Back to Journals » OncoTargets and Therapy » Volume 13
PWP1 Promotes the Malignant Phenotypes of Lung Cancer Cells by Interacting with DVL2 and Merlin
Authors Wei L, Li P, Luo Y, Zhang M, Yan T, Yang Y, Han Y, Liu S, Wang E
Received 2 June 2020
Accepted for publication 15 September 2020
Published 8 October 2020 Volume 2020:13 Pages 10025—10037
DOI https://doi.org/10.2147/OTT.S263815
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 5
Editor who approved publication: Prof. Dr. Nicola Silvestris
Lai Wei, 1 Pengcheng Li, 1 Yuan Luo, 1 Meiyu Zhang, 1 Ting Yan, 1 Yue Yang, 1 Yuchen Han, 2 Shuli Liu, 1 Enhua Wang 1
1Department of Pathology, College of Basic Medical Science, and First Affiliated Hospital of China Medical University, Shenyang, People’s Republic of China; 2Department of Pathology, Shanghai Chest Hospital, Shanghai Jiao Tong University, Shanghai, People’s Republic of China
Correspondence: Enhua Wang Tel +86-24-83282234
Fax +86-24-23261638
Email [email protected]
Shuli Liu Email [email protected]
Purpose: The significance of periodic tryptophan protein 1 (PWP1) expression in human cancer and its molecular mechanism of action have not been reported so far.
Materials and Methods: Immunohistochemistry was performed to analyze the expression of PWP1 in non-small cell lung cancer (NSCLC) tissues and statistical analysis was applied to analyze the relationship between PWP1 expression and the clinicopathological factors. The effects of PWP1 on NSCLC proliferation and invasion were determined by colony formation, transwell and MTT assays. Western blot analysis (WB), dual-luciferase reporter gene assays and immunofluorescence staining were performed to demonstrate whether PWP1 stimulates Wnt pathway and inhibits Hippo pathway. Co-immunoprecipitation (Co-ip) assays were used to confirm the potential role of PWP1 in Wnt and Hippo signaling pathways.
Results: PWP1 expression in NSCLC was higher than that in normal bronchial epithelium and normal submucosal glands. In addition, PWP1 expression had a positive correlation with poor differentiation, high pathological tumor-node-metastasis (TNM) stage, and lymph node metastasis. Kaplan–Meier database demonstrated that the overall survival time of patients with high PWP1 expression was significantly shorter than that of patients with low PWP1 expression. Mechanistically, we found that PWP1 could interact with DVL2 to upregulate β-catenin (thereby activating the Wnt pathway), whereas PWP1 could interact with Merlin (NF2) to downregulate p-MST1 (thereby inhibiting the Hippo signaling pathway). The effects of PWP1 on promoting the Wnt pathway or inhibiting the Hippo pathway were offset in DVL2- or Merlin-knockdown cells transiently overexpressing PWP1.
Conclusion: PWP1 expression in NSCLC was correlated with poor prognosis. PWP1 enhanced the activity of the Wnt pathway by interacting with DVL2, whereas PWP1 inhibited the activity of the Hippo pathway by interacting with Merlin. Together, these two effects promoted the detrimental biological behaviors of NSCLC cells.
Keywords: PWP1, Hippo, Wnt, DVL2, Merlin
Corrigendum for this paper has been published
Introduction
PWP1 is also known as endonuclein, which contains five WD40 repeated domains and belongs to the WD40-repeated superfamily. The gene encoding PWP1 is 1506 bp and is located on chromosome 12q23.3. Data from previous studies have shown that PWP1 has key regulatory effects on various biological functions such as RNA processing,1,2 signal transduction,3 gene expression, vesicle transport, cytoskeleton assembly, and cell cycle progression.4 However, the mechanism(s) whereby PWP1 plays such roles is far from being clearly elucidated. By altering PWP1 expression in mouse embryonic stem cells (ESCs) and embryonic fibroblasts, using gene chip and GSEA-pathway analyses of the resulting differentially expressed genes, Honore et al demonstrated that the Jak/Stat3 and Wnt/β-catenin signaling pathways had undergone significant changes, further investigation showed that PWP1 and H4K20me3 can form a complex that binds to upstream sites of the Stat3 gene and affect Stat3 expression and early ESC differentiation.5 PWP1 was located in the nucleus of human pancreatic cancer cells and was involved in regulating cell cycle progression.6 However, whether high PWP1 expression is ubiquitous in human solid tumors, the relationship between high PWP1 expression and the clinicopathological factors of tumors, and the mechanism played by PWP1 in human solid tumors are still unclear. Although there have been few studies on the role of PWP1 in human malignancies, analysis of PWP1 has shown a possible interaction between PDZ domain in TIP-1 and PWP1.7 We speculated that because DVL2 has a PDZ domain and DVL2 plays an important role in Wnt pathway, PWP1 may regulate this pathway by binding to Dvl2, thereby affecting the malignant phenotype of NSCLC. The overstimulation of Wnt pathway can lead to developmental abnormalities and tumour formation. In tumour cells, Wnts, proteins secreted by autocrine and paracrine cells, bind to the membrane receptor Frizzled and co-receptor LRP5/6 to activate intracellular signalling pathways. The main effector of the Wnt pathway is β-catenin, a transcription factor. When the canonical Wnt pathway is activated, casein kinase 1δ/ε (CK1δ/ε) is activated and phosphorylates Dishevelled (Dvl), which disassembles the destruction complex. Unphosphorylated β-catenin then enters the nucleus, binds to transcription factor Tcf-4, and activates expression of Wnt signalling pathway target genes, such as MYC, CCND1, and MMP7. Recent reports have identified novel links between canonical Wnt signalling and alternative pathways, including the Hippo pathway. Originally discovered in Drosophila, the Hippo pathway is highly conserved and involved in cell proliferation, differentiation, apoptosis, and regulation of organ size. In recent decades, the relationship between the Hippo pathway and tumour cell proliferation, invasion, and metastasis has garnered significant attention. Specifically, the Hippo pathway includes upstream molecules (Fat, NF2, AMOT, and the WWC protein), a central kinase complex (MST1/2-WW45-LATS1/2-MOB), and downstream effectors (YAP/TAZ). Hippo pathway inactivation leads to YAP/TAZ dephosphorylation, which enables YAP nuclear translocation and enhances TEAD transcriptional activity. The targets of nuclear YAP/TAZ include CTGF, CYR61, Cyclin E, and DIAP, and increases in YAP/TAZ target proteins increase cell proliferation and decrease apoptosis.8 There are complicated cross-relationships that also exist between the Wnt and Hippo pathways.9 The Hippo and Wnt pathways interact with each other at the genetic and functional levels.10 Since the occurrence, invasion, and metastasis of lung cancer are closely related to the Wnt11,12 and Hippo pathways.13–15 In this study, we first examined the expression of PWP1 in lung cancer cell lines. Then, we detected the effect of bidirectional regulation of PWP1 on the proliferation and invasion of NSCLC cells. Finally, we detected whether PWP1 interacts with DVL2 and Merlin. This study showed the expression mode, biological functions, and possible molecular mechanism underlying the regulation of Wnt and Hippo pathways via PWP1.
Materials and Methods
Cell Lines
The HBE cell line was obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). The LK2 cell line was a gift from Dr. Hiroshi Kijima (Department of Pathology and Bioscience, Hirosaki University Graduate School of Medicine, Japan); this cell line was approved for use by the Institute Research Ethics Committee of China Medical University. All other cell lines were obtained from the Shanghai Cell Bank (Shanghai, China) and cultured according to the instructions of the ATCC. All cell lines were authenticated by performing short tandem repeat DNA profiling.
Immunohistochemistry
NSCLC samples were collected from 150 patients who were hospitalized at First Affiliated Hospital of China Medical University, as well as paired non-tumor tissues (150 patients; >5 cm distal to the primary edge of each tumor). Informed consent was obtained and the study protocol was approved by the Ethical Committee of China Medical University and conformed to the Declaration of Helsinki. (No. 2015[LS]023, China Medical University). Immunohistochemical staining was performed with the streptavidin-peroxidase method, using a diaminobenzidine kit (MaiXin, Fuzhou, China). Two independent investigators scored the microarray by evaluating the staining intensities and percentages of stained cells blindly and randomly. The staining intensity was scored from 0 to 3 as follows: 0 (negative), 1 (weak staining), 2 (moderate staining), and 3 (strong staining). The percentage of positively stained cells was scored from 0 to 4 as follows: 0 (0%), 1 (1–25%), 2 (26–50%), 3 (51–75%), and 4 (76–100%). Both scores were multiplied to generate final scores, which ranged from 0 to 12. PWP1 expression was regarded as positive if the final score was ≥4, otherwise, it is negative. Phosphate-buffered saline (PBS) and goat serum were used as negative controls. Briefly, tissue sections were incubated with a polyclonal rabbit anti-PWP1 antibody (#EPR16130, 1: 100, Abcam, UK). According to the WHO2015 classification criteria for lung cancer,16 75 patients each presented with either adenocarcinoma or squamous cell carcinoma. According to the International Union of Cancer TNM staging standards of 2015,17 60 tumors were classified as Phase I tumors, 41 tumors were classified as Phase II tumors, and 49 tumors were classified as Phase III tumors.
Protein Extraction and Immunoblotting
Assays were performed as described previously.18 Proteins were electrophoresed via sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis using an appropriate SDS concentration. For WB analysis, primary antibodies against the following target were obtained from Cell Signaling Technology, MYC tag (#2276, 1: 1000), Merlin (#12896, 1: 1000), p-Merlin (#73505, 1: 1000), MST1 (#3682, 1: 1000), p-MST1 (#49332, 1: 500), LATS1 (#9153, 1: 1000), p-LATS1 (#8654, 1: 500), YAP (#4912, 1: 1000), p-YAP (S127) (#4911, 1: 1000), β-catenin (#8480, 1: 1000), and Non-phospho (Active) β-catenin (#8814, 1: 1000). In addition, primary antibodies against PWP1 (#SC-390188, 1: 500), DVL2 (#SC-166303, 1: 500) were purchased from Santa Cruz Biotechnology, Inc. GAPDH (ZSGB-BIO, China, #TA309157, 1: 1000) was also used. Peroxidase-coupled secondary antibodies were purchased from Santa Cruz Biotechnology, Inc. The rest of the antibodies were purchased from Cell Signaling Technology. The target proteins were transferred to a polyvinylidene fluoride membrane (EMD Millipore, Billerica, MA, USA) and visualized using an ECL Kit (Thermo Fisher Scientific, Waltham, MA, USA). Images were obtained using a Bio-Rad Imaging System (Bio-Rad Laboratories, Hercules, CA, USA).
Immunoprecipitation
Protein extraction and quantitation were performed as described previously.18 Proteins were taken from each sample and mixed with IgG and an appropriate amount of A/G beads (30 μL). Next, the samples were shaken gently for 4 h on ice. The mixtures were centrifuged at 1000 rpm for 5 min at 4°C, and the supernatant was transferred to a new 1.5-mL Eppendorf tube. Each sample was divided into two equal parts, which were incubated with a primary antibody against PWP1 (Santa Cruz Biotechnology, Inc., #SC-390188, 3 µg/IP), DVL2 (Santa Cruz Biotechnology, Inc., #SC-166303, 3 µg/IP), or NF2 (Santa Cruz Biotechnology, Inc., #SC-55575, 3 µg/IP), or an isotype-matched control IgG. The samples were shaken on ice for 4 h, 40 µL Sepharose A/G beads were added, and the samples were shaken on ice overnight. Each precipitate was washed three times on ice (with NP-40 protein lysate and protease inhibitors added), and each supernatant was discarded after centrifugation at 1000 rpm for 5 min at 4°C. Next, 2× loading buffer (diluted in NP-40) was added, and each sample was boiled and stored at −20°C. Electrophoresis was performed within 1 week.
Plasmid Construction and Transfection
The empty PCMV6 vector (#PS-100001), MYC/DDK-PCMV6-PWP1 plasmid (#RC200892), and shPWP1 plasmid (#TR310031) were purchased from Origene (Rockville, MD, USA). A control siRNA (#siN0000001-1-5) siRNA-DVL2 (#stB0006196B), and siRNA-MERLIN (#stB0002007B) were purchased from RiboBio (Guangzhou, China). Cells were cultured in serum-deprived medium for 3 h, after which they were transfected with plasmids using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA), per the manufacturer’s instructions.
Dual-Luciferase Assay
Recombinant human Wnt3a (#5036-WN, R&D Systems, France) was reconstituted in PBS containing 0.1% BSA to a concentration of 10 μg/mL and used in experiments at a final concentration of 50 ng/mL. YAP/TEAD transcriptional activity was measured using a luciferase assay based on the pGL3b_8xGTIIC-luciferase plasmid (#34615; Addgene, Cambridge, MA, USA). β-catenin/TCF4 transcriptional activity was measured using the Super 8xTOPflash plasmid (Addgene plasmid #124560). The cells were transfected to express the indicated proteins and Renilla luciferase was used as a control for signal normalization. The dual-luciferase assays were performed according to the manufacturer’s protocol (Promega, Madison, Wisconsin, USA). Three independent transfections were performed for each experiment. The data were normalized to an empty-vector control and are presented as the mean ± SD. The assays were performed in the dark.
Colony Formation, Matrigel Invasion, and MTT Assays
Colony Formation
At 48 h post-transfection with PWP1 plasmids or shPWP1, cells were seeded in three 6-cm cell culture dishes (1000 cells/dish) and incubated for 12 days in RPMI-1640 medium containing 10% fetal bovine serum (FBS). The plates were then washed with PBS and incubated with Giemsa stain (Beyotime Biotechnology, Shanghai, China) before counting the number of colonies consisting of >50 cells.
Matrigel Invasion
Cell-invasion assays were performed using 24-well transwell chambers with an 8-μm pore size (Thermo Fisher Scientific), and the inserts were coated with 20 μL Matrigel (1: 3 dilution, BD Bioscience, New Jersey, USA). LK2 and SK cells were transfected with the PWP1-overexpression plasmid for 48 h, trypsinized, transferred to the upper Matrigel chamber in 100 μL of serum-free DMEM and MEM medium containing 8 × 105 cells, and incubated for 16 h. H1299 and A549 cells were transfected with the shPWP1 for 48 h, trypsinized, transferred to the upper Matrigel chamber in 100 μL of serum-free 1640 medium containing 5×105 cells, and incubated for 16 h. Medium containing 10% FBS was added to the lower chambers as the chemoattractant. The number of invading cells was counted in 10 randomly selected high-power fields under a TE300 microscope (Nikon, Melville, NY, USA).
3-(4,5-Dimethylthiazol-2-Yl)-2,5-Diphenyltetrazolium Bromide (MTT) Assays
At 48 h post-transfection, cells were plated in 96-well plates in medium containing 10% FBS at a density of approximately 3000 cells/well, and cell viabilities were determined in MTT assays. Briefly, each well was incubated with 20 μL of MTT solution (5 mg/mL; Sigma, St. Louis, MO, USA) for 4 h at 37°C. Then, the medium was removed from each well and the resultant MTT formazan was solubilized in 150 μL of dimethyl sulfoxide (Sigma). The results were quantified spectrophotometrically (Bio-Rad Laboratories, Inc.) by measuring the absorbance at 490 nm.
Immunofluorescence Staining
Cells were seeded in 24-well plates. After reaching the desired confluency, they were fixed with 4% paraformaldehyde for 15 min, washed three times with PBS, incubated with 0.1% Triton-X for 5 min, and washed three times with PBS. Next, each well was incubated with 200 μL of goat serum for 2 h at room temperature. Then, the serum was removed and each well was incubated overnight in a at 4°C humidifier with a primary antibody against PWP1 (Santa Cruz Biotechnology, Inc., #SC-390188, 1: 100), DVL2 (Proteintech, USA, #12037-1-AP, 1: 100), or Merlin (Proteintech, USA, #21686-1-AP, 1: 100). Subsequently, the primary antibodies were removed, and the wells were washed three times with PBS. Then, the wells were incubated for 2 h at room temperature with FITC- or TRITC-conjugated secondary antibodies in the dark, the unbound secondary antibody was discarded, and the wells were washed three times with PBS. Cell nuclei were dyed with 4¢, 6-diamidino-2-phenylindole for 10 min, washed with PBS, and then embedded with 40% glycerol. After sealing, the samples were stored at −20°C in the dark and observations were made within 1 week. The cells were observed using a confocal microscope and microscope.
Statistical Analysis
SPSS software, version 22.0 (SPSS, Chicago, IL, USA) was used for all statistical analyses. The chi-squared test was used to assess correlations between PWP1 expression and clinicopathological factors. Differences between the groups were tested with Student’s t-test. P < 0.05 was considered to reflect a statistically significant difference.
Results
PWP1 Was Highly Expressed in NSCLC and Was Correlated with Poor Prognosis
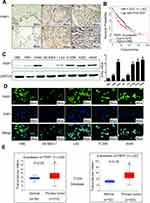
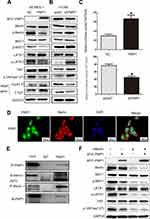
To investigate the expression pattern of PWP1 in NSCLC, we collected 150 NSCLC specimens. Immunohistochemical staining showed that PWP1 was mainly localized in the nucleus and cytoplasm of cancer cells. Most patients (76.7%, 115/150) showed negative PWP1 expression in normal bronchial epithelial cells, submucosal glands, and alveolar cells (Figure 1A, magnification 400×), with a positive expression rate of only 23.3%, whereas the lung cancer tissues of 120/150 patients (80%) were positive for PWP1 expression. In addition, Kaplan–Meier survival analysis was performed, using the Kaplan Meier Plotter (http://kmplot.com/analysis/index), which showed that patients with low PWP1 expression survived significantly longer than patients with high PWP1 expression (p = 0.0057, Figure 1B). We analyzed the expression of PWP1 in NSCLC cell lines (H661, H460, SK-MES-1, LK2, H1299, H292, A549) and a human bronchial epithelial (HBE) cell line in Western blot (WB) analysis. Except for the SK-MES-1 cell line, the PWP1 expression levels in all six remaining lung cancer cell lines were higher than those in the HBE cell line (Figure 1C). Immunofluorescence analysis showed that PWP1 was mainly located in the nucleus and cytoplasm of NSCLC cells. This finding was consistent with our immunohistochemical findings (Figure 1D, magnification 400×). We statistically analyzed data from 101 patients with complete p-TNM staging information and demonstrated that high PWP1 expression positively correlated with low differentiation (p = 0.007), lymph node metastasis (p = 0.001), and high p-TNM staging (p = 0.002) (Table 1). Data from The Cancer Genome Atlas (TCGA) database (http://ualcan.path.uab.edu/cgl-bin/tcgaexres) indicated that PWP1 mRNA-expression levels were significantly higher (P < 0.05) in lung adenocarcinoma and squamous cell lung carcinoma tissues than in normal lung tissues (Figure 1E).
 |
Table 1 The Relationship Between PWP1 Expression and Clinicopathological Factors in NSCLC |
PWP1 Promotes Proliferation and Invasion of NSCLC Cells
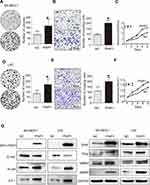
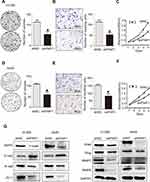
To further investigate the effects of PWP1 on the malignant phenotypes of NSCLC cells, we first transfected PWP1 into SK-MES-1 and LK2 cells (which have low PWP1 expression) and investigated the colony formation, MTT assay and invasive ability of the cells. The results were confirmed that ectopic expression of PWP1 promoted the proliferation (SK-MES-1 cells: control vs PWP1 group, 85.75 ± 15.40 vs 165.80 ± 30.41, p = 0.0220; LK2 cells: control vs PWP1 group, 83.33 ± 17.40 vs 166.30 ± 32.37, p = 0.0312) and invasion (SK-MES-1 cells: control vs PWP1 group, 95.67 ± 5.239 vs 203.00 ± 15.72, p = 0.0194; LK2 cells: control vs PWP1 group, 109.00 ± 23.54 vs 197.70 ± 21.11, p = 0.0246) of NSCLC cells (Figure 2A–F). In addition, after transfected PWP1, the EMT progress was promoted. The expression of E-cadherin (E-cad) protein was decreased, whereas N-cadherin (N-cad), ZO-1, Snail, Slug, MMP2, and MMP9 expressions were significantly increased (Figure 2G). In contrast, after knocking down PWP1 in H1299 and A549 cells (which originally showed the relatively high expression of PWP1), the proliferation (H1299 cells: control vs shPWP1 group, 110.30 ± 5.84 vs 53.00 ± 7.371, p = 0.0442; A549 cells: control vs shPWP1 group, 109.70 ± 2.028 vs 59.67 ± 6.888, p = 0.0131) and invasion (H1299 cells: control vs shPWP1 group, 112.70 ± 5.044 vs 52.67 ± 5.04, p = 0.0206; A549 cells: control vs shPWP1 group, 149.70 ± 3.930 vs 82.67 ± 7.881, p = 0.02490) of cancer cells were inhibited significantly (Figure 3A–F). In addition, after transfecting shPWP1, the EMT progression was inhibited, the expression of E-cad was significantly enhanced (Figure 3G).
PWP1 Activates the Wnt Pathway by Interaction withDVL2
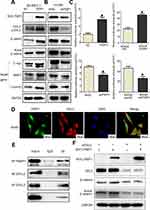
According to the previous reports,5 PWP1 may change the activity of Wnt pathway. To gain insights into the mechanism underlying the promotion of NSCLC cell proliferation and invasion by PWP1 and its possible effects on the Wnt signaling pathway, we transfected PWP1 into SK-MES-1 and LK2 cell lines. WB showed that the phosphorylation of DVL2 and the active β-catenin were upregulated significantly. Furthermore, the expression of target proteins of the Wnt pathway such as Cyclin D, MMP7, and C-myc18 were also upregulated (Figure 4A; Figure S1A). In contrast, transfecting shPWP1 into H1299 and A549 cell lines, the phosphorylation of DVL2 and the active β-catenin were downregulated significantly. Furthermore, the expression of target proteins of the Wnt pathway such as Cyclin D, MMP7, and C-myc were also downregulated (Figure 4B; Figure S1B). We tested the effect of PWP1 on the activities of Wnt pathway in HEK293T cell line by using luciferase reporter assays. The luciferase activity of β-cat/TCF4 was enhanced after transfecting PWP1, to easily observe these differences, we used Wnt3A to activate the Wnt signaling pathway,8,19 the luciferase activity was enhanced more obviously. In contrast, the luciferase activity was inhibited after knocking down PWP1, the activity was inhibited more obviously by using Wnt3A (Figure 4C). Given the previous report, PWP1 could interact with the PDZ domain7 and DVL2 has PDZ domain. Then, we found that the extremely relevant relationship between PWP1 and DVL2 was existed by using GEPIA database (Figure S1C). Immunofluorescence analysis showed that PWP1 and DVL2 co-localized to the cytoplasm (Figure 4D). At the same time, co-ip experiments indicated that PWP1 could interact with DVL2 (Figure 4E). To confirm that PWP1 activates Wnt pathway through interacting with DVL2, we knocked down DVL2 and transfected PWP1, the effects of PWP1 on promoting Wnt pathway were offset (Figure 4F).
PWP1 Inhibits the Hippo Signaling Pathway by Interaction with Merlin
The above experiments showed that PWP1 could interact with DVL2, and a previous report suggested that a factor operating upstream of the Hippo signaling pathway (MTS1 kinase) can inhibit DVL2 phosphorylation.20 Therefore, we speculated that PWP1 may also influence the activity of the Hippo signaling pathway. In order to confirm the effect, first of all, we transfected PWP1 into SK-MES-1 and LK2 cell lines, wb showed that the phosphorylation of Merlin, MST1, LATS1 and YAP were downregulated. At the same time, CTGF and Cyclin E were increased, which are the target genes of Hippo pathway21 (Figure 5A; Figure S2A). In contrast, the phosphorylation of Merlin, MST1, LATS1 and YAP were upregulated after knocking down PWP1 in H1299 and A549 cell lines, nevertheless, CTGF and Cyclin E were decreased (Figure 5B; Figure S2B). The luciferase gene-reporter assays were performed to test whether PWP1 could inhibit the activity of the Hippo signaling pathway. The luciferase activity of YAP/TEAD was enhanced after transfecting PWP1 in HEK293T cells, the opposite effect was showed after knocking down PWP1 in HEK293T cells (Figure 5C). The cytoplasmic co-localization of PWP1 and Merlin were indicated by immunofluorescence (Figure 5D). Although the results above demonstrated that PWP1 inhibited the activity of the Hippo signaling pathway, the underlying mechanism(s) was not immediately clear. After searching the GEPIA database, we discovered an extremely significant correlation between PWP1 and an upstream regulator of the Hippo signaling pathway, Merlin (NF2) (Figure S2C). Hence, we performed co-immunoprecipitation experiments and confirmed the interaction between endogenous PWP1 and Merlin protein in A549 cells (Figure 5E). To confirm that PWP1 inhibited Hippo pathway through interacting with Merlin, we transfected PWP1 and simultaneously knocked down Merlin and found that the effects of PWP1 on inhibiting Hippo pathway were offset (Figure 5F).
Discussion
The expression and functional mechanism(s) of PWP1 in human malignant tumors remain unclear. Through immunohistochemistry, we examined PWP1 protein expression in lung cancer resection specimens and found that PWP1 expression was high in NSCLC. This finding was consistent with data retrieved from the UALCAN database suggesting that PWP1 expression was higher in LUAD and LUSC than in normal lung tissue. Statistical analysis showed that high PWP1 expression correlated positively with poor differentiation of lung cancer tissue, high p-TNM stage, and lymph node metastasis. TCGA database-survival analysis showed that the survival time of patients with high PWP1 expression was significantly shorter than that of patients with low PWP1 expression. Collectively, these findings showed that PWP1 exerts tumor-promoting effects in NSCLC and can potentially serve as a prognostic factor for lung cancer. The possible tumor-promoting effect exerted by PWP1 in lung cancer was also demonstrated in vitro. Transfecting PWP1 into lung cancer cell lines significantly promoted proliferation and invasion, whereas transfecting shPWP1 showed the opposite effects.
Although no reports have described the mechanism of PWP1 in lung cancer cells, previous data showed that PWP1 can alter the activity of the Wnt/β-catenin signaling pathway5 and that the Wnt signaling pathway is related to the Hippo signaling pathway.9,10 Hence, we started by investigating the effects of PWP1 on the activities of the Wnt and Hippo signaling pathways in terms of a potential mechanism whereby PWP1 promotes cancer cell proliferation and invasion. Our luciferase gene-reporter assays and WB results demonstrated that PWP1 promoted the proliferative and invasive abilities of lung cancer cells by enhancing the activity of the Wnt pathway and inhibiting the activity of the Hippo pathway. Co-immunoprecipitation experiments showed that PWP1 could interact with DVL2, possibly stabilizing DVL2 and upregulating its phosphorylation level to thereby upregulate β-catenin and its activation level. Increased nuclear translocation of β-catenin activated the Wnt signaling pathway, which initiated Cyclin D1, MMP7, and C-myc transcription. In addition, PWP1 interacted with Merlin (NF2) to downregulate the level of p-MST1 and, thus, downregulate the p-LATS1 and p-YAP levels. Increased nuclear translocation of YAP drove CTGF and Cyclin E transcription.22,23 By combining PWP1 overexpression with DVL2 or Merlin down-regulation, we offset the effects of PWP1 in promoting Wnt signaling activation and inhibiting the Hippo signaling pathway (Figure 6). These combined effects promoted increased lung cancer cell proliferation and invasion.
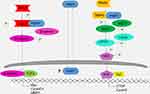 |
Figure 6 Schematic diagram of activating Wnt and inhibiting Hippo pathway by cytosolic PWP1. |
However, our experimental results are still preliminary and several questions remain. For example, it is unclear why PWP1 is highly expressed in NSCLC, whether PWP1 mainly promotes lung cancer cell proliferation and invasion mainly by activating the Wnt pathway or inhibiting the Hippo pathway, whether PWP1 interacts with DVL2 and Merlin by direct binding or by forming a multi-protein complex, and how the interaction of PWP1 with DVL2 and Merlin alters protein phosphorylation. Answers to all of these questions will investigate and validate in our further experiment.
Conclusions
Our study is the first to confirm the high expression of PWP1 in NSCLC and its correlation with a poor prognosis of NSCLC patients. PWP1 promotes proliferation and invasion of cancer cells by activating the Wnt signaling pathway and inhibiting the Hippo pathway. This function of PWP1 is related to its interaction with a key molecule in the Wnt pathway, DVL2, and an upstream factor of the Hippo pathway, Merlin. The findings of this study provide an experimental foundation for further investigating the functional mechanism of PWP1 in malignant cancers, discovering therapeutic targets for lung cancer, and developing new targeted drugs.
Abbreviations
PWP1, periodic tryptophan protein 1; NSCLC, non-small cell lung cancer; ESC, embryonic stem cell; ATCC, American Type Culture Collection; FBS, fetal bovine serum; GEPIA, gene expression profiling interactive analysis; HBE, human bronchial epithelial; WB, Western Blot; PVDF membrane, polyvinylidene fluoride membrane; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; p-TNM, pathological tumor-node-metastasis; PBS, phosphate-buffered saline; SDS, sodium dodecyl sulfate; NC, negative control; EMT, epithelial mesenchymal transition; LATS1, large tumor suppressor-1; MST1, mammalian Ste20-like kinase 1; YAP, yes-associated protein; NF2, neurofibromin; DVL2, dishevelled; CTGF, connective tissue growth factor; IP, immunocoprecipitatition; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; IB, immunoblotting; PDZ domain, PSD95/DLG/ZO1 domain; SPSS, statistical program for social sciences.
Data Sharing Statement
All data generated or analysed during this study are included in this published article and its supplementary information files. Further details were available from the corresponding author upon request.
Ethics Approval and Informed Consent
The patient and healthy volunteer consents were written informed consent and the study protocol was approved by the Ethical Committee of China Medical University and conformed to the Declaration of Helsinki.
Acknowledgment
We thank Dr Hiroshi Kijima for providing the LK2 cell line.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study was supported by the National Natural Science Foundation of China (No. 81772489 to Enhua Wang).
Disclosure
The authors declare that they have no competing interests.
References
1. Liu Y, Matos C, Heino TI, Hietakangas V. PWP1 promotes nutrient-responsive expression of 5S ribosomal RNA. Biol Open. 2018;7(11). doi:10.1242/bio.037911
2. Talkish J, Campbell IW, Sahasranaman A, Jakovljevic J, Woolford JL
3. Zhang C, Lin J, Liu W, Chen X, Chen R, Ye K. Structure of Utp21 tandem WD domain provides insight into the organization of the UTPB complex involved in ribosome synthesis. PLoS One. 2014;9(1):e86540.
4. Neer EJ, Schmidt CJ, Nambudripad R, Smith TF. The ancient regulatory-protein family of WD-repeat proteins. Nature. 1994;371(6495):297–300. doi:10.1038/371297a0
5. Shen J, Jia W, Yu Y, et al. Pwp1 is required for the differentiation potential of mouse embryonic stem cells through regulating Stat3 signaling. Stem Cells. 2015;33(3):661–673. doi:10.1002/stem.1876
6. Honoré B, Baandrup U, Nielsen S, Vorum H. Endonuclein is a cell cycle regulated WD-repeat protein that is up-regulated in adenocarcinoma of the pancreas. Oncogene. 2002;21(7):1123–1129. doi:10.1038/sj.onc.1205186
7. Ludvigsen M, Østergaard M, Vorum H, Jacobsen C, Honoré B. Identification and characterization of endonuclein binding proteins: evidence of modulatory effects on signal transduction and chaperone activity. BMC Biochem. 2009;10:34. doi:10.1186/1471-2091-10-34
8. Han Q, Lin X, Zhang X, et al. WWC3 regulates the Wnt and Hippo pathways via Dishevelled proteins and large tumour suppressor 1, to suppress lung cancer invasion and metastasis. J Pathol. 2017;242(4):435–447.
9. Tsai BP, Hoverter NP, Waterman ML. Blending hippo and WNT: sharing messengers and regulation. Cell. 2012;151(7):1401–1403. doi:10.1016/j.cell.2012.12.007
10. Imajo M, Miyatake K, Iimura A, Miyamoto A, Nishida E. A molecular mechanism that links Hippo signalling to the inhibition of Wnt/beta-catenin signalling. EMBO J. 2012;31(5):1109–1122. doi:10.1038/emboj.2011.487
11. Reguart N, He B, Taron M, You L, Jablons DM, Rosell R. The role of Wnt signaling in cancer and stem cells. Future Oncol. 2005;1(6):787–797. doi:10.2217/14796694.1.6.787
12. Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127(3):469–480. doi:10.1016/j.cell.2006.10.018
13. Hu C, Sun J, Du J, et al. The Hippo-YAP pathway regulates the proliferation of alveolar epithelial progenitors after acute lung injury. Cell Biol Int. 2019;43(10):1174–1183. doi:10.1002/cbin.11098
14. Gao L, Cao H, Cheng X. A positive feedback regulation between long noncoding RNA SNHG1 and YAP1 modulates growth and metastasis in laryngeal squamous cell carcinoma. Am J Cancer Res. 2018;8(9):1712–1724.
15. Lehmann W, Mossmann D, Kleemann J, et al. ZEB1 turns into a transcriptional activator by interacting with YAP1 in aggressive cancer types. Nat Commun. 2016;7:10498. doi:10.1038/ncomms10498
16. Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol. 2015;10(9):1243–1260. doi:10.1097/JTO.0000000000000630
17. Asamura H, Chansky K, Crowley J, et al. The international association for the study of lung cancer lung cancer staging project: proposals for the revision of the N descriptors in the forthcoming 8th edition of the TNM classification for lung cancer. J Thorac Oncol. 2015;10(12):1675–1684. doi:10.1097/JTO.0000000000000678
18. Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997;16(13):3797–3804. doi:10.1093/emboj/16.13.3797
19. Bryja V, Schulte G, Arenas E. Wnt-3a utilizes a novel low dose and rapid pathway that does not require casein kinase 1-mediated phosphorylation of Dvl to activate beta-catenin. Cell Signal. 2007;19(3):610–616. doi:10.1016/j.cellsig.2006.08.011
20. Xu F, Wang YL, Chang JJ, et al. Mammalian sterile 20-like kinase 1/2 inhibits the Wnt/beta-catenin signalling pathway by directly binding casein kinase 1epsilon. Biochem J. 2014;458(1):159–169.
21. Hilman D, Gat U. The evolutionary history of YAP and the hippo/YAP pathway. Mol Biol Evol. 2011;28(8):2403–2417. doi:10.1093/molbev/msr065
22. Rong X, Han Q, Lin X, Kremerskothen J, Wang E. FRMPD1 activates the Hippo pathway via interaction with WWC3 to suppress the proliferation and invasiveness of lung cancer cells. Cancer Manag Res. 2019;11:3395–3410. doi:10.2147/CMAR.S194512
23. Rong X, Liang Y, Han Q, et al. Molecular mechanisms of tyrosine kinase inhibitor resistance induced by membranous/cytoplasmic/nuclear translocation of epidermal growth factor receptor. J Thorac Oncol. 2019;14(10):1766–1783. doi:10.1016/j.jtho.2019.06.014
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.