Back to Journals » Cancer Management and Research » Volume 11
PUF60 accelerates the progression of breast cancer through down-regulation of PTEN expression
Authors Sun D , Lei W , Hou X, Li H , Ni W
Received 14 July 2018
Accepted for publication 19 November 2018
Published 17 January 2019 Volume 2019:11 Pages 821—830
DOI https://doi.org/10.2147/CMAR.S180242
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Dongying Sun,* Wei Lei,* Xiaodong Hou, Hui Li, Wenlu Ni
Department of Medical Imaging, Henan University First Affiliated Hospital, Kaifeng, Henan, China
*These authors contributed equally to this work
Background: PUF60 is a splicing variant of far upstream element binding protein 1-interacting repressor, which is abnormally expressed in a variety of tumors and is closely involved in their progression. However, whether PUF60 participates in the occurrence and development of breast cancer remains unknown. Therefore, the objective of the current study is to explore the effects and mechanism of PUF60 in the progression of breast cancer.
Methods: PUF60 expression patterns in breast cancer tissues and cells were determined by RT-PCR and Western blotting. The relationship between PUF60 expression and patients’ clinical features and outcome was evaluated to assess the potential of PUF60 as a marker for progression and prognosis prediction. CCK-8, flow cytometry, transwell and in vivo tumor formation assays were used to detect cell proliferation, apoptosis, migration, invasion and tumorigenesis. The effects of PUF60 on the activation of PTEN/PI3K/AKT were also evaluated by Western blotting and immunofluorescence assays.
Results: The expression of PUF60 was elevated in breast cancer tissue samples and cell lines, and its high expression was closely associated with the high incidence of lymph node metastasis and advanced TNM stage. Besides, upregulation of PUF60 with lentivirus infection significantly increased the growth, migration, and invasion and repressed the apoptosis of breast cancer HCC1937 and MDA-MB-231 cells, while silencing of PUF60 with shRNA showed the opposite results. Moreover, PUF60 upregulation promoted the expression of p-AKT, PI3K, and mTOR, while decreased PTEN expression through inhibiting its stability and enhancing its ubiquitination. Furthermore, upregulation of PUF60 promoted the tumorigenesis in vivo, whereas this effect was impaired when PTEN expression was upregulated in MDA-MB-231 and HCC1937 cells.
Conclusion: This study demonstrates that PUF60 is highly expressed in breast cancer; upregulation of PUF60 accelerates the progression of breast cancer through PTEN inhibition.
Keywords: PUF60, PTEN, PI3K/AKT, breast cancer, proliferation, tumorigenesis
Introduction
Breast cancer is one of the most commonly diagnosed malignant cancers and a leading cause of cancer-related mortality in women worldwide.1–3 Every year, there are ~1.4 million new cases and 450,000 deaths caused by breast cancer.2 Although big progress has been made in the diagnosis and therapy recently, the prognosis of patients with breast cancer is still dim.4 Therefore, it is necessary to further explore the pathogenesis of breast cancer, hoping to find new potent therapeutic schedule for patients’ with breast cancer.
It’s well documented that precursor mRNAs (pre-mRNAs) need to go through a series of complex processes prior to becoming an active and mature mRNA, in which splicing plays a crucial role.5 Through selective splicing of different 5′ or 3′ end splicing sites of the same gene, different gene transcripts can be produced followed by the formation of proteins with different functions. Note that 95% of pre-mRNAs are alternatively spliced in humans,6 leading to at least 100,000 distinct proteins despite only 20,000 protein-coding genes.7 Once disorders, abnormal protein subtype will be formed and deregulate multiple biological processes, such as cell proliferation, differentiation, apoptosis, cycle, metabolism, invasion, migration and angiogenesis,8 and has been considered to be involved in the development of many kinds of tumors, including ovarian cancer,9 lung cancer,10 osteosarcoma,11 gastric cancer12 and breast cancer,13 and so on. In recent years, studies have found that PUF60, a splicing factor, is involved in the progression of several various kinds of tumors.14 In breast cancer, Gumireddy et al15 demonstrated that downregulation of PUF60 expression partially rescued the E-cadherin suppression induced by translational regulatory lncRNA, suggesting that PUF60 might involve in the malignant progression of breast cancer.
Therefore, this study was carried out to explore the function of PUF60 in the development and progression of breast cancer, as well as study its molecular mechanism.
Materials and methods
Ethic statement
All patients enrolled in this study had provided the written informed consent. Experiments related to human or human samples were approved by the Human Research Committee of Henan University First Affiliated Hospital and were conducted in accordance with the Declaration of Helsinki; the animal experiments were approved by Animal Ethics Committee of Henan University First Affiliated Hospital and carried out in accordance with the principles and procedures outlined in the National Institutes of Health Guide for the Care and Use of Laboratory Animals. All efforts were made to relief the suffering of animals during the whole operation procedure.
Patients and tissue samples
Thirty patients with breast cancer and 30 gender- and age-matched healthy individuals were enrolled in this study for analysis of the blood level of PUF60. At the same time, 48 patients with PUF60 high expression and 30 patients with PUF60 low expression determined by immunohistochemistry score of PUF60 were chosen as the study subjective of this study.
Immunohistochemistry
Fifteen matched breast cancer tissues and the adjacent normal tissues were selected from the 78 breast cancer patients with PUF60 high or low expression randomly. After being formalin fixed and paraffin embedded, the tissues were cut into 5 µm slides and then received immunohistochemistry staining according to previous study.16 Briefly, the slides were dewaxed with xylene and ethanol, and antigen repaired with EDTA buffer (pH 8.0) in a microwave oven. After being incubated with 5% goat serum diluted with TBS buffer containing 1% Tween-20, the slides were probed with the primary antibody against PUF60 (no. ab22819; Abcam, Cambridge, MA, USA) overnight at 4°C and the corresponding second antibody for 1 hour at room temperature.
To analyze the expression level of PUF60 in different breast tissue samples, we graded the staining of PUF60 in 15 paired cancer tissues and normal tissues by multiplying the staining extent and intensity referring to previous study.17 The staining extent was scored according to the percentage of the positively stained area with the following scale: 0 (<5%), 1 (5%–25%), 2 (25%–50%), 3 (50%–75%), and 4 (>75%). The staining intensity was scored as: 0 (no staining), 1 (weak), 2 (distinct), and 3 (strong). The scoring was determined by two evaluators who did not know the pathological and clinical characteristics of the patients.
Cell lines and treatments
Human normal breast cell line MCF-10A and breast cancer cell lines HCC1937, MDA-MB-231 and MCF-7 were obtained from BeNa Culture Collection (Beijing, China). MCF-10A and HCC1937 cells were cultured in the completely culture medium containing 90% RPMI-1640 (Thermo Fisher Scientific, Waltham, MA, USA) and 10% FBS (Thermo Fisher Scientific); MDA-MB-231 cells were cultured in 90% L-15 cell culture medium (Thermo Fisher Scientific) and 10% FBS; MDA-MB-231 cells and MCF-7 cells were cultured in minimum essential medium (Thermo Fisher Scientific) supplemented with 10% FBS. All cells were maintained in a humidified incubator with 5% CO2 in a constant temperature of 37°C.
To upregulate or downregulate the expression of PUF60, lentivirus containing PUF60 open reading frame (OE-PUF60) and shRNA targeting human PUF60 gene were purchased from OriGene (Beijing, China). And lentivirus (OE-PTEN) used to upregulate PTEN expression was designed and synthesized by GenePharma (Shanghai, China).
Real-time PCR analysis (RT-PCR)
Total RNA was extracted from tissues and cells with RiboPure™ RNA Purification Kit (Invitrogen™, Carlsbad, CA, USA) according to the manufacturer’s description. Then, a total of 1 µg of RNA was reverse-transcribed into cDNA with SuperScript® VILO™ cDNA kit (Thermo Fisher Scientific). And the cDNA samples from different groups were amplified in triplicate using the commercially available SYBR Green PCR Master Mix (Thermo Fisher Scientific) with ABI7000 PCR instrument. GAPDH was used as an internal reference to normalize the expression of other mRNAs. Sequences used in this study were listed as follows: AKT forward (F): 5′-CCAGCCTGGGTCAAAGAAGT-3′, AKT reverse (R): 5′-TCTCCTCCTCCTCCTGCTTC-3′; mTOR (F): 5′- CTTAGAGGACAGCGGGGAAG-3′, mTOR (R): 5′-TCCAAGCATCTTGCCCTGAG-3′; PTEN (F): 5′-CTGCAGAAAGACTTGAAGGCG-3′, PTEN (R): 5′-GGGAATAGTTACTCCCTTTTTGTC-3′; PI3K (F): 5′-TGGAGAGAGAGCAGTTCCAA-3′, PI3K (R): 5′- GCAAAGCTGTTGCATTTTCAGT-3′; PUF60 (F): 5′-AATGGAAACCTCCACAGGGC-3′, PUF60 (R): 5′- GGCGTACTTCTTGGCCTTCT-3′; GAPDH (F): 5′-GAGAGAAACCCGGGAGGCTA-3′, GAPDH (R): 5′- GACTCCACGACGTACTCAGC-3′.
Western blotting analysis
Protein samples obtained from cells and tissues using RIPA lysis buffer (Beyotime, Jiangsu, China) were centrifuged at 12,000 g for 30 minutes at 4°C. After quantified with BCA Kit (Thermo Fisher Scientific), equal protein samples (20–30 µg) from each group were separated by 10% SDS-PAGE and transferred to the polyvinylidene fluoride membranes (Millipore Sigma, Burlington, MA, USA). Next, the membranes were incubated with primary antibody against PUF60 (no. ab22819; Abcam), AKT (no. ab8805; Abcam), phosphorylated (p)-AKT (no. ab38449; Abcam), mTOR (no. ab2732; Abcam), PTEN (no. ab31392; Abcam), PI3K (no. ab86714; Abcam), Bax (no. 2772; Cell Signaling Technology, Danvers, MA, USA), Bcl-2 (no. 2872; Cell Signaling Technology), and Ub (no. 3933; Cell Signaling Technology), followed by being probed with the corresponding horseradish peroxidase-conjugated secondary antibodies (Cell Signaling Technology). Protein-expressing signal was detected by using enhanced chemiluminescence and detected by Gel imaging system (Shanghai Bio-Tech Co., Ltd. Shanghai, China).
Immunoprecipitation (IP) assay
Immunoprecipitation (IP) assay was performed to evaluate the interaction between two proteins.18 First, cell lysates were incubated with protein A/G-agarose beads (Millipore) for 1 hour at 4°C. Then, the lysates were transferred into a new tube and incubated with antibody against PUF60 or Flag at 4°C overnight. Subsequently, the immune complex was washed with 1× IP buffer for three times and subjected to Western blot using PTEN, AKT, mTOR, or Ub antibodies.
Cell proliferation detection
CCK-8 regent purchased from Dojindo (Tokyo, Japan) was recruited to detect cell proliferation in this study. Briefly, MDA-MB-231 or HCC1937 cells at a concentration 2×103 were grown into 96-well culture plates, and 100 µL culture medium containing 10% FBS was added to it. Then, different treatments (sh-NC, sh-PUF60, OE-NC, and OE-PUF60) were executed to the cells before growth detection. After 1–5 days of the treatments, 10 µL of CCK-8 reagent was added and incubated for further 3 hours at 37°C with 5% CO2. The absorbance measured at 450 nm was used to draw cell growth curve.
Cell apoptosis detection
The apoptosis of MDA-MB-231 and HCC1937 cells treated with sh-NC, sh-PUF60, OE-NC, or OE-PUF60 was determined by flow cytometry assay with Annexin V (FITC)/propidium iodide (PI) apoptosis detection kit (KeyGEN Biotech, Jiangsu, China) according to previous study.19
Transwell assay
The migration and invasion potentials of MDA-MB-231 and HCC1937 cells were determined by using 24-well Transwell chambers (Costar, MA, USA) coated with Matrigel (invasion detection) or not (migration detection). After 48 hours of different treatments, 1×105 cells suspended with 200 µL serum-free medium were digested by trypsin and seeded into the upper chamber of an 8 μm insert. At the same time, 600 µL cell culture medium supplemented with 15% FBS was added into the lower chamber. After 24 hours (migration detection) or 48 hours (invasion detection), cells on the surface of the upper chamber were wiped with cotton tips, and the invasive cells sticking to the lower membrane surface were fixed with precold methyl alcohol for 20 minutes and stained with crystal violet for 5 minutes, successively. The number of invasive cells was counted in six randomly selected fields under a microscope.
Immunofluorescence technique
Cells seeded on coverslips were washed thrice with PBS and fixed with cold methanol, washed with PBST thrice and sealed with 5% goat serum (diluted with PBS) for 1 hour in succession. Next, the cells were probed with primary antibodies against PTEN (1:100) and PUF60 (1:150) overnight at 4°C, followed by incubation with the corresponding fluorescent second antibodies including IgG AF488 (green) and IgG AF568 (red) (Invitrogen) in the dark for 1 hour. Eventually, cells were incubated with DAPI solution (1:1×104 dilution with PBS) for 5 minutes and covered with the antifade mounting medium. The expression and subcellular location of PUF60 and PTEN were visualized by an Olympus Confocal FV100 microscope.
Xenograft transplantation in vivo
Four-week-old male athymic Balb/C mice were purchased from Jackson Laboratory (Beijing, China) and maintained in a specific pathogen-free condition. One million of MDA-MB-231 or HCC1937 cells with PUF60 or PUF60+PTEN stably overexpressing (selected by 7 µg/mL puromycin or 100 µg/mL G418) were subcutaneously injected in the armpit area of these mice. Mice were observed over 4 weeks, and then the tumors were took out and weighted.
Data analysis
Experiments in the current study were performed at least thrice and the results are presented as the mean ± SD. Statistical analysis was executed by SPSS23.0 or GraphPad software using the Student’s t-test or one-way ANOVA test. The association of PUF60 expression and the clinicopathological features of breast cancer patients was analyzed by chi-squared test. The relationship between PUF60 expression and patients’ survival time was analyzed by Kaplan–Meier analysis. A P-value <0.05 was identified statistically significant.
Results
PUF60 expression is increased in breast cancer tissues and cell lines
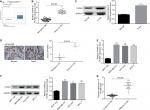
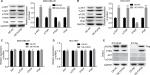
To determine the roles of PUF60 in the development of breast cancer, we first assessed its expression patterns in the breast cancer tissues and cell lines. First, from the data of Oncomine, we found that the expression of PUF60 was higher in the breast cancer patients than that of the control individuals (Figure 1A). Second, we performed real-time PCR, Western blotting, and immunohistochemistry staining to evaluate the mRNA and protein levels of PUF60 in breast cancer tissues and the corresponding adjacent nontumor tissues. Compared to the adjacent nontumor tissue samples, the mRNA and protein levels of PUF60 were all increased in the breast cancer tissue samples (Figure 1B and C), as well as the average score of immunohistochemistry staining of PUF60 (Figure 1D). Subsequently, we compared the expression pattern of PUF60 among breast cancer cell lines and normal breast cell line. As shown in Figure 1E and F, both the mRNA and protein expressions of PUF60 were increased in breast cancer cell lines, including MDA-MB-231, HCC1937, and MCF-7 when compared with the normal breast cell line MCF-10A. Moreover, PUF60 was also significantly upregulated in the blood samples of patients with breast cancer as compared with that of the healthy individuals (Figure 1G). These above results suggest that PUF60 might involve in the progression of breast cancer.
High expression of PUF60 closely associates with the malignant clinical progress and poor prognosis of breast cancer patients
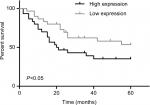
Next, we explored the relationship between PUF60 expression and patients’ clinical progress and outcome. As Table 1 showed, 48 cases of breast cancer patients with PUF60 high expression and 30 cases with PUF60 low expression were enrolled in this study. And from Table 1, we could find that patients in PUF60 high-expression group had a higher incidence of lymph node metastasis (P=0.018) and higher TNM stage (P=0.01) than those of PUF60 low-expression group. Besides, we performed Kaplan–Meier analysis to assess the effect of PUF60 expression on the overall survival of breast cancer patients. Result showed that the average overall survival in PUF60 high-expression patients was shorter than patients with PUF60 low expression (Figure 2). And the HR of the expression of PUF60 was ~1.46 (63.3%/43.3%). These findings demonstrate that high expression of PUF60 predicts an advanced clinical progress and poor prognosis in patients with breast cancer.
Overexpression of PUF60 promotes the malignant phenotypic transformation of breast cancer cells
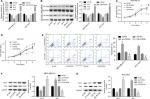
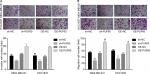
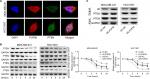
Then, we explored the influence of PUF60 in cell function of breast cancer MDA-MB-231 and HCC1937 cells. sh-2 targeting human PUF60 gene showed the best knockdown efficiency among the three shRNAs, which obviously decreased PUF60 expression and was chosen for further study, and OE-PUF60 markedly increased PUF60 expression both in mRNA and protein levels (Figure 3A and B). Upregulation of PUF60 with OE-PUF60 infection obviously increased the growth (Figure 3C and D) and inhibited the apoptosis (Figure 3E) of MDA-MB-231 and HCC1937 cells, while knockdown of PUF60 with sh-PUF60 infection obviously inhibited cell growth and promoted cell apoptosis (Figure 3C–E). And the apoptosis inhibition role of PUF60 was also confirmed by the Western blotting analysis that PUF60 upregulation increased the expression of Bcl-2 and decreased Bax expression, and vice versa (Figure 3F and G). Furthermore, upregulation of PUF60 enhanced the migration and invasion of MDA-MB-231 and HCC1937 cells, while knockdown of PUF60 caused the opposite results (Figure 4A and B). These results indicate that PUF60 promotes the malignant phenotypic transformation of breast cancer, suggesting that PUF60 functions as an oncogene in breast cancer.
Upregulation of PUF60 promotes the activation of PI3K/AKT/PTEN signaling pathway in breast cancer
We then assessed if PI3K/AKT/PTEN signaling pathway takes part in the process in which PUF60 accelerated the malignant phenotypic transformation of breast cancer cells. Results from Western blotting analysis demonstrated that the expression of p-AKT and PI3K and mTOR were upregulated while PTEN protein level was reduced in MDA-MB-231 (Figure 5A) and HCC-1975 cells (Figure 5B) with no obvious change in mRNA levels (Figure 5B–D). Besides, IP assay showed that PUF60 protein could bind to PTEN protein but not AKT or mTOR (Figure 5E). In addition, we performed immunofluorescence staining to verify the combination of PUF60 protein and PTEN protein in MDA-MB-231 and HCC1937 cells. And results demonstrated that there was a colocation of PUF60 and PTEN proteins in nuclear (Figure 6A). Moreover, upregulation of PUF60 speeded up the degradation of PTEN protein (Figure 6B) and enhanced its ubiquitination (Figure 6C), while knockdown of PUF60 made PTEN protein more stable and reduced its ubiquitination in both MDA-MB-231 and HCC1937 cells (Figure 6B and C). Overall, these findings make clear that overexpression of PUF60 promotes the activation of PI3K/AKT/PTEN signaling pathway in breast cancer.
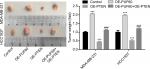
Upregulation of PUF60 promotes the tumorigenesis of breast cancer cells through downregulation of PTEN in vivo
Finally, we recruited the in vivo mouse tumor-bearing assay to evaluate the effect of PUF60 on the tumorigenesis of MDA-MB-231 and HCC1937 cells. Results showed that upregulation of PUF60 increased the average tumor weight of six nude mice when compared with the OE-NC group, while PTEN upregulation impaired this effect (Figure 7), indicating that PUF60 promotes the tumorigenesis of breast cancer cells through downregulating PTEN.
Discussion
Almost 60% of all human genes have at least one alternative splice variant, and alternative splicing has been identified to play a vital role in human diseases, including multiple categories of cancers.14,20 In this study, we first focused on the association of PUF60 expression and the clinical and pathological features of breast cancer. We found that PUF60 was highly expressed in breast cancer tissue samples and cell lines, and its high expression closely associated with the lymph node metastasis (P=0.018) and advanced clinical stage (P=0.01) of patients with breast cancer. Similarly, Muller et al revealed that overexpression of PUF60 also significantly correlated with lymph node metastasis in non-small cell lung cancer (NSCLC).21 All findings indicate the important value of PUF60 in the diagnosis and targeted therapy for cancers.
To explore the effects of PUF60 in the progression of breast cancer, we carried out gain-/loss-of-function assays. Results showed that PUF60 overexpression significantly enhanced the proliferation, migration, invasion, and tumorigenesis of breast cancer cells, as well as repressed cell apoptosis, indicating that PUF60 functions as an oncogene, which promotes the malignant progression of breast cancer. PUF60 is a splicing variant of FIR that lacks the exon 5, which consists of 17 amino acids.22 Identically to FIR, PUF60 is identified to be a transcriptional repressor of c-Myc gene, which is critically activated in tumorigenesis in various tumors,23–25 and the exon 2 of FIR/PUF60 plays an indispensable role in this process.26 However, in several kinds of cancers such as NSCLC, colorectal cancer, and hepatocellular carcinoma,7,21,27–29 FIR is alternatively spliced lacking exon 2 (FIRΔexon2), which functions as a dominant negative form of authentic FIR.7 Additionally, FIR/PUF60 and FIRΔexon2 have been reported to be upregulated in various malignant tumors, including colon cancer,28 hepatocellular carcinoma,29 acute lymphoblastic leukemia,30 and lung cancer.21 For instance, high expressed FIR was significantly enriched at the invasion type of primary NSCLCs and siRNA-mediated knockdown of all FIR variants obviously decreased the speed and directional movement of tumor cells, as well as the sprouting efficiency and single cell invasiveness.21 Noticeably, silencing of FIRΔexon2 alone was sufficient to reduce cell migration and invasion.21 And in the present study, we explored the function of PUF60 only (not FIRΔexon2) in breast cancer progression and found it functioned as an oncogene in breast cancer.
Importantly, FIR/PUF60 is a multifunctional protein through alternative splicing, resulting in c-Myc gene transcriptional repression, RNA splicing, and DNA damage repair.31 According to this, we speculate that the deregulation of PUF60/FIR might affect the activation of cell survival-related signaling through regulating the process of RNA splicing or DNA damage of the crucial genes. Compared with the control group, upregulation of PUF60 with lentivirus obviously increased the expression of p-AKT, mTOR, and PI3K while decreased PTEN expression, suggesting that the deregulation of PUF60 activated the PI3K/AKT signaling pathway in breast cancer. The PI3K/AKT/mTOR pathway is a key intracellular signaling transduction pathway that drives cellular growth and survival and represses cell apoptosis. Hyperactivation of this pathway is generally implicated in the tumorigenesis of breast cancer.32,33 Phosphatase and tensin homolog deleted on chromosome 10 (PTEN), as a key member of this pathway, is also well documented as a tumor-suppressor gene and frequently mutated or silenced in breast cancer.34 And in the current study, we also observed that there was colocation in nucleus of PUF60 and PTEN proteins, and overexpression of PUF60 could significantly repress the protein stability of PTEN and enhance its ubiquitination. Furthermore, the promotion of tumorigenesis of breast cancer MDA-MB-231 and HCC1937 cells induced by PUF60 upregulation was weakened when PTEN was upregulated in vivo, indicating that PTEN was involved in the process in which PUF60 accelerates the malignant development of breast cancer.
However, we are failed to uncover if the expression form of PUF60/FIR is FIRFIRΔexon2 or not and that if FIRFIRΔexon2 alone is sufficient to stimulate breast cancer progression. One possible explanation is that FIR (or FIRFIRΔexon2) facilitates its protumourigenic properties through the activation of c-Myc transcription. Except for c-Myc, additional factors might be involved in the oncogenic effects of PUF60/FIR in human carcinogenesis. Besides, if RNA splicing or DNA damage of the main genes in PI3K/AKT signaling pathway is involved in PUF60-induced PI3K/AKT activation remains unknown. We would like to explore them in our further study.
Overall, the present study demonstrates that PUF60 is highly expressed in breast cancer, and PUF60 overexpression accelerates the progression of breast cancer partly depend on PTEN inhibition.
Acknowledgment
This work was supported by key scientific research projects in Henan colleges and universities, China (grant no. 19A320001).
Disclosure
The authors report no conflicts of interest in this work.
References
Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66(4):271–289. | ||
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. | ||
Mittal S, Brown NJ, Holen I. The breast tumor microenvironment: role in cancer development, progression and response to therapy. Expert Rev Mol Diagn. 2018;18(3):227–243. | ||
Corona SP, Sobhani N, Ianza A, et al. Advances in systemic therapy for metastatic breast cancer: future perspectives. Med Oncol. 2017; 34(7):119. | ||
Zhou Z, Fu XD. Regulation of splicing by SR proteins and SR protein-specific kinases. Chromosoma. 2013;122(3):191–207. | ||
Liu J, He L, Collins I, et al. The FBP interacting repressor targets TFIIH to inhibit activated transcription. Mol Cell. 2000;5(2):331–341. | ||
Matsushita K, Tomonaga T, Shimada H, et al. An essential role of alternative splicing of c-myc suppressor FUSE-binding protein-interacting repressor in carcinogenesis. Cancer Res. 2006;66(3):1409–1417. | ||
Sveen A, Kilpinen S, Ruusulehto A, Lothe RA, Skotheim RI. Aberrant RNA splicing in cancer; expression changes and driver mutations of splicing factor genes. Oncogene. 2016;35(19):2413–2427. | ||
Lee AW, Bomkamp A, Bandera EV, et al. A splicing variant of TERT identified by GWAS interacts with menopausal estrogen therapy in risk of ovarian cancer. Int J Cancer. 2016;139(12):2646–2654. | ||
Zhou B, Wang GZ, Wen ZS, et al. Somatic mutations and splicing variants of focal adhesion kinase in non-small cell lung cancer. J Natl Cancer Inst. 2018;110(2):195–204. | ||
Pruksakorn D, Teeyakasem P, Klangjorhor J, et al. Overexpression of KH-type splicing regulatory protein regulates proliferation, migration, and implantation ability of osteosarcoma. Int J Oncol. 2016;49(3):903–912. | ||
Li Y, Yuan Y. Alternative RNA splicing and gastric cancer. Mutat Res. 2017;773:263–273. | ||
Read A, Natrajan R. Splicing dysregulation as a driver of breast cancer. Endocr Relat Cancer. 2018;25(9):R467–R478. | ||
Chen J, Weiss WA. Alternative splicing in cancer: implications for biology and therapy. Oncogene. 2015;34(1):1–14. | ||
Gumireddy K, Li A, Yan J, et al. Identification of a long non-coding RNA-associated RNP complex regulating metastasis at the translational step. Embo J. 2013;32(20):2672–2684. | ||
Xin B, He X, Wang J, et al. Nerve growth factor regulates CD133 function to promote tumor cell migration and invasion via activating ERK1/2 signaling in pancreatic cancer. Pancreatology. 2016;16(6):1005–1014. | ||
Wei W, Sun HH, Li N, et al. WNT5A modulates cell cycle progression and contributes to the chemoresistance in pancreatic cancer cells. Hepatobiliary Pancreat Dis Int. 2014;13(5):529–538. | ||
Cai J, du S, Wang H, et al. Tenascin-C induces migration and invasion through JNK/c-Jun signalling in pancreatic cancer. Oncotarget. 2017;8(43):74406–74422. | ||
Wang H, Cai J, du S, et al. Fractalkine/CX3CR1 induces apoptosis resistance and proliferation through the activation of the AKT/NF-κB cascade in pancreatic cancer cells. Cell Biochem Funct. 2017;35(6):315–326. | ||
Brinkman BM. Splice variants as cancer biomarkers. Clin Biochem. 2004;37(7):584–594. | ||
Müller B, Bovet M, Yin Y, et al. Concomitant expression of far upstream element (FUSE) binding protein (FBP) interacting repressor (FIR) and its splice variants induce migration and invasion of non-small cell lung cancer (NSCLC) cells. J Pathol. 2015;237(3):390–401. | ||
Rahmutulla B, Matsushita K, Satoh M, et al. Alternative splicing of FBP-interacting repressor coordinates c-Myc, P27Kip1/cyclinE and Ku86/XRCC5 expression as a molecular sensor for bleomycin-induced DNA damage pathway. Oncotarget. 2014;5(9):2404–2417. | ||
Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8(12):976–990. | ||
Yang L, Zhu JY, Zhang JG, et al. Far upstream element-binding protein 1 (FUBP1) is a potential c-Myc regulator in esophageal squamous cell carcinoma (ESCC) and its expression promotes ESCC progression. Tumour Biol. 2016;37(3):4115–4126. | ||
Liu ZH, Hu JL, Liang JZ, et al. Far upstream element-binding protein 1 is a prognostic biomarker and promotes nasopharyngeal carcinoma progression. Cell Death Dis. 2015;6:e1920. | ||
Matsushita K, Tamura M, Tanaka N, et al. Interactions between SAP155 and FUSE-binding protein-interacting repressor bridges c-Myc and P27Kip1 expression. Mol Cancer Res. 2013;11(7):689–698. | ||
Kajiwara T, Matsushita K, Itoga S, et al. SAP155-mediated c-myc suppressor far-upstream element-binding protein-interacting repressor splicing variants are activated in colon cancer tissues. Cancer Sci. 2013;104(2):149–156. | ||
Malz M, Bovet M, Samarin J, et al. Overexpression of far upstream element (FUSE) binding protein (FBP)-interacting repressor (FIR) supports growth of hepatocellular carcinoma. Hepatology. 2014;60(4):1241–1250. | ||
Matsushita K, Kitamura K, Rahmutulla B, et al. Haploinsufficiency of the c-myc transcriptional repressor FIR, as a dominant negative-alternative splicing model, promoted p53-dependent T-cell acute lymphoblastic leukemia progression by activating Notch1. Oncotarget. 2015;6(7):5102–5117. | ||
Müller B, Bovet M, Yin Y, et al. Concomitant expression of far upstream element (FUSE) binding protein (FBP) interacting repressor (FIR) and its splice variants induce migration and invasion of non-small cell lung cancer (NSCLC) cells. J Pathol. 2015;237(3):390–401. | ||
Page-Mccaw PS, Amonlirdviman K, Sharp PA. PUF60: a novel U2AF65-related splicing activity. RNA. 1999;5(12):1548–1560. | ||
Sharma VR, Gupta GK, Sharma AK, et al. PI3K/Akt/mTOR intracellular pathway and breast cancer: factors, mechanism and regulation. Curr Pharm Des. 2017;23(11):1633–1638. | ||
Basho RK, Gilcrease M, Murthy RK, et al. Targeting the PI3K/AKT/mTOR pathway for the treatment of mesenchymal triple-negative breast cancer: evidence from a Phase 1 trial of mTOR inhibition in combination with liposomal doxorubicin and bevacizumab. JAMA Oncol. 2017;3(4):509–515. | ||
Chiang KC, Chen HY, Hsu SY, et al. PTEN insufficiency modulates ER+ breast cancer cell cycle progression and increases cell growth in vitro and in vivo. Drug Des Devel Ther. 2015;9:4631–4638. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.