17-5P expression, but the actual values may be discrepant due to different algorithms and result in some heterogeneity. Diversity of anatomical locations can be considered as another potential reason for heterogeneity. Fourth, given the limited availability of included studies, for DFS/PFS/RFS, the included studies were not stratified. Finally, the influence of adjuvant therapies on the prognostic effect of malignant tumor was not assessed in our study due to scant data. In spite of these limitations, the study about the relationship between miR-17/17-5P and prognosis of cancers is certainly warranted. In summary, the high expression of miR-17/17-5P was significantly associated with poor survival in patients with cancer. In addition, overexpression of miR-17/17-5P was associated with ethnicity, cancer subtypes, and sample types. These findings suggested that miR-17/17-5P might be a novel prognostic indicator of cancer. Given its limitations, the results of the present analysis should be interpreted with caution. Further clinical investigations are needed to determine the association between miR-17/17-5P and cancer prognosis.
Acknowledgment
This study was supported by the National Natural Science Foundation of China (No. 81701536) and Zhengzhou University Joint First Affiliated Hospital Cultivation Fund (No. 2016-BSTDJJ-15).
Disclosure
The authors report no conflicts of interest in this work.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. | ||
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7. | ||
Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. | ||
Sun K, Lai EC. Adult-specific functions of animal microRNAs. Nat Rev Genet. 2013;14(8):535–548. | ||
Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. | ||
Ambros V. The functions of animal microRNAs. Nature. 2004; 431(7006):350–355. | ||
Winbanks CE, Ooi JY, Nguyen SS, McMullen JR, Bernardo BC. MicroRNAs differentially regulated in cardiac and skeletal muscle in health and disease: Potential drug targets? Clin Exp Pharmacol Physiol. 2014;41(9):727–737. | ||
Vorvis C, Koutsioumpa M, Iliopoulos D. Developments in miRNA gene signaling pathways in pancreatic cancer. Future Oncol. 2016;12(9):1135–1150. | ||
Lin CW, Lin PY, Yang PC. Noncoding RNAs in tumor epithelial-to-mesenchymal transition. Stem Cells Int. 2016;2016:2732705. | ||
Walter RFH, Vollrecht C, Werner R, et al. MicroRNAs are differentially regulated between MDM2-positive and negative malignant pleural mesothelioma. Oncotarget. 2016;7(14):18713–18721. | ||
Treece AL, Duncan DL, Tang W, et al. Gastric adenocarcinoma microRNA profiles in fixed tissue and in plasma reveal cancer-associated and Epstein–Barr virus-related expression patterns. Lab Invest. 2016;96(6):661. | ||
Gomes BC, Santos B, Rueff J, Rodrigues AS. Methods for studying microRNA expression and their targets in formalin-fixed, paraffin-embedded (FFPE) breast cancer tissues. Methods Mol Biol. 2016;1395:189–205. | ||
Wen J, Ye F, He X, et al. Development and validation of a prognostic nomogram based on the log odds of positive lymph nodes (LODDS) for breast cancer. Oncotarget. 2016;7(15):21046–21053. | ||
Koga Y, Yamazaki N, Yamamoto Y, et al. Fecal miR-106a is a useful marker for colorectal cancer patients with false-negative results in immunochemical fecal occult blood test. Cancer Epidemiol Biomarkers Prev. 2013;22(10):1844–1852. | ||
Tang Y, Zhang Y, Chen Y, Xiang Y, Shen C, Li Y. The role of miR-19b in the inhibition of endothelial cell apoptosis and its relationship with coronary artery disease. Sci Rep. 2015;5:15132. | ||
Fang Y, Xu C, Fu Y. MicroRNA-17-5p induces drug resistance and invasion of ovarian carcinoma cells by targeting PTEN signaling. J Biol Res (Thessalon). 2015;22(12):1–10. | ||
Cohen R, Greenberg E, Nemlich Y, Schachter J, Markel G. miR-17 regulates melanoma cell motility by inhibiting the translation of ETV1. Oncotarget. 2015;6(22):19006–19016. | ||
Jin YY, Andrade J, Wickstrom E. Non-specific blocking of miR-17-5p guide strand in triple negative breast cancer cells by amplifying passenger strand activity. PloS one. 2015;10(12):e0142574. | ||
Stroup DF, Berlin JA, Morton SC, et al. Meta-analysis of observational studies in epidemiology: A proposal for reporting. Meta-analysis of observational studies in epidemiology (MOOSE) group. JAMA. 2000;283(15):2008–2012. | ||
Moher D, Liberati A, Tetzlaff J, Altman DG; PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009;151(4):264–269. | ||
Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med. 1998;17(24):2815–2834. | ||
Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8(1):16. | ||
Wells G, Shea B, O’Connell D, et al. The Newcastle-Ottawa Scale (NOS) for Assessing the Quality of Nonrandomised Studies in Meta-Analyses. Ottawa, ON, Canada: The Ottawa Hospital Research Institute; 2011. | ||
Hayden JA, van der Windt DA, Cartwright JL, Côté P, Bombardier C. Assessing bias in studies of prognostic factors. Ann Intern Med. 2013;158(4):280–286. | ||
DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. | ||
Thompson SG, Higgins JP. How should meta-regression analyses be undertaken and interpreted? Stat Med. 2002;21(11):1559–1573. | ||
Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–748. | ||
Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–1101. | ||
Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. | ||
Yang F, Yuan L, Xu L, et al. miR-17 as a diagnostic biomarker regulates cell proliferation in breast cancer. Onco Targets Ther. 2017;10:543–550. | ||
Li S, Gao Y, Wang Y, et al. Serum microRNA-17 functions as a prognostic biomarker in osteosarcoma. Oncol Lett. 2016;12(6):4905–4910. | ||
Robaina MC, Faccion RS, Mazzoccoli L, et al. miR-17-92 cluster components analysis in Burkitt lymphoma: overexpression of miR-17 is associated with poor prognosis. Ann Hematol. 2016;95(6):881–891. | ||
Xi Y, Li J, Zhang P, et al. Upregulation of miRNA-17 and miRNA-19 is associated with unfavorable prognosis in patients with T-cell lymphoblastic lymphoma. Exp Mol Pathol. 2015;99(2):297–302. | ||
Xu XL, Jiang YH, Feng JG, Su D, Chen PC, Mao WM. MicroRNA-17, microRNA-18a, and microRNA-19a are prognostic indicators in esophageal squamous cell carcinoma. Ann Thorac Surg. 2014;97(3):1037–1045. | ||
Chang CC, Yang YJ, Li YJ, et al. Corrigendum to “MicroRNA-17/20a functions to inhibit cell migration and can be used a prognostic marker in oral squamous cell carcinoma” [Oral Oncol. 49(9) (2013) 923–931]. Oral Oncol. 2017;72:202–203. | ||
Lu S, Wang S, Geng S, Ma S, Liang Z, Jiao B. Increased expression of microRNA-17 predicts poor prognosis in human glioma. J Biomed Biotechnol. 2012;2012:970761. | ||
Gao X, Zhang R, Qu X, et al. miR-15a, miR-16-1 and miR-17-92 cluster expression are linked to poor prognosis in multiple myeloma. Leukemia Res. 2012;36(12):1505–1509. | ||
Yu G, Tang JQ, Tian ML, et al. Prognostic values of the miR-17-92 cluster and its paralogs in colon cancer. J Surg Oncol. 2012;106(3):232–237. | ||
Saito M, Schetter AJ, Mollerup S, et al. The association of microRNA expression with prognosis and progression in early-stage, non-small cell lung adenocarcinoma: a retrospective analysis of three cohorts. Clin Cancer Res. 2011;17(7):1875–1882. | ||
Valladares-Ayerbes M, Blanco M, Haz M, et al. Prognostic impact of disseminated tumor cells and microRNA-17-92 cluster deregulation in gastrointestinal cancer. Int J Oncol. 2011;39(5):1253–1264. | ||
Komatsu S, Ichikawa D, Tsujiura M, et al. Prognostic impact of circulating miR-21 in the plasma of patients with gastric carcinoma. Anticancer Res. 2013;33(1):271–276. | ||
Chen Q, Si Q, Xiao S, et al. Prognostic significance of serum miR-17-5p in lung cancer. Med Oncol. 2013;30(1):353. | ||
Zheng J, Dong P, Gao S, Wang N, Yu F. High expression of serum miR-17-5p associated with poor prognosis in patients with hepatocellular carcinoma. Hepatogastroenterology. 2013;60(123):549–552. | ||
Chen L, Jiang M, Yuan W, Tang H. miR-17-5p as a novel prognostic marker for hepatocellular carcinoma. J Invest Surg. 2012;25(3):156–161. | ||
Wang M, Gu H, Wang S, et al. Circulating miR-17-5p and miR-20a: Molecular markers for gastric cancer. Mol Med Rep. 2012;5(6):1514–1520. | ||
Yu J, Ohuchida K, Mizumoto K, Fujita H, Nakata K, Tanaka M. MicroRNA miR-17-5p is overexpressed in pancreatic cancer, associated with a poor prognosis, and involved in cancer cell proliferation and invasion. Cancer Biol Ther. 2010;10(8):748–757. | ||
Busacca S, Germano S, De Cecco L, et al. MicroRNA signature of malignant mesothelioma with potential diagnostic and prognostic implications. Am J Respir Cell Mol Biol. 2010;42(3):312–319. | ||
Díaz R, Silva J, García JM, et al. Deregulated expression of miR-106a predicts survival in human colon cancer patients. Genes Chromosomes Cancer. 2008;47(9):794–802. | ||
Cho WC. MicroRNAs: potential biomarkers for cancer diagnosis, prognosis and targets for therapy. Int J Biochem Cell Biol. 2010;42(8):1273–1281. | ||
Cho WC. OncomiRs: the discovery and progress of microRNAs in cancers. Mol Cancer. 2007;6:60. | ||
Browne, G, Taipaleenmäki H, Stein GS, Stein JL, Lian JB. MicroRNAs in the control of metastatic bone disease. Trends Endocrinol Metab. 2014;25(6):320–327. | ||
Bryant RJ, Pawlowski T, Catto JWF, et al. Changes in circulating microRNA levels associated with prostate cancer. Br J Cancer. 2012;106(4):768–774. | ||
Joosse SA, Müller V, Steinbach B, Pantel K, Schwarzenbach H. Circulating cell-free cancer-testis MAGE-A RNA, BORIS RNA, let-7b and miR-202 in the blood of patients with breast cancer and benign breast diseases. Br J Cancer. 2014;111(5):909–917. | ||
Zampetaki A, Kiechl S, Drozdov I, et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res. 2010;107(6):810–817. | ||
Yu Z, Wang C, Wang M, et al. A Cyclin D1/microRNA 17/20 regulatory feedback loop in control of breast cancer cell proliferation. J Cell Biol. 2008;182(3):509–517. | ||
Zheng ZF, Su HF, Zou Y, Peng Z, Wu SX. Expression profiles of microRNAs in radioresistant esophageal cell line. Zhonghua Yi Xue Za Zhi. 2011;91(9):639–642. In Chinese. | ||
Dews M, Homayouni A, Yu D, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38(9):1060–1065. | ||
Hossain A, Kuo MT, Saunders GF. mir-17-5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Mol Cell Biol. 2006;26(21):8191–8201. | ||
Zhang W, Lin J, Wang P, Sun J. miR-17-5p down-regulation contributes to erlotinib resistance in non-small cell lung cancer cells. J Drug Target. 2017;25(2):125–131. | ||
Yang X, Du WW, Li H, et al. Both mature miR-17-5p and passenger strand miR-17-3p target TIMP3 and induce prostate tumor growth and invasion. Nucleic Acids Res. 2013;41(21):9688–9704. | ||
Lynam-Lennon N, Heavey S, Sommerville G, et al. MicroRNA-17 is downregulated in esophageal adenocarcinoma cancer stem-like cells and promotes a radioresistant phenotype. Oncotarget. 2016;8(7):11400–11413. | ||
Diosdado B, Wiel MAVD, Droste JSTS, et al. miR-17-92 cluster is associated with 13q gain and c-myc expression during colorectal adenoma to adenocarcinoma progression. Br J Cancer. 2009;101(4):707–714. | ||
Cuyun Carter G, Barrett AM, Kaye JA, Liepa AM, Winfree KB, John WJ. A comprehensive review of nongenetic prognostic and predictive factors influencing the heterogeneity of outcomes in advanced non-small-cell lung cancer. Cancer Manag Res. 2014;6:437–449. | ||
Barker EV, Cervigne NK, Reis PP, et al. microRNA evaluation of unknown primary lesions in the head and neck. Mol Cancer. 2009;8:127. | ||
Jamali Z, Asl Aminabadi N, Attaran R, Pournagiazar F, Ghertasi Oskouei S, Ahmadpour F. MicroRNAs as prognostic molecular signatures in human head and neck squamous cell carcinoma: a systematic review and meta-analysis. Oral Oncol. 2015;51(4):321–331. |
Supplementary materials
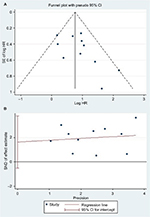  | Figure S1 (A) Begg’s funnel plot of publication bias on the relationship between miR-17 expression and OS.20 (B) Egger’s funnel plot of publication bias on the relationship between miR-17 expression and OS.21 Abbreviations: HR, hazard ratio; miR, microRNA; SND, standard normal deviate. |
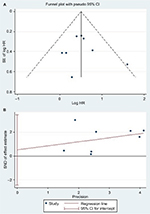  | Figure S2 (A) Begg’s funnel plot of publication bias on the relationship between miR-17-5P expression and OS.20 (B) Egger’s funnel plot of publication bias on the relationship between miR-17-5P expression and OS.21 Abbreviations: HR, hazard ratio; miR, microRNA; SND, standard normal deviate. |
References
Yang F, Yuan L, Xu L, Zhu Y, Gao H, Zhen L, Fan L. miR-17 as a diagnostic biomarker regulates cell proliferation in breast cancer. Onco Targets Ther. 2017;10:543–550. | ||
Li S, Gao Y, Wang Y, et al. Serum microRNA-17 functions as a prognostic biomarker in osteosarcoma. Oncol Lett. 2016;12(6): 4905–4910. | ||
Robaina MC, Faccion RS, Mazzoccoli L, et al. miR-17-92 cluster components analysis in Burkitt lymphoma: overexpression of miR-17 is associated with poor prognosis. Ann Hematol. 2016;95(6):881–891. | ||
Xi Y, Li J, Zhang P, et al. Upregulation of miRNA-17 and miRNA-19 is associated with unfavorable prognosis in patients with T-cell lymphoblastic lymphoma. Exp Mol Pathol. 2015;99(2):297 -302. | ||
Xu XL, Jiang YH, Feng JG, Su D, Chen PC, Mao WM. MicroRNA-17, microRNA-18a, and microRNA-19a are prognostic indicators in esophageal squamous cell carcinoma. Ann Thorac Surg. 2014;97(3):1037–1045. | ||
Chang CC, Yang YJ, Li YJ, et al. Corrigendum to “MicroRNA-17/20a functions to inhibit cell migration and can be used a prognostic marker in oral squamous cell carcinoma” [Oral Oncol. 2013;49(9):923–931]. Oral Oncol. 2017;72:202–203. | ||
Lu S, Wang S, Geng S, Ma S, Liang Z, Jiao B. Increased expression of microRNA-17 predicts poor prognosis in human glioma. J Biomed Biotechnol. 2012;2012:970761. | ||
Gao X, Zhang R, Qu X, et al. MiR-15a, miR-16-1 and miR-17-92 cluster expression are linked to poor prognosis in multiple myeloma. Leukemia Res. 2012;36(12):1505–1509. | ||
Yu G, Tang JQ, Tian ML, et al. Prognostic values of the miR-17-92 cluster and its paralogs in colon cancer. J Surg Oncol. 2012;106(3):232–237. | ||
Saito M, Schetter AJ, Mollerup S, et al. The association of microRNA expression with prognosis and progression in early-stage, non-small cell lung adenocarcinoma: a retrospective analysis of three cohorts. Clin Cancer Res. 2011;17(7):1875–1882. | ||
Valladares-Ayerbes M, Blanco M, Haz M, et al. Prognostic impact of disseminated tumor cells and microRNA-17-92 cluster deregulation in gastrointestinal cancer. Int J Oncol. 2011;39(5):1253–1264. | ||
Komatsu S, Ichikawa D, Tsujiura M, et al. Prognostic impact of circulating miR-21 in the plasma of patients with gastric carcinoma. Anticancer Res. 2013;33(1):271–276. | ||
Chen Q, Si Q, Xiao S, et al. Prognostic significance of serum miR-17-5p in lung cancer. Med Oncol. 2013;30(1):353. | ||
Zheng J, Dong P, Gao S, Wang N, Yu F. High expression of serum miR-17-5p associated with poor prognosis in patients with hepatocellular carcinoma. Hepatogastroenterology. 2013;60(123):549–552. | ||
Chen L, Jiang M, Yuan W, Tang H. miR-17-5p as a novel prognostic marker for hepatocellular carcinoma. J Invest Surg. 2012;25(3):156–161. | ||
Wang M, Gu H, Wang S, et al. Circulating miR-17-5p and miR-20a: molecular markers for gastric cancer. Mol Med Rep. 2012;5(6):1514–1520. | ||
Yu J, Ohuchida K, Mizumoto K, Fujita H, Nakata K, Tanaka M. MicroRNA miR-17-5p is overexpressed in pancreatic cancer, associated with a poor prognosis, and involved in cancer cell proliferation and invasion. Cancer Biol Ther. 2010;10(8):748–757. | ||
Busacca S, Germano S, De Cecco L, et al. MicroRNA signature of malignant mesothelioma with potential diagnostic and prognostic implications. Am J Respir Cell Mol Biol. 2010;42(3):312–319. | ||
Díaz R, Silva J, García JM, et al. Deregulated expression of miR-106a predicts survival in human colon cancer patients. Genes Chromosomes Cancer. 2008;47(9):794–802. | ||
Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–1101. | ||
Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.