Back to Journals » Cancer Management and Research » Volume 12
Proteomics of Sentinel Lymph Nodes in Early Breast Cancer for Identification of Thymidylate Synthase as a Potential Biomarker to Flag Metastasis: A Preliminary Study
Authors Pathania S, Khan MI, Kumar A, Gupta AK, Rani K, Ramesh Parashar T, Jayaram J, Ranjan Mishra P , Srivastava A, Mathur S, Hari S, Hariprasad G
Received 26 March 2020
Accepted for publication 21 May 2020
Published 23 June 2020 Volume 2020:12 Pages 4841—4854
DOI https://doi.org/10.2147/CMAR.S255684
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
Video abstract presented by Gururao Hariprasad.
Views: 241
Sheetal Pathania,1,* Mohd Imran Khan,1,* Abhishek Kumar,1 Ashish Kumar Gupta,1 Komal Rani,1 Tanvi Ramesh Parashar,1 Jnaneshwari Jayaram,2 Piyush Ranjan Mishra,2 Anurag Srivastava,2 Sandeep Mathur,3 Smriti Hari,4 Gururao Hariprasad1
1Department of Biophysics, All India Institute of Medical Sciences, New Delhi 110029, India; 2Department of Surgery, All India Institute of Medical Sciences, New Delhi 110029, India; 3Department of Pathology, All India Institute of Medical Sciences, New Delhi 110029, India; 4Department of Radiology, All India Institute of Medical Sciences, New Delhi 110029, India
*These authors contributed equally to this work
Correspondence: Gururao Hariprasad Tel +91-11-26594240
Fax +91-11-26588663
Email [email protected]
Introduction: Breast cancer is the second most common cancer in women across the world. Some of the patients who present in the early stage of disease are affected by metastasis to the axillary group of lymph nodes. The first among this group that is affected is called as sentinel lymph node, and its diagnosis is crucial for the staging of cancer thereby dictating the type of surgical therapy. Therefore, the sentinel lymph node status provides the most relevant information to the surgeon and patient prognosis. The expanded utilization of breast conservation surgery has declined the morbidity associated with mastectomy and axillary lymph node surgery. Recent interest is, therefore, centered on techniques that allow accurate assessment of the sentinel lymph node metastasis. A current procedure such as sentinel lymph node biopsy (SLNB) that is used to assess axillary lymph node metastasis is neither specific nor sensitive, and besides, it is time-consuming.
Objective: To compare the protein profiles between metastatic and non-metastatic lymph nodes to identify a biomarker that can flag lymph node metastasis.
Materials and Methods: Women with early breast cancer were screened using mammography imaging and recruited to the study. Surgical resection was done to remove the breast tissue, and sentinel lymph node was identified using fluorescein and methylene blue tracer. Lymph node was sliced, and one set was sent for histopathology, which was considered the gold standard to assess the metastatic status of the lymph node. One set of slices was taken for proteomic experiments. Proteins were labelled with fluorescent cyanine tags and were subjected to difference gel electrophoresis experiment. Differentially expressed spots that had at least a twofold relative ratio and consistent pattern across three sets of biological replicate experiments were marked. Gel spots were trypsin digested and identified on mass spectrometry machine. Validation study was done by Western blot experiment on the same set of samples.
Results: Thymidylate synthase has a twofold higher expression in the metastatic sentinel lymph nodes as compared to non-metastatic lymph nodes in early breast cancer patients.
Conclusion: Differential in gel expression proteomics is an ideal platform for the identification of potential protein biomarker candidates that can differentiate metastatic from non-metastatic lymph nodes in early breast cancer. The identification of thymidylate synthase offers a scope to develop an on-table diagnostic kit to assess the status of sentinel lymph nodes during mastectomy procedure to guide surgical management of axillary lymph nodes in early breast cancer.
Keywords: early breast cancer, sentinel lymph node metastasis, gel-based proteomics, difference gel expression, biomarkers, thymidylate synthase, surgical intervention
Introduction
Breast cancer is the most common cancer in women worldwide and distant metastasis is the main reason for patient mortality.1 Worldwide, breast cancer remains the second leading cause of cancer incidence and mortality, with 2,088,849 new breast cancer cases and 626,679 deaths in 2018, representing close to 1 in 4 (6.6%) cancer deaths.2 The incidence rate of breast cancer is estimated to reach 3.2 million by 2050.3 Breast cancer ranks as the number one cancer among Indian females with rate as high as 25.8 per 100,000 women and mortality of 12.7 per 100,000 women.4 Incidence of metastatic breast cancer has been reported to be approximately 5% to 25% from various centres in India.5
The TNM-based classification is used to categorize and assign stages between 0 and IV, where stage 0 describes the non-invasive; stage I, IIA and IIB describe early breast cancer, which is confined to the breast with or without axillary lymph node involvement; and stage IIIA, IIIB and IV describe advanced breast tumor that has spread to distant organs.6,7 Early breast cancer depends on tumor size and extent of its spread to axillary lymph nodes. Its treatment includes mastectomy followed by evaluation of axillary lymph nodes to know the lymphatic spread, and a radical axillary lymph node excision is subsequently done based on its metastatic state.8
Axillary lymph nodes receive the major share of lymphatic drainage from the breast, and the first node in this group to be affected is called as sentinel lymph node, and its involvement is one of the main factors that dictates the extent of surgical dissection and also the prognosis.9,10 Sentinel lymph node biopsy (SLNB) was first introduced as a technique of axillary nodal staging for breast cancer, and is currently the standard of care for axillary staging in clinically node-negative breast cancer.11,12 A properly performed SLNB identifies patients who need further axillary clearance, while sparing others a potentially morbid axillary lymph node dissection.13 Sentinel lymph nodes are typically identified using tracers, and it is excised and tissue is taken for the histopathological evaluation which is the gold standard for lymph node metastasis.14 The concept that the presence or absence of malignant cells in the sentinel node can reflect the status of tumor involvement in the entire lymph node region markedly reduced the need for Axillary Lymph Node Dissection (ALND) and its associated morbidity.15–17 There have been a number of approaches including radiological techniques that have been developed to evaluate sentinel lymph nodes.18–23 Also, protein-based biomarkers have been identified to diagnose axillary lymph node metastasis.24–27 However, these have not been translated into a diagnostic tool as desired by the surgeons due to their poor sensitivity and poor specificity. In the recent past, our team has been actively involved in identifying potential protein biomarkers to understand various clinical phenotypes.28–30 In this study, a 2-D fluorescence difference gel electrophoresis (DIGE) experiment has been carried out using sentinel lymph node tissue to identify a biomarker that can accurately reflect the metastatic lymph node in early breast cancer.
Methodology
Ethics and Patient Recruitment
This study was conducted after approval was obtained from the Institute Ethics committee of All India Institute Medical Science, New Delhi, India (Ref. No. IECPG/77/11/2015). The procedures followed were as per the ethical standards formulated in the Helsinki declaration. Women patients with palpable lumps underwent triple assessment that is comprised of clinical examination, imaging, and core biopsy. Based on these findings, patients who had early breast were recruited in the study and admitted at the Department of Surgery after obtaining written informed consent.
Patient Inclusion and Exclusion Criteria
Staging of the breast cancer was determined as per the American Joint Committee on Cancer (AJCC) cancer staging criteria. Women with early invasive intraductal breast cancer, without prior therapy were recruited to the study. Patients with advanced breast cancer and who had either received chemotherapy or radiotherapy were excluded from the study.
Identification and Excision of Sentinel Lymph Node
The patients scheduled for surgery were injected with radioactive isotope Tc99m-tagged sulphur colloid of 200–300 microcurie in 0.1–0.2 mL normal saline in sub-areolar region 1–3 h before surgery.31 Radiotracer was injected in the afternoon, a day before surgery, if the patient was scheduled as first case next day morning. A gamma-detecting probe was used intra-operatively to identify radioactivity positive nodes. Four millilitres of 1% methylene blue and 0.1 mL fluorescein were injected in the sub-areolar region.32 A hot, blue, fluorescent or palpable node was considered to be the sentinel lymph node and was excised.33,34
Sample Collection
The sentinel lymph node specimen was cleared of adherent adipose tissues, and blood stains were washed thoroughly with 1× PBS (pH 7.4). The nodes were longitudinally sectioned to obtain 2mm thick slices. One set of alternate slices were sent to the Department of Pathology for histopathological assessment, and the remaining slices were taken to clinical proteomics facility and stored at −80 °C to be used at a later date for proteomic experiments. For example, slices numbered 1,3,5,7,9 were taken for histopathology laboratory, and slices numbered 2,4,6,8,10 were taken for proteomics work.
Histopathology
Lymph node slices were fixed into formalin and then embedded in paraffin blocks. These were further sectioned into 4 µm poly-L-lysine-coated slides.35 These paraffin sections were de-paraffinized with three subsequent washes in xylene and then rehydrated by washing them stepwise in 100% ethanol, 90% ethanol, 70% ethanol and distilled water. The sections were stained with hematoxylin and washed in running water for 5 minutes. The slides were stained in eosin solution for two minutes and then rinsed with 95% ethanol. The slides were then subjected to 95% ethanol and 100% ethanol for two minutes, twice. The tissue slice was mounted with a drop of Distyrene Plasticizer Xylene (DPX) after exposure to xylene. The slides were examined and sentinel lymph node tissue samples were annotated as either metastatic sentinel lymph node (M) or non-metastatic sentinel lymph node (NM). This histopathological evaluation was considered as the gold standard in the assessment of lymph node metastasis.36–38
Sample Phenotyping and Protein Isolation
The sentinel lymph node tissue sections were stored at −80 °C and labeled as either metastatic (M) or non-metastatic (NM) based on histopathological reporting of the adjacent slice sections. The tissue samples were minced and the proteins were solubilized in 100 µL of lysis buffer containing 8M urea, 2M thiourea and 4% 3-[(3-cholamidopropyl)dimethylammonio]1-propanesulfonate (CHAPS). The tissue was homogenized by sonication for 10 cycles, followed by vortexing for 2 minutes. The samples were centrifuged at 12000 rpm for 15 minutes at 4 °C, and the supernatant was collected. Tissue impurities such as salts and lipids were minimized using in-house protocols.39,40 Protein quantification was done by Bradford method using bovine serum albumin as standard (GeNei, Bengaluru, Karnataka, India)
DIGE
Fifty micrograms of protein from each of the three M and NM tissues were labeled with cyanine dye flours according to minimal labeling protocol provided by the manufacturer (Amersham Biosciences, USA). Two of the metastatic samples were labeled with Cy3 dye and two non-metastatic samples were labeled with Cy5 dye. Dye swapping was done for the third set wherein the metastatic sample was labeled with Cy5 dye and non-metastatic with Cy3 dye. Fifty micrograms of protein from each of the six samples were mixed to generate an internal standard, and 50 μg of this protein was labeled with Cy2 dye and used in each of the three gels for normalization. Labeled samples from M, NM and internal standard were pooled together, and final volume was made up to 250 µL using rehydration solution (8M urea, 2M thiourea, 4% CHAPS and 0.002% bromophenol blue). Dithiothreitol (DTT) and Immobilized pH gradient (IPG) buffer (pH 3–10) were added at final concentrations of 0.003% and 0.5%, respectively. The samples were centrifuged at 4000 rpm for 2 minutes at 4°C to remove any particulate matter. The solution was then loaded on a reswelling tray (Amersham Biosciences, USA). Plastic cover of Immobiline Dry Gel Strip (pH 3–10) of 13 cm was carefully removed, and the gel surface was placed over the sample in the tray with forceps. The gel strip was overlaid with IEF cover fluid (Amersham Biosciences, USA) and was kept overnight for 15 hours for optimum rehydration. Rehydrated IPG strip was kept in a strip holder and subjected for isoelectric focusing in an Ettan IPGphor 3 IEF system (Amersham Biosciences, USA) as per the following program. 1. Step mode, 150V for 30 minutes; 2. Step mode, 500V for 30 minutes; 3. Step mode, 1000V for 30 minutes; 4. Gradient mode, 4000V for 2 hr; 5. Step mode, 4000V for 2 hr; 6. Gradient mode, 6000V for 2 hr; 7. Step mode, 6000V till total volt-hours of 28,000 were achieved. The electro-focused strip was equilibrated, first with 10mL of SDS buffer containing 10 mg/mL DTT for 15 minutes, and second with 10 mL of SDS buffer containing 25 mg/mL iodoacetamide for 15 minutes. The strip was then transferred onto 10% homogeneous polyacrylamide gels cast on SE 600 Ruby gel apparatus (GE Healthcare, Piscataway, NJ). The strip was overlaid with 0.5% agarose sealing solution (0.5% agarose, 0.002% bromophenol blue in Tris-glycine electrode buffer). Separation in SDS-PAGE was carried out with constant running current set at 15 mA per gel at 20 °C for 30 minutes, followed by 30 mA per gel at 20 °C until the bromophenol blue dye front ran off from the bottom of the gel. Two more such gels were run as biological replicate experiments.
Gel Imaging, Spot Detection, and Statistical Analysis
DIGE gels were scanned using a Typhoon TRIO Variable Mode Imager (GE Healthcare). Cy2 images were scanned with 488 nm/520 nm band pass, Cy3 images were scanned with 532 nm/580 nm band pass, and Cy5 images were scanned with 633 nm/670 nm band pass. All gels were scanned with a photon multiplier tube setting of 700 V with 100 μm/pixel resolution and high sensitivity. Three images were acquired from each gel corresponding to metastatic lymph node, non-metastatic lymph node, and internal standard. 2D-DIGE gels images we reanalyzed using Progenesis Same Spots software (Non-Linear Dynamics, UK). Image quality evaluation was done. Reference image gel was selected automatically, and image area was chosen manually for analyses. All gels were automatically aligned onto one reference gel. Cy3 dye and Cy5 dye gel images were selected into either M or NM. Statistical analysis of protein expression levels was determined for every spot based on log normalized spot volume, and differences in protein expression between two groups were assessed by analysis of variance (ANOVA). Spots with ANOVA p-value ≤ 0.05 and maximum fold change ≥2 were selected as spots of interest. Correlation analysis was performed on log normalized volume spot expression levels between two group spots together according to similarities in their expression profiles. A final report was created showing analyzed spots on the gel together with chosen ANOVA p-value, ranks, and expression profiles and log normalized volume graphs for each spot.
Mass Spectrometric Analysis and Protein Identification
Gels were stained with colloidal coomassie blue-G250 stain. Spots of interest were picked manually from the gel. These spots were subjected to in-gel trypsinization according to the manufacturer’s protocol (Promega Corporation, Fitchburg, WI, USA). After overnight digestion, buffer containing the peptides was collected. Additional extraction of peptides was carried out with 100μL of 50% acetonitrile in 1% formic acid. The extracts were pooled, vacuum-dried and stored in 4°C refrigerator. At the time of analysis, the peptide extract was reconstituted in the 10µL of 60% acetonitrile and 0.1% formic acid (TFA). The peptides were mixed with the α-cyano-4-hydroxycinnamic acid matrix in a ratio of 1:1 and spotted on the Matrix-assisted laser desorption/ionization (MALDI) plate. MALDI-MS data were acquired over a mass range of 800–3500 Da in the reflector ion mode on a 4800 MALDI-TOF/TOF Analyzer (Applied Biosystems, Foster City, CA, USA) with 4000 Series Explorer v3.5 software, using a fixed laser intensity for 1500 shots/spectrum, with a uniformly random spot search pattern. The potential difference between acceleration voltage and floating collision cell defines the collision energy, which was 1keV in all experiments. Re-optimized instrument settings were employed to achieve optimal sensitivity. The air was used as the collision gas such that nominally single collision conditions were achieved. In each MS spectrum, the 10 most abundant MS peaks were selected for MS/MS using an acquisition method that excluded ions with S/N less than 50, and which filtered out identical peaks detected in nearby spots, selecting only the strongest precursor. The precursor ions with the weakest S/N were acquired first to achieve the maximum signal intensity for low-abundance peptides. A 1kV MS/MS operating mode was used, the relative precursor mass window was set at 250 (full-width half mass), with metastable suppression enabled. MS/MS acquisition of selected precursors was set to a maximum of 1250 shots with 50 shots per sub-spectrum using fixed laser intensity. The stop condition criteria were established to a minimum of 100 S/N on more than seven peaks within the spectrum of the minimum 1000 shots. Obtained masses of the peptide from the mass spectrometric analysis were searched in MASCOT search engine with NCBI database for the identification of proteins. Detected protein threshold was fixed at a confidence score of 99.9%.
Bioinformatics Pathway Analysis
The entire raw interaction information of genes corresponding to the identified proteins, their related functions were obtained from UniProt, BioGrid, Kyoto Encyclopedia of Genes and Genomes (KEGG) and from published literature in PubMed. Using this information, proteins were analysed for their biological interactions in lymph node metastasis in breast cancer. Cytoscape v3.2.0 software was used for constructing an integrated model of protein–protein interaction network and visualizing the interaction of identified proteins with its interactors.41,42 Michigan Molecular Interactions plugin was used to collect the human gene regulatory interactome obtained from the public databases including STRING, Uniprot, IntAct, and IMEx and merge the information.43–46 From this complete network, sub-networks for breast cancer were obtained up to the first neighboring nodes using the plugin BiNoM v2.5. The resulting PPI network was analysed and the interactions of the identified proteins were noted.
Western Blot
Isolated protein samples from three metastatic (M1, M2, M3) and three non-metastatic (NM1, NM2, NM3) sentinel lymph nodes were quantified by the Bradford method (Bio-Rad Laboratories) with bovine serum albumin as a standard (Bradford assay). A total of 10 μg/lane protein were electrophoresed in 12% sodium dodecyl sulfate-polyacrylamide gels and transferred onto a nitrocellulose membrane. After transfer, the blot was blocked with 3% BSA in Tris-buffered saline, and blots were incubated with the anti-thymidylate synthase antibody (Elabsciences; 1:10,000) and anti-β-actin antibody (Elabsciences; 1:10,000), at 4 °C overnight. Membranes were incubated with HRP-conjugated secondary antibodies (rabbit and mouse from Thermo Fisher Scientific) (1:10,000) in 1.5% BSA for 2 hrs at room temperature. Protein bands were visualized using an enhanced chemiluminescence system with ECL Western blot detection reagents (GBiosciences). A semi-quantitative analysis was performed to quantify the thymidylate synthase and β-actin bands intensity in blot images using ImageJ software. Data analysis was carried out by using Graph Pad Prism 7 software and P value was calculated. An unpaired t-test was performed to compare the band intensity ratios of thymidylate synthase and β- actin proteins among both the groups. All the data were represented as median ± standard error of mean. A P-value of less than 0.05 was considered statistically significant.
Results
Clinical Profile
The clinical data of six patients in the study group are provided in Table 1. All patients had early breast invasive carcinoma as documented by mammography (Figure 1). Patients’ progesterone receptor and estrogen receptor and human epidermal growth factor receptor 2 (HER2) statuses are also provided. However, progesterone, estrogen and Her2 status had no correlation with either the type, staging or lymph node status of the breast cancer.
 |
Table 1 Clinical Profile of Early Breast Cancer Patients Recruited in the Study |
Histopathology
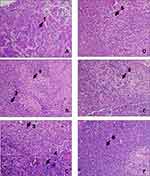
Once the patients were confirmed to be having early-stage invasive intraductal breast carcinoma, it was mandatory to assess the sentinel lymph node in the axilla of the side affected by disease. Using the method of methylene and fluorescein-based imaging technique developed by Srivastava and his group, sentinel lymph nodes were identified, excised and sent for histopathological analysis. Hematoxylin and Eosin stained sections of the sentinel lymph nodes are shown in Figure 2. Sentinel lymph nodes of SR42, PD45, and SD42 showed classical features of metastasis like either complete effacement of architecture, reactive lymphoid tissue or infiltration by metastatic cancer cells. On the other hand, sentinel lymph nodes of FJ30, SP49 and KK49 showed features of inflammation like reactive changes of reactive follicular hyperplasia, sinus histiocytosis and reactive lymphadenitis confirming the absence of metastasis. The reactive changes represent local immunological response in lymph nodes that are benign in nature.
2D-DIGE
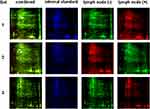
Proteins were isolated from M and NM tissues for comparative gel-based proteomic experiments. The experimental design for cyanine-based fluorescent label DIGE is shown in Table 2. The proteins isolated from six samples were run on three biological replicate gels. Nine images corresponding to three colored channels representing M, NM and internal standard generated for each of the three gels are shown in Figure 3. The images show a uniform and comparable distribution of spots consistently present across three gels. Number of spots ranged between 300 and 500 in each of the three gels. Twenty signals were marked by the software as shown in Figure 4. Based on the criteria for selection only spot #790 was chosen for detailed analysis. Differential expression, spot volume comparison and log normalized volume graph clearly showed a statistically significant twofold higher expression in M as compared NM. The spot was manually picked, subjected to trypsin digestion and mass spectrometric analysis to identify the protein on MASCOT software. A snapshot of mass spectrometric data is shown in Supplementary Figure 1, and identification profile is presented in Table 3.
 |
Table 2 Difference Gel Electrophoresis Experimental Design Showing Labeling of Clinical Phenotypes |
 |
Table 3 Identification Profile of Differentially Expressed Protein |
Western Blot
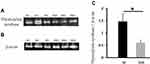
Expression of thymidylate synthase was validated by Western blot experiment using metastatic and non-metastatic sentinel lymph node tissue samples from early invasive breast cancer patients. β-Actin, a housekeeping protein was used for the sake of normalization. The expression of thymidylate synthase was found significantly higher (p=0.015) in patients with lymph node metastasis as compared to patients without lymph node metastasis in early invasive breast cancer. The thymidylate synthase expression is 2.41 times up-regulated in positive samples as compared to negative samples as shown in Figure 5.
Bioinformatic Analysis
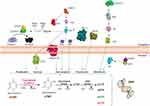
Bioinformatic analysis was done to get a detailed understanding of the various pathways in cancer metastasis and the interactions between the molecules (Figure 6). Based on these results, the plausible hypothesis relating the up-regulation of thymidylate synthase with clinical phenotype of sentinel lymph node metastasis is explained below and its pictorial representation is shown in Figure 7.
 |
Figure 6 Protein interaction pathway analysis in metastasis. |
Discussion
We have shown that thymidylate synthase has a twofold expression in metastatic state as compared to non-metastatic state of the sentinel lymph nodes. Thymidylate synthase belongs to the ThyA family of enzymes which is located on 18p11.32. It catalyzes the first step in the de novo biosynthesis of thymine, and an essential enzyme for DNA synthesis.47,48 This enzyme catalyzes the conversion of 2-deoxyuridine-5-monophosphate in conjunction with its cofactor 5,10-methylenetetrahydrofolate to thymidine monophosphate and dihydrofolate through reductive methylation.49
There are many biochemical pathways at play in the phenomenon of metastasis. They are 1) Erb-B2 receptor tyrosine kinase 2 (ERBB2), a cell surface-bound protein receptor tyrosine kinase (RTK) activates PI3 kinase and PI(3,4,5)P3whichsubsequently causes phosphorylation of Akt and mTOR that is responsible for cell proliferation, survival and metastasis50; 2) activated AKT kinase phosphorylates and inactivates BAD protein freeing BCL2 to suppresses apoptosis51,52; 3) activated Akt also phosphorylates Bax protein and inactivates it, causing inhibition of apoptosis and leading to cell survival.53,54 4) MCL-1 is a pivotal pro-survival BCL2 family protein which further inhibits Bax and causes anti-apoptosis and cell-survival55; 5) MAPK pathway increases the expression of transcriptional regulatory protein Myc that is responsible for the phosphorylation of Rb family of proteins and subsequently activation of the G1/S genes responsible for cell proliferation56,57; 6) Overexpression of SUMO-activating enzyme (SAE1) induces an increase of the SUMOylation and phosphorylation of AKT that causes tumour progression, metastasis, and is associated with poor survival58; 7) Activated ribosomal protein S6 kinase 2 (RPS6K) promotes cell proliferation, cell growth, cell cycle progression and survival. Akt protein pathway59,60; and 8) integrin subunit alpha 4 (ITGA4) is also activated by critical pathways like Src, FAK, Akt and ERK which promotes cancer cell metastasis and survival.61 The cells in metastatic lymph nodes are in a state of multiplication necessitating an increased DNA turnover and in turn its synthesis. For DNA synthesis to proceed normally, a balanced pool of the four deoxyribonucleotides, dCMP, dGMP, dAMP and dTMP is an essential requirement.62 These four deoxyribonucleotides are synthesised from ribonucleotides (AMP, GMP, CMP and UMP) by a same enzyme called ribonucleotide reductase that catalyzes the reduction of these four ribonucleotides to their corresponding deoxyribonucleotides.63 Of these four deoxyribonucleotides, only dUMP requires further metabolism for its conversion to dTMP. This methylation step is done by thymidylate synthase, which is the sole cellular enzyme capable of de novo synthesis of dTMP, and is, therefore, the rate-limiting step in thymidine biosynthesis.64,65 It implies that: 1) suppression of thymidylate synthase expression in cells causes a deficiency in the intracellular concentration of thymidylate, bringing DNA reproduction to a halt and thus causing death to cells66; 2) inhibition of thymidylate synthase will result in both a depletion of dTTP and an accumulation of dUTP which can be mis-incorporated into DNA resulting in DNA breakage and cell death.67,68 Thymidylate synthase is a S-phase-dependent enzyme undergoing a 5- to 10-fold increase as cells progress from G1- to S-phase.64,69 The over-expression of thymidylate synthase in metastatic lymph node cells is significantly related to the cell doubling time and can therefore be regarded as an important parameter for cell metastasis and proliferation.70,71 The increased expression of thymidylate synthase in the metastatic sentinel lymph node cells in our experiments complements the above observations and places them in the right perspective. Till date, there have been several studies that have shown the significant correlation between high expression of thymidylate synthase and cancer. It has been the most extensively investigated as a biomarker for the detection of metastasis in colorectal cancer, glioma, pancreatic, breast, melanoma.72–76 The results of this study therefore complement the previous observations that advocate thymidylate synthase as a biomarker for cancer metastasis.
Conclusions
Differential in gel expression proteomics is an ideal platform for the identification of potential protein biomarker candidates for metastasis of early breast cancer to axillary sentinel lymph nodes. Thymidylate synthase has a twofold higher expression in metastatic lymph nodes as compared to non-metastatic lymph nodes in early breast cancer. This study offers scope for the design of an 'on-table diagnostic kit' to assess the status of axillary sentinel lymph nodes during mastectomy procedure to guide surgical management of early breast cancer.
Acknowledgments
GH acknowledges All India Institute of Medical Sciences for the grant A590. The work was carried out at the Proteomics Division of Central Core Research Facility at AIIMS, New Delhi, India.
Disclosure
The authors report no conflicts of interest in this work.
References
1. McAnena P, Tanriverdi K, Curran C, et al. Circulating microRNAs miR-331 and miR-195 differentiate local luminal a from metastatic breast cancer. BMC Cancer. 2019;19(1):436. doi:10.1186/s12885-019-5636-y
2. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
3. Hortobagyi GN, de la Garza Salazar J, Pritchard K, et al. The global breast cancer burden: variations in epidemiology and survival. Clin Breast Cancer. 2005;6(5):391–401. doi:10.3816/cbc.2005.n.043
4. Malvia S, Bagadi SA, Dubey US, Saxena S. Epidemiology of breast cancer in Indian women. Asia Pac J Clin Oncol. 2017;13(4):289–295. doi:10.1111/ajco.12661
5. Gogia A, Deo SVS, Sharma D, et al. Discordance in biomarker expression in breast cancer after metastasis: single center experience in India. J Glob Oncol. 2019;5:1–8. doi:10.1200/JGO.18.00184
6. Koh J, Kim MJ. Introduction of a new staging system of breast cancer for radiologists: an emphasis on the prognostic stage. Korean J Radiol. 2019;20(1):69–82. doi:10.3348/kjr.2018.0231
7. Singletary SE, Allred C, Ashley P, et al. Revision of the American Joint Committee on Cancer staging system for breast cancer. J Clin Oncol. 2002;20(17):3628–3636. doi:10.1200/JCO.2002.02.026
8. Blackburn HL, Ellsworth DL, Shriver CD, Ellsworth RE. Breast cancer metastasis to the axillary lymph nodes: are changes to the lymph node “soil” localized or systemic? Breast Cancer. 2017;11:1178223417691246. doi:10.1177/1178223417691246
9. Blumgart EI, Uren RF, Nielsen PM, Nash MP, Reynolds HM. Lymphatic drainage and tumour prevalence in the breast: a statistical analysis of symmetry, gender and node field independence. J Anat. 2011;218(6):652–659. doi:10.1111/j.1469-7580.2011.01362.x
10. Giuliano AE, Kirgan DM, Guenther JM, Morton DL. Lymphatic mapping and sentinel lymphadenectomy for breast cancer. Ann Surg. 1994;220(3):391–401. doi:10.1097/00000658-199409000-00015
11. Kumar R, Bozkurt MF, Zhuang H, Alavi A. Sentinel lymph node biopsy in the management of breast cancer. Indian J Cancer. 2003;40(2):60–66.
12. Lyman GH, Temin S, Edge SB, et al. Sentinel lymph node biopsy for patients with early-stage breast cancer: American Society of Clinical Oncology clinical practice guideline update. J Clin Oncol. 2014;32(13):1365–1383. doi:10.1200/JCO.2013.54.1177
13. Rao R, Euhus D, Mayo HG, Balch C. Axillary node interventions in breast cancer: a systematic review. JAMA. 2013;310(13):1385–1394. doi:10.1001/jama.2013.277804
14. Barranger E, Dubernard G, Fleurence J, et al. Subjective morbidity and quality of life after sentinel node biopsy and axillary lymph node dissection for breast cancer. J Surg Oncol. 2005;92(1):17–22. doi:10.1002/jso.20343
15. Borgstein PJ, Pijpers R, Comans EF, et al. Sentinel lymph node biopsy in breast cancer: guidelines and pitfalls of lymphoscintigraphy and gamma probe detection. J Am Coll Surg. 1998;186(3):275–283. doi:10.1016/s1072-7515(98)00011-8
16. Wedgwood KR, Benson EA. Non-tumour morbidity and mortality after modified radical mastectomy. Ann R Coll Surg Engl. 1992;74(5):314–317.
17. Lin PP, Allison DC, Wainstock J, et al. Impact of axillary lymph node dissection on the therapy of breast cancer patients. J Clin Oncol. 1993;11(8):1536–1544. doi:10.1200/JCO.1993.11.8.1536
18. Mokhtar M, Tadokoro Y, Nakagawa M, et al. Triple assessment of sentinel lymph node metastasis in early breast cancer using preoperative CTLG, intraoperative fluorescence navigation and OSNA. Breast Cancer. 2016;23(2):202–210. doi:10.1007/s12282-014-0551-1
19. Zhao QL, Xia XN, Zhang Y, et al. Elastosonography and two-dimensional ultrasonography in diagnosis of axillary lymph node metastasis in breast cancer. Clin Radiol. 2018;73(3):312–318. doi:10.1016/j.crad.2017.09.013
20. Solon JG, Power C, Al-Azawi D, Duke D, Hill AD. Ultrasound-guided core biopsy: an effective method of detecting axillary nodal metastases. J Am Coll Surg. 2012;214(1):12–17. doi:10.1016/j.jamcollsurg.2011.09.024
21. Nandu VV, Chaudhari MS. Efficacy of sentinel lymph node biopsy in detecting axillary metastasis in breast cancer using methylene blue. Indian J Surg Oncol. 2017;8(2):109–112. doi:10.1007/s13193-016-0616-z
22. Harada T, Tanigawa N, Matsuki M, Nohara T, Narabayashi I. Evaluation of lymph node metastases of breast cancer using ultra-small superparamagnetic iron oxide-enhanced magnetic resonance imaging. Eur J Radiol. 2007;63(3):401–407. doi:10.1016/j.ejrad.2007.02.010
23. Roumen RM, Valkenburg JG, Geuskens LM. Lymphoscintigraphy and feasibility of sentinel node biopsy in 83 patients with primary breast cancer. Eur J Surg Oncol. 1997;23(6):495–502. doi:10.1016/s0748-7983(97)92885-7
24. Zeng L, Deng X, Zhong J, et al. Prognostic value of biomarkers EpCAM and αB-crystallin associated with lymphatic metastasis in breast cancer by iTRAQ analysis. BMC Cancer. 2019;19(1):831. doi:10.1186/s12885-019-6016-3
25. Liu AN, Qu HJ, Yu CY, et al. Serum biomarkers for lymph node metastasis in patients with triple-negative breast cancer by proteomics. Int J Clin Exp Pathol. 2017;10(3):3377–3384.
26. Harms W, Malter W, Krämer S, et al. Clinical significance of urokinase-type plasminogen activator (uPA) and its type-1 inhibitor (PAI-1) for metastatic sentinel lymph node involvement in breast cancer. Anticancer Res. 2014;34(8):4457–4462.
27. Monsalve-Lancheros A, Ibáñez-Pinilla M, Ramírez-Clavijo S. Detection of mammagloblin by RT-PCR as a biomarker for lymph node metastasis in breast cancer patients: a systematic review and meta-analysis. PLoSOne. 2019;14(5):e0216989. doi:10.1371/journal.pone.0216989
28. Gupta AK, Pokhriyal R, Khan MI, et al. Cerebrospinal fluid proteomics for identification of α2-macroglobulin as a potential biomarker to monitor pharmacological therapeutic efficacy in dopamine dictated disease states of parkinson’s disease and schizophrenia. Neuropsychiatr Dis Treat. 2019;15:2853–2867. doi:10.2147/NDT.S214217
29. Sehrawat U, Pokhriyal R, Gupta AK, et al. Comparative proteomic analysis of advanced ovarian cancer tissue to identify potential biomarkers of responders and nonresponders to first-line chemotherapy of carboplatin and paclitaxel. Biomark Cancer. 2016;8:43–56. doi:10.4137/BIC.S35775
30. Pokhriyal R, Hariprasad R, Kumar L, Hariprasad G. Chemotherapy resistance in advanced ovarian cancer patients. Biomark Cancer. 2019;11:1179299X19860815. doi:10.1177/1179299X19860815
31. Challa VR, Seenu V, Srivastava A, et al. Sentinel lymph node mapping in early breast cancer - Our experience. Indian J Surg Oncol. 2010;1(1):52–58. doi:10.1007/s13193-010-0012-z
32. Kataria K, Srivastava A, Qaiser D. What is a false negative sentinel node biopsy: definition, reasons and ways to minimize it? Indian J Surg. 2016;78(5):396–401. doi:10.1007/s12262-016-1531-9
33. Challa VR, Srivastava A, Dhar A, et al. Role of fluorine-18-labeled 2-fluoro-2-deoxy-D-glucose positron emission tomography-computed tomography in the evaluation of axillary lymph node involvement in operable breast cancer in comparison with sentinel lymph node biopsy. Indian J Nucl Med. 2013;28(3):138–143. doi:10.4103/0972-3919.119542
34. Seenu V, Suhani S, Srivastava A, et al. Optimization of sentinel lymph node identification techniques in the Indian setting: a randomized clinical trial. Indian J Cancer. 2019;56(2):114–118. doi:10.4103/ijc.IJC_163_18
35. Zeng J, Xie H, Lu Y, Feng Z, Li F. Clinical implications of micrometastasis detection in internal mammary nodes of breast cancer patients. Breast Care. 2012;7(3):216–219. doi:10.1159/000339686
36. Delgado-Bocanegra RE, Millen EC, Nascimento CMD, Bruno KA. Intraoperative imprint cytology versus histological diagnosis for the detection of sentinel lymph nodes in breast cancer treated with neoadjuvant chemotherapy. Clinics. 2018;73:e363. doi:10.6061/clinics/2018/e363
37. Cserni G. Axillary staging of breast cancer and the sentinel node. J Clin Pathol. 2000;53(10):733–741. doi:10.1136/jcp.53.10.733
38. vanDiest PJ, Peterse HL, Borgstein PJ, Hoekstra O, Meijer CJ. Pathological investigation of sentinel lymph nodes. Eur J Nucl Med. 1999;26(4 Suppl):S43–S49. doi:10.1007/s002590050577
39. Hariprasad G, Srinivasan A, Vedakumari Y. Electrical conductivity as a tool to detect salt in clinical proteomics samples. Indian J Clin Biochem. 2011;26(1):82–83. doi:10.1007/s12291-010-0086-y
40. Pokhariyal R, Basnet B, Gupta AK, et al. Effect of lipid on gel based electrophoretic proteomic experiments. J Proteins Proteom. 2014;5(2):121–124.
41. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi:10.1101/gr.1239303
42. Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27(3):431–432. doi:10.1093/bioinformatics/btq675
43. Gao J, Ade AS, Tarcea VG, et al. Integrating and annotating the interactome using the MiMI plugin for cytoscape. Bioinformatics. 2009;25(1):137–138. doi:10.1093/bioinformatics/btn501
44. Jensen LJ, Kuhn M, Stark M, et al. STRING 8–a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37(Database issue):D412–D416. doi:10.1093/nar/gkn760
45. Orchard S, Ammari M, Aranda B, et al. The MIntAct project–IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014;42(Database issue):D358–D363. doi:10.1093/nar/gkt1115
46. Orchard S, Kerrien S, Abbani S, et al. Protein interaction data curation: the International Molecular Exchange (IMEx) consortium. Nat Methods. 2012;9(4):345–350. doi:10.1038/nmeth.1931
47. Pozzi C, Lopresti L, Tassone G, Mangani S. Targeting methyltransferases in human pathogenic bacteria: insights into Thymidylate Synthase (TS) and Flavin-Dependent TS (FDTS). Molecules. 2019;24(8):1638. doi:10.3390/molecules24081638
48. Wang X, Wei H, Wu Y, Tian Y, Luo L. Thymidylate synthase gene variation is associated with the risk for conotruncal heart defects in Chinese population. Cardiol Young. 2018;1–6. doi:10.1017/S1047951118002184
49. Pacifico S, Santucci M, Luciani R, et al. cyclic peptides acting as allosteric inhibitors of human thymidylate synthase and cancer cell growth. Molecules. 2019;24(19):3493. doi:10.3390/molecules24193493
50. Zoi I, Karamouzis MV, Xingi E, et al. Combining RANK/RANKL and ERBB-2 targeting as a novel strategy in ERBB-2-positive breast carcinomas. Breast Cancer Res. 2019;21(1):132. doi:10.1186/s13058-019-1226-9
51. Pandey V, Wang B, Mohan CD, et al. Discovery of a small-molecule inhibitor of specific serine residue BAD phosphorylation. Proc Natl Acad Sci U S A. 2018;115(44):E10505–E10514. doi:10.1073/pnas.1804897115
52. Yamamoto K, Ichijo H, Korsmeyer SJ. BCL-2 is phosphorylated and inactivated by an ASK1/Jun N-terminal protein kinase pathway normally activated at G(2)/M. Mol Cell Biol. 1999;19(12):8469–8478. doi:10.1128/mcb.19.12.8469
53. Niemi NM, MacKeigan JP. Mitochondrial phosphorylation in apoptosis: flipping the death switch. Antioxid Redox Signal. 2013;19(6):572–582. doi:10.1089/ars.2012.4982
54. Zhou H, Li XM, Meinkoth J, Pittman RN. Akt regulates cell survival and apoptosis at a postmitochondrial level. J Cell Biol. 2000;151(3):483–494. doi:10.1083/jcb.151.3.483
55. Lee EF, Harris TJ, Tran S, et al. BCL-XL and MCL-1 are the key BCL-2 family proteins in melanoma cell survival. Cell Death Dis. 2019;10(5):342. doi:10.1038/s41419-019-1568-3
56. Zhang W, Liu HT. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 2002;12(1):9–18. doi:10.1038/sj.cr.7290105
57. Meloche S, Pouysségur J. The ERK1/2 mitogen-activated protein kinase pathway as a master regulator of the G1- to S-phase transition. Oncogene. 2007;26(22):3227–3239. doi:10.1038/sj.onc.1210414
58. Yang Y, Liang Z, Xia Z, et al. SAE1 promotes human glioma progression through activating AKT SUMOylation-mediated signaling pathways. Cell Commun Signal. 2019;17(1):82. doi:10.1186/s12964-019-0392-9
59. Slattery ML, Lundgreen A, Herrick JS, Wolff RK. Genetic variation in RPS6KA1, RPS6KA2, RPS6KB1, RPS6KB2, and PDK1 and risk of colon or rectal cancer. Mutat Res. 2011;706(1–2):13–20. doi:10.1016/j.mrfmmm.2010.10.005
60. Pearce LR, Komander D, Alessi DR. The nuts and bolts of AGC protein kinases. Nat Rev Mol Cell Biol. 2010;11(1):9–22. doi:10.1038/nrm2822
61. Young SA, McCabe KE, Bartakova A, et al. Integrin α4 enhances metastasis and may be associated with poor prognosis in MYCN-low neuroblastoma. PLoS One. 2015;10(5):e0120815. doi:10.1371/journal.pone.0120815
62. Thomas WC, Brooks FP, Burnim AA, et al. Convergent allostery in ribonucleotide reductase. Nat Commun. 2019;10(1):2653. doi:10.1038/s41467-019-10568-4
63. Jonna VR, Crona M, Rofougaran R, et al. Diversity in overall activity regulation of ribonucleotide reductase. J Biol Chem. 2015;290(28):17339–17348. doi:10.1074/jbc.M115.649624
64. Le François BG, Maroun JA, Birnboim HC. Expression of thymidylate synthase in human cells is an early G1 event regulated by CDK4 and p16INK4A but not E2F. Br J Cancer. 2007;97(9):1242–1250. doi:10.1038/sj.bjc.6604020
65. Kanaan N, Martí S, Moliner V, Kohen A. QM/MM study of thymidylate synthase: enzymatic motions and the temperature dependence of the rate limiting step. J Phys Chem A. 2009;113(10):2176–2182. doi:10.1021/jp810548d
66. Islam Z, Gurevic I, Strutzenberg TS, et al. Bacterial versus human thymidylate synthase: kinetics and functionality. PLoS One. 2018;13(5):e0196506. doi:10.1371/journal.pone.0196506
67. Aschele C, Lonardi S, Monfardini S. Thymidylate synthase expression as a predictor of clinical response to fluoropyrimidine-based chemotherapy in advanced colorectal cancer. Cancer Treat Rev. 2002;28(1):27–47. doi:10.1053/ctrv.2002.0253
68. Chen D, Jansson A, Sim D, Larsson A, Nordlund P. Structural analyses of human thymidylate synthase reveal a site that may control conformational switching between active and inactive states. J Biol Chem. 2017;292(32):13449–13458. doi:10.1074/jbc.M117.787267
69. Nagarajan M, Johnson LF. Regulation of thymidylate synthase gene expression in mouse fibroblasts synchronized by mitotic selection. Exp Cell Res. 1989;181(1):289–297. doi:10.1016/0014-4827(89)90203-6
70. Derenzini M, Montanaro L, Treré D, et al. Thymidylate synthase protein expression and activity are related to the cell proliferation rate in human cancer cell lines. Mol Pathol. 2002;55(5):310–314. doi:10.1136/mp.55.5.310
71. Pestalozzi B, McGinn C, Kinsella T, et al. Increased thymidylate synthase protein levels are principally associated with proliferation but not cell cycle phase in asynchronous human cancer cells. Br J Cancer. 1995;71(6):1151–1157. doi:10.1038/bjc.1995.225
72. Lu Y, Zhuo C, Cui B, et al. TYMS serves as a prognostic indicator to predict the lymph node metastasis in Chinese patients with colorectal cancer. Clin Biochem. 2013;46(15):1478–1483. doi:10.1016/j.clinbiochem.2013.06.017
73. Ding B, Gao M, Li Z, et al. Expression of TYMS in lymph node metastasis from low-grade glioma. Oncol Lett. 2015;10(3):1569–1574. doi:10.3892/ol.2015.3419
74. Kang M, Zheng W, Chen Q, et al. Thymidylate synthase prompts metastatic progression through the dTMP associated EMT process in pancreatic ductal adenocarcinoma. Cancer Lett. 2018;419:40–52. doi:10.1016/j.canlet.2018.01.026
75. Cabibi D, Calascibetta A, Martorana A, et al. Different expression of thymidylate synthase in primary tumour and metastatic nodes in breast cancer patients. Anticancer Res. 2007;27(4B):2227–2230.
76. Vlaykova T, Jekunen AP, Kesomaa M, et al. Increased thymidylate synthase gene expression in metastatic melanoma. Oncology. 1997;54(2):146–152. doi:10.1159/000227679
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.