Back to Journals » Infection and Drug Resistance » Volume 14
PspA Diversity, Serotype Distribution and Antimicrobial Resistance of Invasive Pneumococcal Isolates from Paediatric Patients in Shenzhen, China
Authors Jiang H, Meng Q, Liu X, Chen H, Zhu C, Chen Y
Received 19 October 2020
Accepted for publication 17 December 2020
Published 11 January 2021 Volume 2021:14 Pages 49—58
DOI https://doi.org/10.2147/IDR.S286187
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Hanfang Jiang,1 Qing Meng,2 Xiaorong Liu,2 Hongyu Chen,2 Chunqing Zhu,2 Yunsheng Chen2
1Clinical Laboratory, Institute of Pediatrics, Shenzhen Children’s Hospital, Shenzhen, Guangdong, People’s Republic of China; 2Clinical Laboratory, Shenzhen Children’s Hospital, Shenzhen, Guangdong, People’s Republic of China
Correspondence: Yunsheng Chen
Clinical Laboratory, Shenzhen Children’s Hospital, Shenzhen, Guangdong, People’s Republic of China
Email [email protected]
Introduction: To determine the phenotypes and genotypes of invasive Streptococcus pneumoniae (S. pneumoniae), 108 strains were isolated from paediatric patients with invasive pneumococcal diseases (IPDs) in Shenzhen from 2014 to 2018.
Methods: Serotype profiles were defined by multiplex PCR of the capsule gene. Pneumococcal surface protein A (PspA) classification was performed through pspA gene sequencing. Antimicrobial resistance was examined by broth microdilution. Multilocus sequence typing (MLST) was determined based on next-generation sequencing data.
Results: Eighty-one S. pneumoniae of 17 serotypes were finally collected. The coverage of the 13-conjugated polysaccharide vaccine (PCV13) was 88.9%. After the introduction of PCV13, the nonvaccine serotypes were added by serotypes 15b, 16F and 20. Vaccine serotype 3 increased by four serious cases. The pspA family 1 and pspA family 2 are predominant. The multiple drug resistance rate is 91.3%. None of the nonmeningitis isolates were resistant to penicillin, while 98.8% of all the isolates were resistant to erythromycin.
Discussion: This work characterizes the molecular epidemiology of invasive S. pneumoniae in Shenzhen. Continued surveillance of serotype distribution and antimicrobial susceptibility is necessary to alert antibiotic-resistant nonvaccine serotypes and highly virulent serotypes.
Keywords: Streptococcus pneumoniae, invasive pneumococcal disease, PspA family, serotype, antimicrobial resistance
Introduction
S. pneumoniae is a human respiratory tract pathogen that colonizes the nasopharynx of healthy carriers.1,2 It can cause otitis media, sinusitis, bronchitis and invasive pneumococcal diseases (IPDs), such as sepsis and meningitis.3 IPDs lead to high morbidity and mortality in young children and elderly people worldwide, posing a huge threat to public health.4 The World Health Organization (WHO) reported that S. pneumoniae is the most common pathogen of pneumonia, accounting for 16% of all deaths in children under 5 years old globally.
S. pneumoniae is capsular Gram-positive diplococcus.1 The capsule is an effective vaccine target. The currently available vaccines are based on pneumococcal capsular polysaccharides, of which 98 serotypes exist.5 The 23-valent polysaccharide vaccine, which covers 23 serotypes, cannot stimulate efficient protective immunity in children under 2 years old.6 The WHO recommended polyvalent conjugate vaccines, which include 7-conjugated polysaccharide vaccine (PCV7), 10-conjugated polysaccharide vaccine (PCV10) and 13-conjugated polysaccharide vaccine (PCV13). They show enhanced immunogenicity.7 Since the introduction of PCVs, the incidence of IPDs caused by nonvaccine serotypes has increased drastically worldwide.8–10 However, the PCVs can`t cover all serotypes. Serotype replacement in the postvaccine era stresses the need for the development of capsule-independent pneumococcal vaccines.
Surface proteins of S. pneumoniae are promising nonpolysaccharide vaccine candidates.11 PspA is able to elicit antibody-mediated protection against pneumococcal infection.12–14 Multiple antigen vaccines containing PspA showed good immunogenicity and protection in a Phase 1 clinical trial.15 PspA is exposed on all pneumococcal strains and highly variable in DNA and protein sequences. It consists of three domains: an α-helical domain (αHD) at the N-terminus, a proline-rich domain and a choline-binding domain with a short hydrophobic tail at the C-terminus.16,17 The C-terminal 100 amino acids of αHD are immunogenic and protective,18 forming a clade-defining region (CDR).16 Based on CDRs, S. pneumoniae is distributed into three families and six clades: family 1 comprises clades 1 and 2; family 2 comprises clades 3, 4 and 5; and family 3 comprises only clade 6. PspA classification offers information about the antigenic/genetic diversity of PspA among prevalent serotypes.19–22
In this study, we will characterize the serotype distribution, PspA diversity and antimicrobial resistance pattern of clinical isolates from children with IPD. We aim to analyse the relevance of serotypes to the PspA family for successful selection of antigens of new vaccines and to monitor the antimicrobial resistance pattern to provide proof for choosing the appropriate antibiotic.
Patients and Methods
Collection of Clinical Isolates of S. pneumoniae
S. pneumoniae isolated from patients with IPDs from 2014 to 2018 was obtained from the biobank of a sentinel paediatric hospital in Shenzhen, China. The frozen samples were removed from the −80°C refrigerator and left at room temperature until completely dissolved. The bacterial solution was mixed and inoculated on a blood agar plate (Columbia agar supplemented with 5% sheep blood) and cultured at 37°C in 5% CO2 for 18 hours to 24 hours. If the colony was small, round, moist, autolyzed with grass green lytic rings and positive in the optochin sensitivity test and biliary lysis test, it was determined to be S. pneumoniae. A total of 81 strains were successfully recovered.
Collection of Clinical Data
Clinical information was obtained from the medical records system of the hospital. Epidemiological information included age, sex, community infection, and antibiotic treatment. Clinical manifestations include underlying disease, pulmonary diseases, extrapulmonary diseases, length of hospital stay, admission to the intensive care unit (ICU), invasive mechanical ventilation and in-hospital death. Privacy of the patients was protected. All procedures performed in studies involving human participants were in accordance with the ethical standards of the ethics committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Definitions
Immunosuppression was defined as cytotoxic chemotherapy or high-dose steroid therapy daily for ≥2 weeks and primary immunodeficiency disease. Pneumoniae were confirmed by clinical and chest CT manifestations. S. pneumoniae meningitis was diagnosed by neurological manifestations and cerebrospinal fluid (CSF) abnormalities accompanied by the growth of S. pneumoniae in CSF. S. pneumoniae osteomyelitis was diagnosed when a positive culture of S. pneumoniae from pyogenic fluids was combined with localized symptoms and imaging changes. Sepsis shock was defined by the 2005 International Pediatric Sepsis Definition Conference.
DNA Extraction, PCR and Sequencing
DNA of each strain was extracted using the QiAamp DNA Minikit. The upstream and downstream primers of PCR were LSM13 and SKH2, of which the sequences were 5-GCAAGCTTATGATATAGAAATTTGTAAC-3ʹ and 5ʹ-CCACATACCGTTTTCTTGTTTCCAGCC-3ʹ, respectively.16 PCR was performed using a TaKaRa TaqTM Hot Start Version kit with a reaction volume of 50 μL. The PCR procedure was as follows: 95°C denaturation for 5 min, cycling consisted of 94°C for 15 seconds, 55°C for 30 seconds, 72°C for 30 seconds, repeated 40 times, extension at 72°C for 5 min, and cooling at 20°C for 10 seconds. The PCR product was obtained using the Axygen® AxyPrep DNA Gel Extraction Kit. The DNA fragments were sent to Invitrogen (Shanghai) for Sanger sequencing.
PspA Sequence Analysis
The pspA gene sequences of 24 strains of S. pneumoniae reported in Hollingshead’s publication were derived from the NCBI database.16 The CDR and the proline-rich domain (PRD) were taken from the PspA amino acid sequence. CDR is the N-terminal 100 amino acids of αHD predicted by SMART.23,24 PRD is considered to start at the first proline of a succession of prolines of succession of prolines interspersed with other amino acids. Mega-X was used to do multiple sequence alignment. Neighbour-joining trees were built from aligned sequences with a bootstrap of 1000. Based on the fasta and nwk files generated by mega-X, the phylogenetic tree was displayed using the ggtree package in R.
Serotyping
Pneumococcal serotypes were assigned using continuous multiplex PCR and PCR-based sequencing methods as described previously.25 This process consists of seven rounds of sequential multiplex PCR. Each round of multiplex PCR includes four pairs of primers for the serotype-specific sequence of the capsule gene (CPS) and one pair for the serotype conservative sequence of CPS. Serotypes 6A and 6B were further distinguished by Sanger sequencing. For S. pneumoniae serotyped as 6A or 6B, PCR was carried out with the upstream primer 6A/B-F: AATTTGTATTTTATTCATGCCTATATCTGG and the downstream primer 6A/B-R: TTAGCGGAGATAATTTAAAATGATGACTA. The PCR products were recovered and sent to Invitrogen for Sangar sequencing. Blastn was used to detect the single nucleotide polymorphism at codon 195 of the cps locus wciP gene as described previously.26
Antimicrobial Susceptibility Testing
Broth microdilution was used to measure the minimum inhibitory concentration (MIC) based on the MIC criteria of 17 antimicrobial agents (penicillin, cefotaxime, ceftriaxone, amoxicillin, ertapenem, meropenem, erythromycin, telithromycin, levofloxacin, moxifloxacin, ofloxacin, tetracycline, tetracycline, vancomycin, linezolid, chloramphenicol, and compound sulfamethoxazole). Multidrug resistance (MDR) is defined as resistance to three or more different antimicrobial classes.
The detailed procedure of broth microdilution is as follows: The pure colonies of S. pneumoniae were suspended in sterile normal saline to prepare bacterial fluid of 0.5 McFarla. Sixty microlitres of the bacterial fluid was diluted into 12 mL CAMHB+LHB. Then, 100 μL of dilution was added to a 96-well plate for pneumococcal antimicrobial susceptibility testing and cultured at 37°C for 22 hrs. The 96-well plate was then read, and the MICs were recorded.
MLST
MLST of S. pneumoniae is based on sequences of internal fragments from the aroE, gdh, gki, recP, spi, xpt and ddl genes. The assembled genomes of four strains of serotype 3 in Shenzhen were gained through next-generation sequencing. MLST1.8 (https://cge.cbs.dtu.dk//services/MLST/) was used to analyse the STs. MLST data of S. pneumoniae serotype 3 from 13 foreign countries were collected from PubMLST (https://pubmlst.org/) and previous publications.27,28 The Eburst algorithm was used to estimate the relationship among isolates. STs that share six identical alleles of the seven MLST loci were grouped as a clone complex (CC).
Statistical Analysis
The chi-square test was used to compare the rates, and Fisher’s exact test was used if the expected value in the grid was less than 5. All statistical analyses were performed using SPSS 19.0. P value<0.05 is considered significant.
Results
Demographical and Clinical Characteristics of Patients with IPDs
Among the 81 isolates, three were collected in 2014, six were collected in 2015, 17 were collected in 2016, 20 were collected in 2017 and 35 were collected in 2018. Eight strains were isolated from CSF, 66 were isolated from blood, and 7 were isolated from pleural fluid and joint cavity fluid. The median age of patients was 1.08 (0.79–3.20). The male-to-female ratio was 1.4:1. The median duration of hospitalization was 8(5–18) days. There were eight children with underlying diseases. The most common IPD was sepsis (41, 50.6%). Among all the patients, 16 were admitted to the ICU, 19.8% and 7.4% of them used invasive mechanical ventilation, and 13.6% of them died. The detailed information is seen in Table 1.
 |
Table 1 Demographical and Clinical Characteristics of 81 Patients |
PspA Diversity
The PspA CDRs were sequenced, and each amino acid sequence was obtained. An NJ tree was generated from 81 Shenzhen isolates and 24 strains reported by Hollingshead et al (Figure 1). The tree supports the same family and clade divisions of the 24 strains as those in Hollingshead’s study. The Shenzhen isolates fell into three families and six clades. There were 24 strains (29.6%) in PspA family 1, 56 (69.1%) strains in family 2 and only one (1.2%) in family 3.
Serotype Distribution
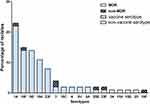
PCV13 was introduced in Shenzhen in June 2017. We defined the period before June 2017 as the period before the introduction of PCV13, during which we collected 32 clinical strains. We defined the period after June 2017 as after the introduction of PCV13, during which we collected 49 strains of S. pneumoniae. Before and after the introduction of PCV13, the proportions of PCV13 serotypes were 87.5% and 89.8%, respectively, with no significant difference (Fisher’s exact test, p=0.734). As shown in Figure 2, before the introduction of PCV13, the nonvaccine serotypes were 35B, 33F, 34,15A. After the introduction of the vaccine, the nonvaccine serotypes were 35B, 33F, 15B, 16F, and 20. Isolates of serotypes 3,6A,34,15A,15B,16F and 20 only had PspA family 1. Isolates of serotypes 19F,14,18C,4, 9V,33F only had PspA family 2. Isolates of serotypes 6B, 19A, and 35B expressed either pspA family 1 or family 2. Isolates of serotype 23F expressed either PspA family 2 or family 3.
Antimicrobial Resistance Pattern and Multiple Drug Resistance
Due to test failure of one serotype 3 strain, the antibiotic activities of only 80 of the 81 S. pneumoniae isolates against 14 antimicrobials are presented in Table 2. According to the revised CLSI breakpoints for penicillin (resistance ≥ 8 mg/mL for nonmeningitis isolates and ≥0.12 mg/mL for meningitis isolates), the prevalence rates of penicillin resistance were 0% and 100% in the nonmeningitis and meningitis isolates, respectively. Antibiotic treatment before blood culture of eight children with pneumococcal meningitis is described as follows: no antibiotic (3), cephalosporins (2), cephalosporins and amoxicillin (1), amoxicillin and erythromycin (1), and meropenem (1). None of the isolates were resistant to amoxicillin/clavulanic acid. The cefuroxime resistance rate was 70%. The proportions of isolates resistant to ceftriaxone were 5.6% in the nonmeningitis isolates and 25% in the meningitis isolates. The cefepime resistance rate was zero for nonmeningitis isolates and 25% for meningitis isolates. As high as 8.8% of isolates were resistant to Meropenem. All of the isolates were susceptible to rifampicin, levofloxacin, linezolid and vancomycin. A total of 97.5% and 98.9% of these isolates were resistant to clindamycin and erythromycin, respectively. A total of 91.3% of these isolates were resistant to at least three classes of antibiotics. The MDR ratios were 94.4% and 66.7% in vaccine serotypes and nonvaccine serotypes, respectively. The MDR ratio of each serotype is shown in Figure 3.
 |
Table 2 Susceptibility and MIC of Antibiotics for IPD Isolates in Shenzhen |
MLST of Serotype 3
Serotype 3 of S. pneumoniae is highly virulent29 and not protected against by immunization with PCV13,30,31 even though serotype 3 polysaccharide is in the vaccine. Serotype 3 is widely spread in Europe, Japan, North America and South America. The majority of this serotype is a single clonal complex 180CC.32 Serotype 3 was rarely isolated from children in China before 2017.33 We isolated serotype 3 S. pneumoniae from four inpatients admitted in 2018, two of whom developed necrotizing pneumonia. MLST results are ST505(1), ST4655(1) and ST12902(2). ST4655 with serotype 3 was reported in Shanghai in 2019.34 ST12902 is a novel ST. PHYLOViZ showed that ST505 belongs to CC180 (Figure 4).
Discussion
IPDs in infants and children lead to high mortality. Vaccination is an effective preventive measure. PCVs, which combine capsular polysaccharides of epidemic serotypes and protein vectors, can cause an effective immune response in infants younger than 2 years old. S. pneumoniae has 98 serotypes, and the proportion of serotypes changes with the passage of time and use of vaccines. The present vaccine based on capsular polysaccharides cannot cover all serotypes. Therefore, the recombinant protein vaccine is a better choice.
PspA is the most promising candidate vaccine antigen. Both CDR and PRD sequences in PspA can induce cross-reactive antibodies and confer protective immunity of S. pneumoniae with PspA of the family. The families of PspA in adult isolates are mainly family 1 and family 2 in the United States, Japan and other foreign countries,16,19–21 while the data from children are limited. In our study, invasive isolates of children fell into PspA family 1, family 2 and family 3. Accordingly, in the preparation of a successful recombinant PspA vaccine, both family 1 and family 2 should be incorporated.
In addition, associations between particular serotypes and the PspA family were observed in our work (eg, serotype 3, PspA family 1; serotype 19F, PspA family 2), as previously reported in another study with a small sample size.22 However, a study including 251 isolates of S. pneumoniae from upper respiratory tract infections drew a different conclusion that serotypes were generally not exclusively associated with certain PspA families, although some capsular types showed a much higher proportion of either family 1 or 2.35 Therefore, more strains are needed to determine the association.
Antibiotic resistance of S. pneumoniae is a worldwide concern.36,37 Penicillin is the first-line antibiotic for treating S. pneumoniae infection.38 According to the revised CLSI breakpoints for penicillin, the penicillin nonsensitivity rate of nonmeningitis strains was 27.8%, and the penicillin resistance rate of meningitis strains was 100%. The resistance rate of meropenem was 8.8%. One meningitis isolate (No. 320, serotype 19A) showed extensive resistance to the spectrum of common antibiotics, including penicillin and meropenem. In addition, we paid attention to the antimicrobial resistance of nonvaccine serotypes. The MDR rate of 9 nonvaccine serotypes was lower than that of vaccine serotypes. This is possibly because vaccine serotypes are dominant in prevalence and are receiving antibiotic screening. We also found that a serotype 34 strain was resistant to penicillin, and we sequenced the whole genome to further study its resistance mechanism. The limitation of this study is the small sample size. We will perform continuous surveillance in Shenzhen to draw a comparative analysis of the molecular epidemiological characteristics and drug resistance trends of invasive S. pneumoniae.
Conclusion
Our work demonstrates that PCV13 is more effective in protecting Chinese children than PCV7 and PCV10. After the introduction of PCV13, the proportion of serotypes of invasive S. pneumoniae changed. Existing PspA family 3 indicates diverse antigens of Shenzhen’s isolates. Pneumococcal antibiotic-resistant nonvaccine serotypes and highly virulent serotype 3 are likely to be an expanding threat to children with IPD.
Ethics Statement
All procedures performed in studies involving human participants were approved by the ethics committee of Shenzhen Children’s hospital and in accordance with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Written informed consent for using the clinical isolates of S. pneumoniae isolated in this study was signed by a parent and/or legal guardian of each participant.
Acknowledgments
The authors sincerely thank all members who took part in this study, especially Peipei Chen, Zijie Wang, Huihua Jiang, Peijin Dai and other co-workers in Shenzhen Children’s Hospital, for their valuable assistance in collecting the isolates.
Funding
This work was funded by Shenzhen Development and Reform Commission Grant [(2019)986], Science Technology and Innovation Commission of Shenzhen Municipality Grant (JCYJ20170413094034978), Health and Family Planning Commission of Shenzhen Municipality Grant (SZLY2017016) and National Natural Science Foundation of China (31670742).
Disclosure
The authors declare that there is no conflict of interest regarding the publication of this paper.
References
1. Kadioglu A, Weiser JN, Paton JC, Andrew PW. The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat Rev Microbiol. 2008;6(4):288–301. doi:10.1038/nrmicro1871
2. Subramanian K, Henriques-Normark B, Normark S. Emerging concepts in the pathogenesis of the Streptococcus pneumoniae: from nasopharyngeal colonizer to intracellular pathogen. Cell Microbiol. 2019;21(11):e13077. doi:10.1111/cmi.13077
3. Engholm DH, Kilian M, Goodsell DS, et al. A visual review of the human pathogen Streptococcus pneumoniae. FEMS Microbiol Rev. 2017;41(6):854–879. doi:10.1093/femsre/fux037
4. Li Q, Li Y, Yi Q, et al. Prognostic roles of time to positivity of blood culture in children with Streptococcus pneumoniae bacteremia. Eur J Clin Microbiol Infect Dis. 2019;38(3):457–465. doi:10.1007/s10096-018-03443-5
5. Paton JC, Trappetti C. Streptococcus pneumoniae capsular polysaccharide. Microbiol Spectr. 2019;7(2). doi:10.1128/microbiolspec.GPP3-0019-2018
6. Dorosti H, Eslami M, Negahdaripour M, et al. Vaccinomics approach for developing multi-epitope peptide pneumococcal vaccine. J Biomol Struct Dyn. 2019;37(13):3524–3535. doi:10.1080/07391102.2018.1519460
7. Marra F, Vadlamudi NK. Efficacy and safety of the pneumococcal conjugate-13 valent vaccine in adults. Aging Dis. 2019;10(2):404–418. doi:10.14336/ad.2018.0512
8. Balsells E, Guillot L, Nair H, Kyaw MH. Serotype distribution of Streptococcus pneumoniae causing invasive disease in children in the post-PCV era: a systematic review and meta-analysis. PLoS One. 2017;12(5):e0177113. doi:10.1371/journal.pone.0177113
9. Hanquet G, Krizova P, Valentiner-Branth P, et al. Effect of childhood pneumococcal conjugate vaccination on invasive disease in older adults of 10 European countries: implications for adult vaccination. Thorax. 2019;74(5):473–482. doi:10.1136/thoraxjnl-2018-211767
10. Savulescu C, Krizova P, Lepoutre A, et al. Effect of high-valency pneumococcal conjugate vaccines on invasive pneumococcal disease in children in SpIDnet countries: an observational multicentre study. Lancet Respir Med. 2017;5(8):648–656. doi:10.1016/S2213-2600(17)30110-8
11. Bogaert D, Hermans PW, Adrian PV, et al. Pneumococcal vaccines: an update on current strategies. Vaccine. 2004;22(17–18):2209–2220. doi:10.1016/j.vaccine.2003.11.038
12. McDaniel LS, Sheffield JS, Delucchi P, Briles DE. PspA, a surface protein of Streptococcus pneumoniae, is capable of eliciting protection against pneumococci of more than one capsular type. Infect Immun. 1991;59(1):222–228. doi:10.1128/IAI.59.1.222-228.1991
13. Nabors GS, Braun PA, Herrmann DJ, et al. Immunization of healthy adults with a single recombinant pneumococcal surface protein A (PspA) variant stimulates broadly cross-reactive antibodies to heterologous PspA molecules. Vaccine. 2000;18(17):1743–1754. doi:10.1016/S0264-410X(99)00530-7
14. Briles DE, Hollingshead SK, King J, et al. Immunization of humans with recombinant pneumococcal surface protein A (rPspA) elicits antibodies that passively protect mice from fatal infection with Streptococcus pneumoniae bearing heterologous PspA. J Infect Dis. 2000;182(6):1694–1701. doi:10.1086/317602
15. Entwisle C, Hill S, Pang Y, et al. Safety and immunogenicity of a novel multiple antigen pneumococcal vaccine in adults: a Phase 1 randomised clinical trial. Vaccine. 2017;35(51):7181–7186. doi:10.1016/j.vaccine.2017.10.076
16. Hollingshead SK, Becker R, Briles DE. Diversity of PspA: mosaic genes and evidence for past recombination in Streptococcus pneumoniae. Infect Immun. 2000;68(10):5889–5900. doi:10.1128/IAI.68.10.5889-5900.2000
17. Mukerji R, Hendrickson C, Genschmer KR, et al. The diversity of the proline-rich domain of pneumococcal surface protein A (PspA): potential relevance to a broad-spectrum vaccine. Vaccine. 2018;36(45):6834–6843. doi:10.1016/j.vaccine.2018.08.045
18. Briles DE, Hollingshead SK, Nabors GS, et al. The potential for using protein vaccines to protect against otitis media caused by Streptococcus pneumoniae. Vaccine. 2000;19(Suppl 1):S87–95. doi:10.1016/S0264-410X(00)00285-1
19. Hollingshead SK, Baril L, Ferro S, et al. Pneumococcal surface protein A (PspA) family distribution among clinical isolates from adults over 50 years of age collected in seven countries. J Med Microbiol. 2006;55(2):215–221. doi:10.1099/jmm.0.46268-0
20. Pimenta FC, Ribeiro-Dias F, Brandileone MC, et al. Genetic diversity of PspA types among nasopharyngeal isolates collected during an ongoing surveillance study of children in Brazil. J Clin Microbiol. 2006;44(8):2838–2843. doi:10.1128/JCM.00156-06
21. Rolo D, Ardanuy C, Fleites A, et al. Diversity of pneumococcal surface protein A (PspA) among prevalent clones in Spain. BMC Microbiol. 2009;9(1):80. doi:10.1186/1471-2180-9-80
22. Hotomi M, Togawa A, Kono M, et al. PspA family distribution, antimicrobial resistance and serotype of Streptococcus pneumoniae isolated from upper respiratory tract infections in Japan. PLoS One. 2013;8(3):e58124. doi:10.1371/journal.pone.0058124
23. Letunic I, Bork P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018;46(D1):D493–D496. doi:10.1093/nar/gkx922
24. Letunic I, Doerks T, Bork P. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 2015;43(D1):D257–260. doi:10.1093/nar/gku949
25. Pai R, Gertz RE, Beall B. Sequential multiplex PCR approach for determining capsular serotypes of Streptococcus pneumoniae isolates. J Clin Microbiol. 2006;44(1):124–131. doi:10.1128/JCM.44.1.124-131.2006
26. Ronaghi M, Elahi E. Pyrosequencing for microbial typing. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;782(1–2):67–72. doi:10.1016/s1570-0232(02)00693-1
27. Echániz-Aviles G, Guerreiro SI, Silva-Costa C, et al. Streptococcus pneumoniae serotype 3 in mexico (1994 to 2017): decrease of the unusual clonal complex 4909 lineage following PCV13 introduction. J Clin Microbiol. 2019;57(1). doi:10.1128/JCM.01354-18
28. Mothibeli KM, Du Plessis M, von Gottberg A, et al. An unusual pneumococcal sequence type is the predominant cause of serotype 3 invasive disease in South Africa. J Clin Microbiol. 2010;48(1):184–191. doi:10.1128/JCM.01011-09
29. Martens P, Worm SW, Lundgren B, et al. Serotype-specific mortality from invasive Streptococcus pneumoniae disease revisited. BMC Infect Dis. 2004;4(1):21. doi:10.1186/1471-2334-4-21
30. Andrews NJ, Waight PA, Burbidge P, et al. Serotype-specific effectiveness and correlates of protection for the 13-valent pneumococcal conjugate vaccine: a postlicensure indirect cohort study. Lancet Infect Dis. 2014;14(9):839–846. doi:10.1016/S1473-3099(14)70822-9
31. Choi EH, Zhang F, Lu YJ, Malley R. Capsular polysaccharide (CPS) release by serotype 3 pneumococcal strains reduces the protective effect of anti-type 3 CPS antibodies. Clin Vaccine Immunol. 2016;23(2):162–167. doi:10.1128/CVI.00591-15
32. Azarian T, Mitchell PK, Georgieva M, et al. Global emergence and population dynamics of divergent serotype 3 CC180 pneumococci. PLoS Pathog. 2018;14(11):e1007438. doi:10.1371/journal.ppat.1007438
33. Chen K, Zhang X, Shan W, et al. Serotype distribution of Streptococcus pneumoniae and potential impact of pneumococcal conjugate vaccines in China: a systematic review and meta-analysis. Hum Vaccin Immunother. 2018;14(6):1453–1463. doi:10.1080/21645515.2018.1435224
34. Li -X-X, Xiao S-Z, Gu -F-F, et al. Serotype distribution, antimicrobial susceptibility, and Multilocus Sequencing Type (MLST) of from adults of three hospitals in Shanghai, China. Front Cell Infect Microbiol. 2019;9:407. doi:10.3389/fcimb.2019.00407
35. Kawaguchiya M, Urushibara N, Aung MS, et al. Association between pneumococcal surface protein a family and genetic/antimicrobial resistance traits of non-invasive pneumococcal isolates from adults in Northern Japan. Microb Drug Resist. 2019;25(5):744–751. doi:10.1089/mdr.2018.0267
36. Wang CY, Chen YH, Fang C, et al. Antibiotic resistance profiles and multidrug resistance patterns of Streptococcus pneumoniae in pediatrics: a multicenter retrospective study in mainland China. Medicine. 2019;98(24):e15942. doi:10.1097/md.0000000000015942
37. Zhang Z, Chen M, Yu Y, et al. Antimicrobial susceptibility among and collected globally between 2015 and 2017 as part of the Tigecycline Evaluation and Surveillance Trial (TEST). Infect Drug Resist. 2019;12:1209–1220. doi:10.2147/idr.s203121
38. Chiba N, Murayama SY, Morozumi M, et al. Genome evolution to penicillin resistance in serotype 3 streptococcus pneumoniae by capsular switching. Antimicrob Agents Chemother. 2017;61(9). doi:10.1128/AAC.00478-17
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.