Back to Journals » International Journal of Nanomedicine » Volume 12
Proximity hybridization-mediated isothermal exponential amplification for ultrasensitive electrochemical protein detection
Authors Yu Y, Su G, Zhu H, Zhu Q, Chen Y, Xu B, Li Y, Zhang W
Received 17 May 2017
Accepted for publication 13 July 2017
Published 17 August 2017 Volume 2017:12 Pages 5903—5914
DOI https://doi.org/10.2147/IJN.S142015
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Linlin Sun
Yanyan Yu, Gaoxing Su, Hongyan Zhu, Qing Zhu, Yong Chen, Bohui Xu, Yuqin Li, Wei Zhang
School of Pharmacy, Nantong University, Nantong, People’s Republic of China
Abstract: In this study, we fabricated a novel electrochemical biosensing platform on the basis of target-triggered proximity hybridization-mediated isothermal exponential amplification reaction (EXPAR) for ultrasensitive protein analysis. Through rational design, the aptamers for protein recognition were integrated within two DNA probes. Via proximity hybridization principle, the affinity protein-binding event was converted into DNA assembly process. The recognition of protein by aptamers can trigger the strand displacement through the increase of the local concentrations of the involved probes. As a consequence, the output DNA was displaced, which can hybridize with the duplex probes immobilized on the electrode surface subsequently, leading to the initiation of the EXPAR as well as the cleavage of duplex probes. Each cleavage will release the gold nanoparticles (AuNPs) binding sequence. With the modification of G-quadruplex sequence, electrochemical signals were yielded by the AuNPs through oxidizing 3,3',5,5'-tetramethylbenzidine in the presence of H2O2. The study we proposed exhibited high sensitivity toward platelet-derived growth factor BB (PDGF-BB) with the detection limit of 52 fM. And, this method also showed great selectivity among the PDGF isoforms and performed well in spiked human serum samples.
Keywords: electrochemical biosensor, proximity hybridization, PDGF-BB, isothermal exponential amplification, G-quadruplex
Introduction
The precise detection of biomarkers is rather important for early cancer diagnosis and treatment in clinic.1,2 In general, the common detection methods rely on commercialized antibody-based assays, for example, enzyme-linked immune sorbent assay (ELISA), whereas the moderate sensitivity and tedious procedures of ELISA make it urgent to develop better methods for the detection of biomarkers. Aptamers, since its first report in 1990s, have received extensive interests as a rival of antibody due to the high affinity and specificity.3–6 As there are not much limitations on the targets of aptamers such as proteins, amino acids, small chemicals, and even metal ions up to now, numerous aptamers have been identified.7 In comparison with antibodies and enzymes, aptamer-based biosensors have superiorities of high flexibility and convenience, enabling to develop various novel aptasensors.8 By combination with amplification strategies, aptamers have been successfully applied to develop ultrasensitive biosensors.9
The advances in DNA proximity binding have inspired exciting developments of nanosensors and ultrasensitive biosensors.10,11 Different from the toehold-mediated strand displacement,12,13 which has been extensively employed to design DNA assembly, a proximity hybridization-induced DNA assembly homogeneous assay has been proposed.14,15 The method was based on the target-binding process to promote strand displacement and reinforce the hybridization of two short DNA sequences, which cannot form stable duplex at normal circumstances. Through converting the target binding event into detectable DNA assembly, proximity hybridization strategy can be combined with various signal amplification strategies, such as enzyme-free cyclic assembly of hairpins,16 ligation-mediated strand recycling,17 and binding-induced self-assembly of DNAzyme18 to build ultrasensitive biosensors. Up to now, there have been many novel applications of proximity hybridization-based strategy to detect proteins,19,20 nucleic acids,17,21 and small molecules.22,23
DNAzymes are derived from catalytic DNA sequences obtained by in vitro selection. Compared with natural enzymes, artificial enzymes, which have been extensively utilized for bioanalytical applications, have several merits owing to the flexible base sequences as well as nonspecific binding properties.24,25 Among them, the G-quadruplex–hemin complex, formed by hemin intercalated into the guanine-rich nucleic acid sequences,26–28 can catalyze H2O2-mediated oxidation of reductive substrates and has been successfully employed for the electrochemical,29,30 colorimetric, and chemiluminescent determination of nucleotides.31–35 By combined application with hybridization chain reaction (HCR), isothermal exponential amplification reaction (EXPAR), and rolling circle amplification (RCA) strategy, G-quadruplex–hemin complexes have been successfully employed to fabricate biosensors.36–39
Herein, we developed an ultrasensitive electrochemical biosensor for protein on the basis of proximity hybridization-induced EXPAR strategy. Platelet-derived growth factor BB (PDGF-BB) was a disease marker, directly related to several cancer transformation and tumor growth process,40,41 and was chosen as the target protein. In this design, the aptamers for PDGF-BB were integrated with two sequences. Binding of PDGF-BB to the aptamers triggered the strand displacement process, releasing an output DNA at the same time. The duplex probes fixed on the electrode surface were arched structured. After hybridization with the output, the probes cleaved, resulting in the initiation of EXPAR process and the free of the gold nanoparticles (AuNPs) binding sequence. In this study, AuNPs were employed to load G-quadruplex sequences due to their unique properties, such as large specific surface area, facial to prepare, and easy to modify.42 By combination of proximity hybridization-mediated DNA assembly and DNAzyme-modified AuNPs, this new strategy achieved ultrasensitive assay of PDGF-BB with the detection limit of 52 fM.
Methods
Chemicals and materials
DNA sequences, hemin, and PDGF-BB (human) were from Sangon Biotechnology Inc. (Shanghai, People’s Republic of China). The Bst 2.0 DNA polymerase (large fragment), Nb.BbvCI nicking endonuclease, and deoxynucleotide (dNTP) solution mixture were obtained from New England Biolabs (Beijing, People’s Republic of China). Tris (2-carboxyethy) phosphine hydrochloride (TCEP) and dimethyl sulfoxide (DMSO) were purchased from Aladdin Reagent Corporation (Shanghai, People’s Republic of China). 6-Mercapto-1-hexanol (MCH) was obtained from J&K Scientific Ltd. (Guangzhou, People’s Republic of China). MCH solution was prepared by diluting the MCH with DNA hybridization buffer (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], 500 mM NaCl, pH 7.4) to 1 mM. The stock solution of hemin (10 mM) was prepared with DMSO and, then, stored at −20°C. DNA sequences were dissolved with 1× tris-EDTA (TE) buffer (pH 8.0) to 100 μM. Sequences of the DNA (Table 1) were designed with the help of the Integrated DNA Technologies (IDT; online software).
  | Table 1 Sequences of involved oligonucleotides |
Preparation of serum samples
Serum samples from whole blood were prepared by centrifugation at 500× g for 15 min and, then, were diluted 100 times with 20 mM HEPES buffer (500 mM NaCl, pH 7.4).
Biosensor fabrication
Firstly, the gold electrode (3 mm in diameter) was cleaned by freshly prepared piranha solution (a 3:1, v/v, mixture of concentrated H2SO4 and 30% H2O2) for 20 min. And then, the electrode was rinsed with ultrapure water (caution: piranha solution reacts violently with organic materials; it must be handled with extreme care). After polishing with alumina (0.05 μm), the electrodes were treated with ultrasonic with ethanol and ultrapure water before electrochemically cleaned with 0.5 M H2SO4 over the potential −0.2 to 1.5 V (100 mV/s). Ultimately, the pretreated electrode was rinsed with water and dried with nitrogen.
Recognition probes were prepared by annealing the same amount of P1 and Output in hybridization buffer. The solution was incubated at 90°C for 5 min before cooling to room temperature for 2 h. The S1–S2 probes were prepared through the same way and reacted with 1 mM TCEP for 1 h to reduce disulfide bonds. Certain amounts of duplex probes (3 μL) were dropped onto the electrode surface and capped for incubation overnight. After rinsing, the modified electrode was blew-dried with nitrogen and immersed in MCH solution subsequently for 1 h to remove the nonspecific adsorption.
Preparation of DNA functionalized nanoparticles
The DNAzyme-modified AuNPs were prepared as reported in literature with little modification.43 Briefly, 100 μL of capture and DNAzyme mixtures (10 μM each) were incubated with 20 μM TCEP for 1 h and, then, were dropped to 200 μL of citrate-capped 15 nm AuNPs. After incubating for 16 h in dark, the mixed solution was treated with 0.1 M NaCl in Tris-HAc (5 mM, pH 8.2) with gentle hand shaking and incubated for another day before use. The AuNPs’ solution was centrifuged twice (15,000 rpm, 30 min) to remove excessive DNA and finally dispersed in 10 mM HEPES buffer (pH 7.4, 50 mM KCl and 500 mM NaCl). At last, 5 μL hemin (400 μM) was added to the as-prepared AuNPs to form G-quadruplex–hemin-modified AuNPs.
Electrochemical measurement and apparatus
CHI660E electrochemical workstation (CH Instruments, Shanghai, People’s Republic of China) was used to conduct electrochemical experiments. A three-electrode system was employed in all electrochemical measurements. The modified electrode was incubated with 20 mM HEPES buffer (pH 6.5, 50 mM KCl and 500 mM NaCl) containing TMB (4 mM) and H2O2 (1 mM) to conduct the enzymatic reaction. The differential pulse voltammetry (DPV) was conducted after 1 min of reaction. Ultraviolet-visible (UV–vis) spectra were obtained with an UV1800PC spectrophotometer (Jinghua, Shanghai, People’s Republic of China). Transmission electron microscope (TEM; 1200 EX; JEOL, Tokyo, Japan) was used to characterize the morphology of the as-prepared AuNPs. The detailed procedures and parameters for TEM, UV–vis spectroscopy, native polyacrylamide gel electrophoresis (PAGE), electrochemical impedance spectroscopy (EIS), and DPV are shown in the Supplementary materials.
Proximity hybridization-induced EXPAR
The oligonucleotide-modified electrode was incubated in 50 μL CutSmart Buffer reaction system, containing 32 U Bst DNA polymerase, 20 U nicking endonuclease, 400 μM dNTPs, 2 μM primer, 1 μM P1–Output, 1 μM P2, and different amounts of PDGF-BB, for 60 min at 30°C. Then, the electrode was carefully rinsed with hybridization buffer before subjected to 50 μL G-quadruplex–hemin-modified AuNPs for 30 min to promote the immobilization of AuNPs.
Results and discussion
Principle of the biosensor for PDGF-BB detection
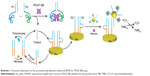
The newly proposed electrochemical biosensor based on proximity hybridization-induced EXPAR is shown in Scheme 1. The aptamers for PDGF-BB were integrated within P1 and P2. Binding of the two aptamers to the same target assembled P1–Output and P2 together. In this proximity hybridization process, the local concentrations of P1 and P2 were increased, thus raising the melting temperature for the intramolecular hybridization (P1 and P2), favoring the strand displacement reaction between P2 and Output.15 As a consequence, the Output was released from P1, triggering the strand displacement cycle. And, the subsequent amplification process was triggered meanwhile. The duplex probe used to modify the electrode surface consisted of S1 and S2 (Table 1). Both S1 and S2 acted as blockers to each other. S2 prevented the hybridization of primer with S1, and S1 stopped the binding of AuNPs to S2. The loop region of S1 was the binding site for the released Output from proximity hybridization. The nicking site, located in the loop region of S1, was unable to bind restriction endonuclease. After hybridization of Output with S1, the target–substrate complex can be formed through toehold-mediated strand displacement. The separation of one stem of the arched structure through the formation of Output–S1 resulted in the thermal melting of the remaining duplex. In this way, Output can displace S2 from S1 successfully even it was partially complementary with S1.44,45 After the S1–Output complex was released into the solution, in the presence of primer, the polymerase and nickase initiated EXPAR, during which Output was recycled and regenerated circularly. Meanwhile, S2 was free to hybridize with the capture-modified AuNPs. Electrochemical signals were yielded by the reduction current of oxidized TMB in the presence of H2O2.
Characterization of the nanomaterials
TEM and UV–vis spectroscopy were utilized to characterize the as-prepared nanoparticles. As illustrated in Figure 1A, the diameters of prepared AuNPs were ~15 nm. The AuNPs exhibited similar sizes and good monodispersity. There was characteristic surface plasmon absorption at 520 nm (Figure 1B, curve b), typical for AuNPs ~15 nm in diameter. The DNA-modified AuNPs, however, red shifted to 523 nm (Figure 1B, curve a). In addition, compared to the blueviolet aggregation exhibited by AuNPs (b, inset), the DNA-functionalized ones maintained red (a, inset) in the solution with 0.1 M NaCl. These results confirmed the successful preparation and functionalization of AuNPs.
Gel analysis
PAGE was further used to analyze the proximity hybridization-mediated EXPAR process. For the proof-of-concept experiment, we utilized a helper DNA (H1) to mimic the P1-protein-P2 complex in the binding-induced reaction process for native PAGE analysis (Figure 1C). H1 brought P1 and P2 in close to form a cooperative complex. P1–Output, P2, S1–S2, and S1–Output showed individual band (lanes 2–5). In the absence of H1, the mixture of P1–Output, P2, and S1–S2 displayed bands located in the same position as lane 2 to lane 4, indicating that no hybridization occurred between them (lane 7). Upon the addition of H1 to the same mixture, a fresh band appeared with much slower migration speed, as well as the appearance of S1–Output band and the disappearance of S1–S2 bands (lane 8). This result verified the formation of P1–H1–P2 complex and the strand displacement of Output from dsDNA. Moreover, the released Output can hybridize with S1–S2, resulting in the cleavage of duplex probes and the initiation of EXPAR. The PAGE image for subsequent EXPAR results is shown in Figure S1. The enzymatic reaction product band (marked in red box of Figure S1) validated the successful proceeding of EXPAR.
Electrochemical characterization of the proximity hybridization-mediated EXPAR
As an effective method for exploring the interfacial properties, EIS was employed to analyze the fabrication process. Figure 1D shows the EIS results of electrode fabrication at different stages. There was a very small charge transfer resistance (Rct) (curve a, 50.20 Ω) for the bare gold electrode. After self-assembly of duplex probes and MCH, the Rct increased to 2,601.76 Ω (curve b), owing to the repellence to [Fe(CN)6]3−/4− from negative charges on the phosphate backbone. Once treated with Output, Rct decreased to 2,337.11 Ω (curve c), as the Output caused the cleavage of probe, reducing the oligonucleotides of the electrode interface. At last, after assembly of G-quadruplex–hemin AuNPs, the Rct increased remarkably to 3,732.88 Ω (curve d), because of the plentiful negatively charged phosphate backbone linked on the AuNPs. The stepwise variation of the electrochemical impedance verified the successful modification of gold electrode.
DPV was further employed to elucidate the feasibility of the strategy proposed. As depicted in Figure 1E, a reduction peak of ~0.3 V can be obtained in the presence of 1 nM PDGF-BB after proximity hybridization-mediated EXPAR (curve d), which was the reduction current of TMB+ oxidized by DNAzyme-modified AuNPs.46,47 In contrast, compared to curve d, the signal of hybridization to the free G-quadruplex sequence alone (curve c) was much lower, conforming that much more DNAzyme units were able to assemble on the electrode surface along with AuNPs. The DPV peak from 1 nM PDGF-BB with proximity hybridization but without EXPAR (curve b) did not show much difference to curve a, whose response derived from the reduction of original TMB+. As reported, EXPAR was rather efficient and can achieve 106–109-fold signal amplification within minutes.48,49 The Output released from proximity hybridization was amplified greatly through EXPAR and activated more probes to release S2 for the binding of AuNPs, yielding higher signal.
Optimization of the detection strategy
To achieve higher sensitivity, the formation of probe–protein complex and the release of Output should be very crucial, especially for trace analysis. To eliminate the target-independent displacement, we optimized the structure of S1–S2 duplex and the length of P2–P1 complementary parts (sequences involved are listed in Table S1). According to the results shown in Figure S2, 12 bp/10 bp was chosen for the arched structure and 11 bp was chosen for both P1–Output and P1–P2. Afterward, we investigated the effect of incubation time for the probe–target reaction on the DPV signal. As illustrated in Figure 2A, the peak currents rose with the increase in reaction time accordingly and reached plateau after 60 min. This result indicated the feasibility of the proximity hybridization and EXPAR process, while the plateau can be attributed to the end of reaction. In conclusion, 60 min was chosen for optimum condition.
As the immobilization concentration was also a key factor for the rate and efficiency of hybridization on the electrode surface. Various amounts of probes (0–1.0 μM) were evaluated for the electrochemical response toward 1 nM PDGF-BB under the optimized reaction time (Figure 2B). The higher concentration can give rise to higher DNA density, whereas the hybridizing efficiency decreased due to the steric hindrance effect, yielding relatively lower electrochemical response.50 Thus, 0.6 μM was chosen.
Detection performance of the assay
Various amounts of protein were measured under the optimized reaction condition to investigate the analytical performance of the method. Figure 3A illustrates that with the increase of target concentration from 0 to 5 nM, the reduction peak currents increased correspondingly, demonstrating that more PDGF-BB will lead more G-quadruplex–hemin to immobilize on the electrode. Figure 3B shows the relationship of peak currents and the concentrations of target. All measurements were repeated three times. The reduction peak currents were in linear relationship with the logarithm of PDGF-BB from 0.1 pM to 1 nM. The correlation coefficient was 0.9915 (Figure 3B). According to the 3σ method, the limit of detection was 52 fM for PDGF-BB. We also compared our method with other reported strategy, and the results are listed in Table 2. The outstanding performance of our biosensor endowed its great potential in early diagnosis of disease.
To investigate the specificity of the strategy proposed, different concentrations of PDGF-BB and other PDGF isoforms, PDGF-AA homodimer and PDGF-AB heterodimer, were detected under the optimized condition. Results shown in Figure 4 revealed that the peak currents of PDGF-AA and PDGF-AB were close to the value of blank control, which were much lower than that of PDGF-BB. The results confirmed our detecting principle that only PDGF-BB can promote the two probes to form proximity complex to trigger the strand displacement process. Notably, our method also exhibited good specificity even when the concentration of PDGF-BB was down to 10 pM. In addition, the strategy we proposed performed well in both the HEPES buffer and the diluted human serums even challenged with fivefold PDGF-AA and PDGF-AB (Figure S3).
Real sample analysis
We investigated the performance of the strategy proposed for protein detection in serum samples with standard addition method. A series of PDGF-BB solutions with different concentrations (10, 100, 500, and 1,000 pM) were prepared by adding certain amounts of target to the human serums diluted 100 times, and then, the electrochemical detection was conducted under the optimized condition. The results are shown in Table 3. The recovery ranged from 93.92 to 105.31%, and the relative standard deviation (RSD) was ≤5.38%, which implied that the electrochemical platform can be a promising alternate for analytical application in complex samples.
Conclusion
We developed a biosensing platform based on proximity hybridization-induced EXPAR for the sensitive assay of PDGF-BB. The target protein can promote the formation of proximity complex and triggered the EXPAR process for recycling and regenerating Output sequences. After each binding and probe cleavage process, the DNAzyme-functionalized AuNPs can be immobilized on the electrode surface, which subsequently catalyzed the oxidation reaction to generate the electrochemical signal. The protocol we proposed provided a broad dynamic range and good sensitivity. Moreover, the bioassay proposed showed satisfying selectivity and anti-interference capability in complex serum samples. By changing the aptamer-associated sequences, this method could be expanded to detect other analytes, demonstrating great potential for biomarker assay in early clinical diagnosis.
Acknowledgment
This study was supported by the National Natural Science Foundation of China (Grant nos 21405084), the Natural Science Foundation of Jiangsu Province in China (BK20170444), Advanced Research Foundation for Natural Science of Nantong University (16ZY17), Start-Up Funding for Research of Nantong University (15R21), and science and technology plan projects of Nantong (MS12016023).
Disclosure
The authors report no conflicts of interest in this work.
References
Spratlin JL, Serkova NJ, Eckhardt SG. Clinical applications of metabolomics in oncology: a review. Clin Cancer Res. 2009;15(2):431–440. | ||
Park JM, Wu M, Datta K, et al. Hyperpolarized sodium [1-C-13]-glycerate as a probe for assessing glycolysis in vivo. J Am Chem Soc. 2017;139(19):6629–6634. | ||
Liu JW, Cao ZH, Lu Y. Functional nucleic acid sensors. Chem Rev. 2009;109(5):1948–1998. | ||
Tombelli S, Minunni A, Mascini A. Analytical applications of aptamers. Biosens Bioelectron. 2005;20(12):2424–2434. | ||
do Carmo FS, Ricci E, Cerqueira-Coutinho C, et al. Anti-MUC1 nano-aptamers for triple-negative breast cancer imaging by single-photon emission computed tomography in inducted animals: initial considerations. Int J Nanomedicine. 2017;12:53–60. | ||
Tang JZ, Huang N, Zhang X, et al. Aptamer-conjugated PEGylated quantum dots targeting epidermal growth factor receptor variant III for fluorescence imaging of glioma. Int J Nanomedicine. 2017;12:3899–3911. | ||
Willner I, Zayats M. Electronic aptamer-based sensors. Angew Chem Int Ed Engl. 2007;46(34):6408–6418. | ||
Ye SJ, Zhai XM, Wu YY, Kuang SP. Dual-primer self-generation SERS signal amplification assay for PDGF-BB using label-free aptamer. Biosens Bioelectron. 2016;79:130–135. | ||
Ozalp VC, Kavruk M, Dilek O, Bayrac AT. Aptamers: molecular tools for medical diagnosis. Curr Top Med Chem. 2015;15(12): 1125–1137. | ||
Pei H, Zuo XL, Zhu D, Huang Q, Fan CH. Functional DNA nanostructures for theranostic applications. Acc Chem Res. 2014;47(2):550–559. | ||
Li D, Song SP, Fan CH. Target-responsive structural switching for nucleic acid-based sensors. Acc Chem Res. 2010;43(5):631–641. | ||
Wu CC, Cansiz S, Zhang LQ, et al. A nonenzymatic hairpin DNA cascade reaction provides high signal gain of mRNA imaging inside live cells. J Am Chem Soc. 2015;137(15):4900–4903. | ||
Song TJ, Liang HJ. Synchronized assembly of gold nanoparticles driven by a dynamic DNA-fueled molecular machine. J Am Chem Soc. 2012;134(26):10803–10806. | ||
Hu J, Wang T, Kim J, Shannon C, Easley CJ. Quantitation of femtomolar protein levels via direct readout with the electrochemical proximity assay. J Am Chem Soc. 2012;134(16):7066–7072. | ||
Zhang H, Li F, Dever B, Wang C, Li XF, Le XC. Assembling DNA through affinity binding to achieve ultrasensitive protein detection. Angew Chem Int Ed Engl. 2013;52(41):10698–10705. | ||
Guo YH, Wu J, Ju HX. Target-driven DNA association to initiate cyclic assembly of hairpins for biosensing and logic gate operation. Chem Sci. 2015;6(7):4318–4323. | ||
Chen JB, Deng B, Wu P, et al. Amplified binding-induced homogeneous assay through catalytic cycling of analyte for ultrasensitive protein detection. Chem Commun. 2016;52(9):1816–1819. | ||
Wu H, Zhang K, Liu YL, et al. Binding-induced and label-free colorimetric method for protein detection based on autonomous assembly of hemin/G-quadruplex DNAzyme amplification strategy. Biosens Bioelectron. 2015;64:572–578. | ||
Zhang L, Zhang KX, Liu GC, Liu MJ, Liu Y, Li JH. Label-free nanopore proximity bioassay for platelet-derived growth factor detection. Anal Chem. 2015;87(11):5677–5682. | ||
Ren KW, Wu J, Zhang Y, Yan F, Ju HX. Proximity hybridization regulated DNA biogate for sensitive electrochemical immunoassay. Anal Chem. 2014;86(15):7494–7499. | ||
Zong C, Wu J, Liu MM, et al. Proximity hybridization-triggered signal switch for homogeneous chemiluminescent bioanalysis. Anal Chem. 2014;86(11):5573–5578. | ||
Zhu Y, Hu XC, Shi S, et al. Ultrasensitive and universal fluorescent aptasensor for the detection of biomolecules (ATP, adenosine and thrombin) based on DNA/Ag nanoclusters fluorescence light-up system. Biosens Bioelectron. 2016;79:205–212. | ||
Li F, Zhang HQ, Wang ZX, et al. Aptamers facilitating amplified detection of biomolecules. Anal Chem. 2015;87(1):274–292. | ||
Sun DP, Lu J, Zhong YW, et al. Sensitive electrochemical aptamer cytosensor for highly specific detection of cancer cells based on the hybrid nanoelectrocatalysts and enzyme for signal amplification. Biosens Bioelectron. 2016;75:301–307. | ||
Willner I, Shlyahovsky B, Zayats M, Willner B. DNAzymes for sensing, nanobiotechnology and logic gate applications. Chem Soc Rev. 2008;37(6):1153–1165. | ||
Murat P, Singh Y, Defrancq E. Methods for investigating G-quadruplex DNA/ligand interactions. Chem Soc Rev. 2011;40(11):5293–5307. | ||
Zong C, Wu J, Liu MM, Yang LL, Yan F, Ju HX. Chemiluminescence imaging for a protein assay via proximity-dependent DNAzyme formation. Anal Chem. 2014;86(19):9939–9944. | ||
Ko CN, Yang C, Lin S, et al. A long-lived phosphorescence iridium(III) complex as a switch on-off-on probe for live zebrafish monitoring of endogenous sulfide generation. Biosens Bioelectron. 2017;94:575–583. | ||
Wu H, Liu YL, Wang HY, Wu J, Zhu FF, Zou P. Label-free and enzyme-free colorimetric detection of microRNA by catalyzed hairpin assembly coupled with hybridization chain reaction. Biosens Bioelectron. 2016;81:303–308. | ||
Wu CT, Fan DQ, Zhou CY, Liu YQ, Wang EK. Colorimetric strategy for highly sensitive and selective simultaneous detection of histidine and cysteine based on G-quadruplex-Cu(II) metalloenzyme. Anal Chem. 2016;88(5):2899–2903. | ||
Yang JM, Dou BT, Yuan R, Xiang Y. Proximity binding and metal ion-dependent DNAzyme cyclic amplification-integrated aptasensor for label-free and sensitive electrochemical detection of thrombin. Anal Chem. 2016;88(16):8218–8223. | ||
Liu X, Wei M, Liu YJ, et al. Label-free detection of telomerase activity in urine using telomerase-responsive porous anodic alumina nanochannels. Anal Chem. 2016;88(16):8107–8114. | ||
Bi S, Luo BY, Ye JY, Wang ZH. Label-free chemiluminescent aptasensor for platelet-derived growth factor detection based on exonuclease-assisted cascade autocatalytic recycling amplification. Biosens Bioelectron. 2014;62:208–213. | ||
Wang MD, Mao ZF, Kang TS, et al. Conjugating a groove-binding motif to an Ir(III) complex for the enhancement of G-quadruplex probe behavior. Chem Sci. 2016;7(4):2516–2523. | ||
Lin S, Gao W, Tian ZR, et al. Luminescence switch-on detection of protein tyrosine kinase-7 using a G-quadruplex-selective probe. Chem Sci. 2015;6(7):4284–4290. | ||
Wang H, Wang DM, Huang CZ. Highly sensitive chemiluminescent detection of lead ion based on its displacement of potassium in G-Quadruplex DNAzyme. Analyst. 2015;140(16):5742–5747. | ||
Dong J, Cui X, Deng Y, Tang Z. Amplified detection of nucleic acid by G-quadruplex based hybridization chain reaction. Biosens Bioelectron. 2012;38(1):258–263. | ||
Yu YY, Chen ZG, Shi LJ, et al. Ultrasensitive electrochemical detection of microRNA based on an arched probe mediated isothermal exponential amplification. Anal Chem. 2014;86(16):8200–8205. | ||
Lin X, Chen Q, Liu W, Li H, Lin JM. A portable microchip for ultrasensitive and high-throughput assay of thrombin by rolling circle amplification and hemin/G-quadruplex system. Biosens Bioelectron. 2014;56:71–76. | ||
Xu XL, Ling DY, Zhu QY, Fan WJ, Zhang W. The effect of 2,3,4′,5-tetrahydroxystilbene-2-0-beta-D glucoside on neointima formation in a rat artery balloon injury model and its possible mechanisms. Eur J Pharmacol. 2013;698(1-3):370–378. | ||
Huang KJ, Shuai HL, Zhang JZ. Ultrasensitive sensing platform for platelet-derived growth factor BB detection based on layered molybdenum selenide-graphene composites and exonuclease III assisted signal amplification. Biosens Bioelectron. 2016;77:69–75. | ||
Wang LH, Li J, Song SP, Li D, Fan CH. Biomolecular sensing via coupling DNA-based recognition with gold nanoparticles. J Phys D Appl Phys. 2009;42(20):203001. | ||
Liu J, Lu Y. Preparation of aptamer-linked gold nanoparticle purple aggregates for colorimetric sensing of analytes. Nat Protoc. 2006;1(1):246–252. | ||
Li D, Shlyahovsky B, Elbaz J, Willner I. Amplified analysis of low-molecular-weight substrates or proteins by the self-assembly of DNAzyme-aptamer conjugates. J Am Chem Soc. 2007;129(18):5804–5805. | ||
Xiao Y, Rowe AA, Plaxco KW. Electrochemical detection of parts-per-billion lead via an electrode-bound DNAzyme assembly. J Am Chem Soc. 2007;129(2):262–263. | ||
Ahirwal GK, Mitra CK. Gold nanoparticles based sandwich electrochemical immunosensor. Biosens Bioelectron. 2010;25(9):2016–2020. | ||
Chang HH, Zhang HC, Lv J, Zhang B, Wei WL, Guo JG. Pt NPs and DNAzyme functionalized polymer nanospheres as triple signal amplification strategy for highly sensitive electrochemical immunosensor of tumour marker. Biosens Bioelectron. 2016;86:156–163. | ||
Jia HX, Li ZP, Liu CH, Cheng YQ. Ultrasensitive detection of microRNAs by exponential isothermal amplification. Angew Chem Int Ed Engl. 2010;49(32):5498–5501. | ||
Connolly AR, Trau M. Isothermal detection of DNA by beacon-assisted detection amplification. Angew Chem Int Ed Engl. 2010;49(15):2720–2723. | ||
Zhang J, Song SP, Zhang LY, et al. Sequence-specific detection of femtomolar DNA via a chronocoulometric DNA sensor (CDS): effects of nanoparticle-mediated amplification and nanoscale control of DNA assembly at electrodes. J Am Chem Soc. 2006;128(26):8575–8580. | ||
Wang CW, Chang HT. Sensitive detection of platelet-derived growth factor through surface-enhanced Raman scattering. Anal Chem. 2014;86(15):7606–7611. | ||
Wang GF, Zhu YH, Chen L, Zhang XJ. Photoinduced electron transfer (PET) based label-free aptasensor for platelet-derived growth factor-BB and its logic gate application. Biosens Bioelectron. 2015;63:552–557. | ||
Guo LM, Zhao Q. Thrombin-linked aptamer assay for detection of platelet derived growth factor BB on magnetic beads in a sandwich format. Talanta. 2016;158:159–164. | ||
Huang Y, Nie XM, Gan SL, Jiang JH, Shen GL, Yu RQ. Electrochemical immunosensor of platelet-derived growth factor with aptamer-primed polymerase amplification. Anal Biochem. 2008;382(1):16–22. |
Supplementary materials
Experimental conditions and procedures for characterization
Transmission electron microscope (TEM)
TEM images of the gold nanoparticles (AuNPs) samples were taken using a JEOL 1200 EX TEM at 80 kV. AuNPs were dispersed in water and dropped onto copper grid before the images were acquired using an AMT 2K CC Camera.
UV–vis spectroscopy
The as-prepared AuNPs or DNA-modified AuNPs (100 μL) were characterized with UV–vis spectroscopy (UV1800PC) in a quartz cell with wavelength from 350 to 700 nm.
Native polyacrylamide gel electrophoresis (PAGE)
Proximity hybridization-triggered EXPAR was assessed with 18% native PAGE. The gels were freshly prepared. Before loading, DNA samples were mixed with DNA-loading buffer (6×) on a volume ratio of 5:1. A potential of 85 V was applied for gel electrophoresis. After separation, gels were stained using 4S Red Plus for 15 min and imaged by digital imaging system (Thermo Fisher Scientific, Waltham, MA, USA).
Electrochemical impedance spectroscopy (EIS)
The parameters for EIS were as shown below:
Init E (V): 0.247
High frequency (Hz): 100,000
Low frequency (Hz): 0.01
Amplitude (V): 0.005
Quiet time (s): 2
Differential pulse voltammetry (DPV)
The parameters for DPV were as shown below:
Init E (V): 0.45
Final E (V): 0.18
Incr E (V): 0.004
Amplitude (V): 0.05
First pulse width (s): 0.2
Second pulse width (s): 0.05
Sampling width (s): 0.0167
Pulse period (s): 0.5
Quiet time (s): 2
Sensitivity (A/V): 1e-5
Native PAGE characterization
Native PAGE analysis is shown in Figure S1.
Elimination of nonspecific reaction
To eliminate the nonspecific displacement, we optimized the structure of S1–S2 duplex and the length of P1–Output complementary parts. As S1–S2 formed an arched structure with two stems and a loop in the middle, we fixed one stem to be 12 bp and the other stem varied from 8 to 14 bp. After incubation on the gold electrode surface (0.5 μM was chosen), 10 nM Output was added to the reaction system to proceed the duplex cleavage process. At last, the electrode was incubated with G-quadruplex-modified AuNPs and electrochemical signals were obtained from the reduction of oxidized 3,3′,5,5′-tetramethylbenzidine (TMB). Figure S2A shows that 10 bp yielded the largest signal/background ratio. While the shorter stem will lead to the nonspecific cleavage of the arched structure, the longer stem will make it hard to cleavage even after the hybridization of Output. Finally, 12 and 10 bp were chosen for the arched structure in this system.
Another important factor to affect the nonspecific reaction was the length of P1–Output complementary part. First, we fixed the complementary part of P2-P1 11 nt, and increased the length of P1–Output from 9 to 13 bp. In principle, shorter P2 was thermodynamically unfavored to displace a longer Output from P1 and, thus, should be able to suppress background signal. Figure S2B shows that the maximal signal-to-background was obtained at 11 nt.
Sequences of involved oligonucleotides are listed in Table S1.
  | Table S1 Sequences of involved oligonucleotides for the elimination of target-independent reaction |
Specificity investigation
We investigated the selectivity of the method that we proposed in different buffers with the addition of fivefold PDGF-AA or PDGF-AB. The results are shown in Figure S3.
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.