Back to Journals » Journal of Pain Research » Volume 14
Proteomic Analysis of Emodin Treatment in Neuropathic Pain Reveals Dysfunction of the Calcium Signaling Pathway
Authors Chen P, Lin D, Wang C , Song C, Wang W, Qu J, Wu Z
Received 5 November 2020
Accepted for publication 18 February 2021
Published 5 March 2021 Volume 2021:14 Pages 613—622
DOI https://doi.org/10.2147/JPR.S290681
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Krishnan Chakravarthy
Peng Chen,1 Dongsheng Lin,2 Chen Wang,2 Cuiwen Song,1 Wenjing Wang,1 Jinglian Qu,1 Zhibing Wu2
1Basic Medical School, Guizhou University of Traditional Chinese Medicine, Guiyang, Guizhou, People’s Republic of China; 2First Clinical Medical School, Guangzhou University of Chinese Medicine, Guangzhou, Guangdong, People’s Republic of China
Correspondence: Zhibing Wu
Department of Neurology, First Clinical Medical College of Guangzhou University of Chinese Medicine, 12 Jichang Road, Guangzhou, Guangdong, People’s Republic of China
Tel +86 020 36585577
Email [email protected]
Background: Neuropathic pain (NP) is a syndrome of pain mediated by distinct pathophysiological processes, and current treatments are not fully satisfactory. Emodin is an effective component of Chinese traditional medicine and has an alleviating effect on NP, but the pharmacological mechanism is not clear.
Methods: We used isobaric tags for relative and absolute quantitation (iTRAQ) technique integrated with liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis to identify potential targets of emodin in a rat peripheral nerve chronic constriction injury (CCI) model.
Results: A total of 177 differentially expressed proteins were identified among the sham group, CCI group, and emodin group, with a threshold of 1.2-fold change and a P value ≤ 0.05. Among them, 100 differentially expressed proteins (51 up-regulated and 49 down-regulated) were identified in the CCI group compared with sham group. Moreover, 108 differentially expressed proteins (65 up-regulated and 43 down-regulated) were identified in the emodin group with the CCI group as reference. The enrichment analysis of Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) revealed an important role of calcium signaling pathway, neurotransmitter regulation, and long-term potentiation (LTP) in emodin-treated CCI model. Real-time quantitative fluorescence PCR (qRT-PCR) and Western blot analysis revealed that emodin decreased expression of calcium signaling related proteins, including calmodulin (CaM) dependent protein kinase II (CaMK II), phospholipase Cβ 1 (PLCβ 1), protein kinase C (PKC), protein kinase C (PKA), and tropomyosin-related kinase B (TrkB), compared with the CCI group.
Conclusion: Overall, these findings indicated that emodin might alleviate NP by regulating the calcium signaling pathway.
Keywords: neuropathic pain, emodin, CCI, iTRAQ, KEGG
Introduction
Neuropathic pain (NP) refers to pain evoked by lesions or diseases in the somatosensory nervous system with prevalence in the general population of close to 10%.1,2 The primary clinical features of NP include allodynia, hyperalgesia, and spontaneous pain, frequently accompanied by insomnia, depression, and anxiety. These have a substantial impact on the patient’s daily life. Peripheral sensitization, central sensitization, activation of glia and other nonneuronal cells, and impairment of endogenous inhibitory mechanisms have been proven to be the underlying mechanism of NP.3 Currently, the pharmacotherapy of NP primarily involves antidepressants, anticonvulsants, opioids, and tramadol.4 However, the therapeutic effects are not always satisfactory and side effects are common.5–7 Due to the large number of affected individuals and the limited medication options, the development of new analgesics remains an important task.
Emodin (1,3,8-trihydroxy-6-methylanthraquinone) is an effective constituent of the traditional Chinese medicine Rheum palmatum. Many pharmacological studies reported that emodin could be utilized in the treatment of several neurological disorders. For example, emodin inhibited aggregation of amyloid-β peptide 1–42 and ameliorated cognitive deficits in Alzheimer's disease transgenic mice.8 Emodin also served as a neuroprotective agent in an ischemic stroke rat model by activation of the extracellular signal-regulated kinase-1/2 (ERK1/2) signaling pathway.9 In particular, emodin was reported to affect NP in a rodent model. We found that emodin alleviated NP of rats induced by chronic constriction injury (CCI).10 The pharmacological study revealed that emodin could inhibit NP transmission mediated by P2X(2/3) receptor in primary sensory.11 Additionally, emodin suppressed the release of calcitonin-gene-related peptide in trigeminal ganglia of rats with trigeminal neuralgia.12 However, the molecular mechanism underlying the effect of emodin on NP in the spinal cord has been scarcely investigated, especially in a systematic manner.
In recent years, isotope relative and absolute quantitative labeling (iTRAQ) technique has been widely used in the research of proteomics due to its advantages of high throughput, excellent stability, remarkable accuracy, and unrestricted sample source. Therefore, potential mechanisms underlying emodin analgesia could be identified by this new method. The aim of this study was to explore the analgesic targets of emodin in CCI model using proteomic techniques and bioinformatics analysis.
Materials and Methods
Animals
Thirty 7-week-old male Sprague-Dawley rats, weighing 180–220 g, were provided by Animal Experimental Center of Guangzhou University of Chinese Medicine (SCXK 2013–0034), and housed in a pathogen-free animal laboratory with free access to water and food under a 12 h light/dark cycle. The rats were randomly divided into the sham group, CCI group, and emodin group (n=10 for each group). All steps of the experiment were reviewed and approved by the Animal Use and Ethic Committee of the First Affiliated Hospital of Guangzhou University of Chinese Medicine. All experimental procedures were performed according to the Guidance Suggestions for the Care and Use of Laboratory Animals issued by the Ministry of Science and Technology of China.
CCI Model and Emodin Treatment
We prepared the CCI model as previously described by Bennett and Xie.13 The rats in each group were anesthetized with chloral hydrate (300 mg/kg) before surgery. The left nervus ischiadicus was exposed at the middle of the thigh and ligated with four silk ligatures proximal to the trifurcation. In the sham group, the nervus ischiadicus was simply exposed without ligation. In the CCI group, however, the skin of the middle thigh of the left lower limb of rats was incised, and the nervus ischiadicus trunk was exposed. At 5 mm above the trifurcation, four ligations were made using 4–0 silk threads with a distance of 1–2 mm. Furthermore, the rats in the emodin group were administered emodin (Chengdu Must Bio-Technology Corporation, Purity: 99.48%, Lot number: MUST-16110712) via gavage once a day for 15 days (50 mg/kg) on the first day after the nerve ligation. Emodin was dissolved to 6.25 g/L in 0.05% CMC-Na.
Behavioral Tests
The measurement of pain-related behaviors in each group was performed by the researchers blinded to the groups before the CCI surgery and 3, 7, 11, and 15 days post-surgery. Mechanical withdrawal threshold (MWT) and thermal withdrawal latency (TWL) were measures in our study. MWT was determined by the withdrawal threshold of hind paw using the electronic von Frey anesthesiometer (IITC Life Science Instruments, Woodland Hills, CA, USA). The rats were placed in a clear plastic cage (22cm×12cm×22cm) with a wire mesh floor for 30 min prior to testing. The detector with a tip was approached below to the plantar surface of the rats. Lifting or licking the paw was regarded as a positive response and data were written down and averaged as MWT. To assess thermal sensitivity, thermal withdrawal latency (TWL) was measured with the Thermal Paw Stimulation System (IITC Life Science Instruments, Woodland Hills, CA, USA). The rats were acclimatized in transparent plastic enclosures on the surface of an elevated glass platform for 30 minutes. The heat stimulus was focused on the left plantar hind paw. TWL referred to the reaction time for the stimulated animal to lift or lick its paw, with a cut-off time of 20 s to prevent injury. Average of TWL for five trials with an interval of 5 min was calculated.
Sample Preparation
On the 15th day after behavioral tests, the rats in each group were anesthetized by intraperitoneal injection of chloral hydrate (300 mg/kg). The ipsilateral L4-L6 spinal cord was isolated and stored at −80°C. To extract protein of the samples, each tissue sample was added with lysis buffer (8 M urea, 2 mM EDTA, 10 mM DTT, 1% protease inhibitor cocktail) and homogenized adequately with a tissue grinder to collect the supernatant. Afterwards the supernatant was precipitated with cold acetone and redissolved by urea buffer (8 M urea, 100 mM TEAB). The protein concentration was determined by the Modified Bradford Protein Assay Kit (Sangon Biotech, Shanghai, China). The rats were killed by cervical dislocation.
Liquid Chromatography with Tandem Mass Spectrometry (LC-MS-MS) Analysis
Protein mixture was digested and labeled by iTRAQ before HPLC fractionation. One hundred micrograms of protein sample were treated with 10 mM DTT for 60 min at 37°C, alkylated with 55 mM iodoacetamide for 30 min under room temperature, diluted with 100 mM TEAB, and then digested by trypsin with the mass ratio of 1:50 at 37°C overnight. Each sample was labeled with the iTRAQ (AB Sciex, Waltham, MA, USA) reagent as follows: the sham group: 116 tags, the CCI group: 117 tags, and the emodin group: 118 tags.
The labeled peptides were then fractioned using high pH reverse-phase high performance liquid chromatography (HPLC; Waters, Milford, MA, USA) with Waters XBridge Peptide BEH C18 (Waters, Milford, MA, USA). Mobile phase A was 2% acetonitrile (ACN) and mobile phase B was 98% ACN. In short, the peptides were separated into 60 fractions with a gradient of 2% to 98% ACN in pH 10 at a speed of 0.5 mL/min. Then the peptides were combined into 12 fractions, dried by vacuum centrifugation, and desalted by Ziptip C18 (Merck Millipore, Billerica, Massachusetts, USA). Finally, the samples were dried under vacuum and kept at −20°C.
For nano LC-MS/MS, 2 mL of each fraction was used. The peptide mixture was loaded onto C18-reversed phase column (Thermo Fisher Scientific, Waltham, MA, USA) in buffer A of 2% ACN and 0.1% formic acid (FA) and separated with a 90 min linear gradient in buffer B of 98% ACN and 0.1% FA at a flow rate of 300 nl/min. The eluent was automatically processed by Orbitrap Elite Mass Spectrometry (Thermo Fisher Scientific, Waltham, MA, USA).
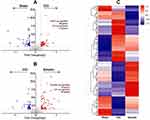
The raw MS/MS data were searched against the Rattus norvegicus protein database downloaded from Uniprot database using Sequest software integration in Proteome Discoverer 1.3 (Thermo Fisher Scientific, Waltham, MA, USA) to identify proteins. The following options were used: peptide mass tolerance of 20 ppm, MS/MS tolerance of 0.1 Da, and the false discovery rate (FDR) of ≤ 0.01. The differentially expressed proteins were selected with the screen criteria of fold change (FC) ≥1.2 and p<0.05. We drew a volcano plot and heat maps with R ggplot2 to display the differentially expressed proteins among the sham group, CCI group, and emodin group.
Bioinformatics Analysis
Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses of the differentially expressed proteins were performed using R package clusterProfiler.14 The most significantly enriched GO terms and KEGG pathways were screened and displayed by P-value.
Real-Time Quantitative Fluorescence PCR (qRT-PCR)
qRT-PCR was applied to examine the gene expression of phospholipase Cβ1 (PLCβ1), protein kinase C (PKC), protein kinase C (PKA), calmodulin (CaM) dependent protein kinase II α (CaMK II α), CaM dependent protein kinase II δ (CaMK II δ), neurotrophic tyrosine kinase receptor type 2 (NTRK2). Based on the manufacturer’s instructions, total RNA was extracted with the TRIzol reagent (Ambion, Texas, USA) and converted to cDNA by the HiScript Reverse Transcriptase (Vazyme, Nanjing, China). Then qRT-PCR was performed under standard conditions. GAPDH was used as internal standard. Primer sequences were listed in Table 1.
 |
Table 1 Sequences of the Primers |
Western Blot Analysis
Western blot analysis was used to examine the protein expression of PLCβ1, PKC, PKA, CaMK II α, CaMK II δ, and tropomyosin-related kinase B (TrkB). Protein was extracted using RIPA lysis buffer from the samples of spinal cord. BCA Protein Quantitation Kit (Beyotime, Beijing, China) was used to detect protein concentrations. The boiled protein supernatant mixed with the sample buffer was subjected to 12% SDS-PAGE electrophoresis and then transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Massachusetts, USA). The rabbit polyclonal anti-PKA (1:1000, Proteintech, Wuhan, China), rabbit polyclonal anti-PKC (1:1000, Proteintech, Wuhan, China), rabbit polyclonal anti-PLCβ1 (1:1000, ABclonal, Wuhan, China), rabbit polyclonal anti-PKA, rabbit polyclonal anti-CaMK II α (1:1000, Cell Signaling Technology, Beverly, MA, USA), rabbit polyclonal anti-CaMK II δ (1:1000, Proteintech, Wuhan, China), rabbit polyclonal anti-TrkB (1:1000, Proteintech, Wuhan, China), and polyclonal anti-GAPDH (1:1000, Hangzhou Xianzhi Biotechnology Co., Hangzhou, China) were applied as primary antibodies and incubated at 4°C overnight. The secondary antibodies, goat anti-rabbit (1:50,000, Boster Biological Technology, China), were incubated for 1 h at room temperature. The PVDF membranes were washed with TBST five times with a 5-min interval, visualized with enhanced chemiluminescent (ECL) reagent (Thermo Fisher Scientific, Waltham, USA), and exposed to X-ray film.
Statistical Analyses
The statistical differences between multiple groups were ascertained using one-way ANOVA test and the post hoc test was analyzed with Bonferroni test. P<0.05 was considered to indicate a statistically significant difference.
Results
Emodin Reversed Alteration of MWT and TWL in CCI Model
The MWT and TWL values were decreased significantly in the CCI group and emodin group compared to the sham group from the 3rd day after operation (P<0.05), suggesting that CCI model was successfully established (Figure 1A and B). However, the MWT and TWL values in the emodin group gradually elevated from the 7th day after operation, the MWT and TWL values in the emodin group were significantly higher than those in the CCI group (P<0.05), which indicated that emodin treatment significantly reversed the changes of MWT and TWL elicited by CCI (Figure 1A and B).
Identification of Differentially Expressed Proteins Associated with Emodin Treatment in NP
Proteomics was used to quantitatively analyze the expression of differentially expressed proteins in the spinal cord samples between the sham group, CCI group, and emodin group. Using a threshold of 1.2-fold change and a p-value of ≤ 0.05, a total of 177 differentially expressed proteins were identified (Figure 2A). Among them, 100 differentially expressed proteins (51 up-regulated and 49 down-regulated) were identified in the CCI group compared with sham group. Moreover, 108 differentially expressed proteins (65 up-regulated and 43 down-regulated) were identified in the emodin group with the CCI group as reference (Figure 2B). The cluster heat map for differentially expressed proteins was displayed in Figure 2C.
Bioinformatics Analysis for Differentially Expressed Proteins
In order to better understand the biological relevance of the differentially expressed proteins, we performed GO enrichment analysis (Figure 3A). The top five enriched biological functions of the differentially expressed proteins between the CCI and emodin groups involved regulation of endopeptidase activity, regulation of peptidase activity, regulation of neurotransmitter transport, regulation of neurotransmitter secretion, and myelination (Figure 3C). The top five enriched molecular functions of the differentially expressed proteins between the CCI and emodin groups involved structural constituent of myelin sheath, endopeptidase inhibitor activity, enzyme inhibitor activity, endopeptidase regulator activity, and peptidase inhibitor activity (Figure 3B).
KEGG pathway enrichment analysis of the differentially expressed proteins between the CCI and emodin groups was also carried out. The differentially expressed proteins participated in several pain-related pathways, including long-term potentiation (LTP), calcium signaling pathway, endocrine and other factor-regulated calcium reabsorption and cholinergic synapse (Figure 3D). Protein-protein interaction (PPI) networks were used to analyze the relationships of the differentially expressed proteins between the CCI and emodin groups (Figure 3E).
qRT-PCR and Western Blot Analysis Confirmed Results of Proteomics Analysis
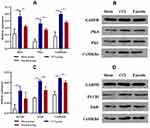
Calcium signaling pathway is the key signaling for initiating and maintaining activity-dependent central sensitization in NP.15 Based on the results of GO and KEGG enrichment analysis, several differentially expressed proteins between the CCI and emodin groups were associated with calcium signaling pathway. To ascertain the applicability of the the results of iTRAQ proteomics screening, we detected the gene and protein expression of PLCβ1, PKC, PKA, CaMK II α, CaMK II δ, and NTRK2 (TrkB) in the sham group, CCI group, and emodin group using qRT-PCR and Western blot. The gene and the corresponding protein expression of PLCβ1, PKC, PKA, CaMK II α, CaMK II δ, and NTRK2 (TrkB) in ipsilateral L4-L6 spinal cord of the CCI rats were significantly regulated compared with the sham rats (P<0.01); while the expression levels of PKC, PKA, CaMK II α, CaMK II δ, and NTRK2 (TrkB) were down-regulated in emodin rats compared with the CCI rats (P<0.05) (Figures 4 and 5). The PLCβ1 gene level in the emodin group also decreased compared with that in the CCI group (P<0.05), but the decreasing trend of PLCβ1 protein level was not statistically significant (P=0.12) (Figures 4 and 5). These results suggested that emodin could downregulate the expression of PLCβ1, PKC, PKA, CaMK II α, CaMK II δ, and NTRK2 (TrkB) in the CCI rats, which was consistent with the results of iTRAQ proteomics screening.
 |
Figure 4 The gene expression of PLCβ1, PKC, PKA, CaMK II α, CaMK II δ, and NTRK2 in spinal cord samples in the sham group, CCI group, and emodin group using qRT-PCR. *P<0.05 and **P<0.01. |
Discussion
Both in vitro and vivo studies revealed the analgesic effect of emodin in NP. However, the molecular mechanism of spinal cord underlying the analgesic effect of emodin has been scarcely investigated. In this study, the iTRAQ based proteomics technique was used to identify the differentially expressed proteins and explore the potential pathways involved in the effects of emodin.
In our study, 177 differentially expressed proteins were identified in the ipsilateral L4-L6 spinal cord samples among the sham group, CCI group, and emodin group. Among them, 100 differentially expressed proteins (51 up-regulated and 49 down-regulated) were identified in the CCI group compared with sham group and 108 differentially expressed proteins (65 up-regulated and 43 down-regulated) were identified in the emodin group with the CCI group as reference. GO and KEGG enrichment analysis showed that the differentially expressed proteins in the CCI group and emodin group were enriched in several pain-related biological processes and signaling pathways, including neurotransmitter regulation, LTP, and calcium signaling pathway.
Calcium signaling pathway is the key signaling for initiating and maintaining activity-dependent central sensitization in NP.15 In this study, we found that calcium signaling pathway could be the potential target of emodin. The association between calcium signaling pathway and emodin has been studied in acute pancreatitis and hepatic steatosis.16,17 However, the association between calcium signaling pathway and emodin in NP has not been reported. In the dorsal horn of the spinal cord, noxious stimuli trigger an increase in intracellular Ca2+ by calcium influx via voltage-gated calcium channels, N-methyl-D-aspartate (NMDA) receptors, and α-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors as well as release from intracellular stores in response to activation of several metabotropic receptors.15 And intracellular Ca2+ further activates downstream signaling molecules, such as calcium/calmodulin (CaM) dependent protein kinase Ⅱ (CaMKⅡ) α and protein kinase C (PKC). CaMKⅡ is a Ca2+/CaM-dependent multifunctional Ser/Thr protein kinase, which can be activated by binding of Ca2+/CaM. CaMKIIa further activates phosphorylated AMPA receptors, enhances excitatory postsynaptic potential, and triggers central sensitization and neuropathic pain.18,19 Several studies have confirmed that inhibition of CaMKII activity eliminated long-term potentiation in spinal cord and alleviated hyperalgesia induced by peripheral tissue and nerve injury.20–22 Conventional PKC are members of phospholipid-dependent serine/threonine kinase, which could regulate cellular events following activation of membrane bound phospholipids and Ca2+.23 Activation of PKC promotes hyperexcitability in nociceptive neurons by facilitating the activated state of NMDA receptors, decreasing inhibitory transmission of gamma-aminobutyric acid (GABA) and glycine, and participating in ERK activation.18 This study showed that emodin significantly decreased the level of total CaMKII α and PKC in the dorsal horn of the spinal cord of the CCI model. We speculated that emodin might alleviate NP by inhibiting the activity of CaMKII α and PKC, which play critical roles in central sensitization and NP.
In addition, we also found that the analgesic effects of emodin might be associated with tropomyosin-related kinase B (TRKB), which is known as neurotrophic tyrosine kinase receptor type 2 (NTRK2). Recently, evidence has emerged of an essential role of TrkB and its ligand brain-derived neurotrophic factor (BDNF) in central sensitization and NP.24,25 BDNF-TrkB pathway induces NMDA receptor activation and downregulation of potassium chloride cotransporter (KCC2), thereby contributing to NP.26,27 And inhibition of the BDNF-TrkB pathway is a promising strategy for pain therapy.28,29 In this study, we confirmed decreased TrkB expression in emodin-treated group and speculated that emodin alleviated NP by the inhibition of TrkB activity.
In conclusion, our study was the first systematic proteomic analysis of NP which highlighted the relationship between calcium signaling pathway and emodin in NP, and further studies could aim at identifying new calcium signaling molecules in emodin treatment. These findings provide insight to discover novel agents targeting calcium signaling pathway for NP therapy.
Data Sharing Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.
Ethics Approval
All experimental protocols were approved by Animal Use and Ethics Committee of the First Affiliated Hospital of Guangzhou University of Chinese Medicine.
Funding
The present study was supported in part by a grant from National Natural Science Fund (nos.81373571).
Disclosure
The authors declare that they have no competing interests.
References
1. Jensen TS, Baron R, Haanpaa M, et al. A new definition of neuropathic pain. Pain. 2011;152(10):2204–2205. doi:10.1016/j.pain.2011.06.017
2. van Hecke O, Austin SK, Khan RA, Smith BH, Torrance N. Neuropathic pain in the general population: a systematic review of epidemiological studies. Pain. 2014;155(4):654–662. doi:10.1016/j.pain.2013.11.013
3. Forouzanfar F, Sadeghnia HR. Fibroblast growth factors as tools in the management of neuropathic pain disorders. Curr Drug Targets. 2020;21(10):1034–1043. doi:10.2174/1389450121666200423084205
4. Bannister K, Sachau J, Baron R, Dickenson AH. Neuropathic pain: mechanism-based therapeutics. Annu Rev Pharmacol Toxicol. 2020;60:257–274. doi:10.1146/annurev-pharmtox-010818-021524
5. Finnerup NB, Attal N, Haroutounian S, et al. Pharmacotherapy for neuropathic pain in adults: a systematic review and meta-analysis. Lancet Neurol. 2015;14(2):162–173. doi:10.1016/S1474-4422(14)70251-0
6. Kamerman PR, Wadley AL, Davis KD, et al. World Health Organization essential medicines lists: where are the drugs to treat neuropathic pain? Pain. 2015;156(5):793–797. doi:10.1097/01.j.pain.0000460356.94374.a1
7. Sommer C. Peripheral neuropathies: new recommendations for neuropathic pain pharmacotherapy. Nat Rev Neurol. 2015;11(5):250–252. doi:10.1038/nrneurol.2015.36
8. Wang L, Liu S, Xu J, et al. Emodin inhibits aggregation of amyloid-beta peptide 1-42 and improves cognitive deficits in Alzheimer’s disease transgenic mice. J Neurochem. 2020. doi:10.1111/jnc.15156
9. Leung SW, Lai JH, Wu JC, et al. Neuroprotective effects of emodin against ischemia/reperfusion injury through activating ERK-1/2 signaling pathway. Int J Mol Sci. 2020;21(8):2899. doi:10.3390/ijms21082899
10. Wang C, Chen P, Lin XD, Wu ZB. The effect of different dosages of emodin on the behavior of CCI rat with neuropathic pain. Guangdong Med J. 2017;38(23):3549–3553.
11. Gao Y, Liu H, Deng L, et al. Effect of emodin on neuropathic pain transmission mediated by P2X2/3 receptor of primary sensory neurons. Brain Res Bull. 2011;84(6):406–413. doi:10.1016/j.brainresbull.2011.01.017
12. Xiong W, Wu RP, Tan MX, et al. Emodin inhibits the expression of receptor and calcitonin-gene-related peptide release in trigeminal ganglia of trigeminal neuralgia rats. Int J Clin Exp Pathol. 2017;10(11):11317–11325.
13. Bennett GJ, Xie YK. A peripheral mononeuropathy in rat that produces disorders of pain sensation like those seen in man. Pain. 1988;33(1):87–107. doi:10.1016/0304-3959(88)90209-6
14. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16(5):284–287. doi:10.1089/omi.2011.0118
15. Liu W, Lv Y, Ren F. PI3K/Akt pathway is required for spinal central sensitization in neuropathic pain. Cell Mol Neurobiol. 2018;38(3):747–755. doi:10.1007/s10571-017-0541-x
16. Wang GJ, Wang Y, Teng YS, et al. Protective effects of emodin-induced neutrophil apoptosis via the Ca(2+)-caspase 12 pathway against SIRS in rats with severe acute pancreatitis. Biomed Res Int. 2016;2016:1736024. doi:10.1155/2016/1736024
17. Wang S, Li X, Guo H, et al. Emodin alleviates hepatic steatosis by inhibiting sterol regulatory element binding protein 1 activity by way of the calcium/calmodulin-dependent kinase kinase-AMP-activated protein kinase-mechanistic target of rapamycin-p70 ribosomal S6 kinase signaling pathway. Hepatol Res. 2017;47(7):683–701. doi:10.1111/hepr.12788
18. Latremoliere A, Woolf CJ. Central sensitization: a generator of pain hypersensitivity by central neural plasticity. J Pain. 2009;10(9):895–926. doi:10.1016/j.jpain.2009.06.012
19. Incontro S, Diaz-Alonso J, Iafrati J, et al. The CaMKII/NMDA receptor complex controls hippocampal synaptic transmission by kinase-dependent and independent mechanisms. Nat Commun. 2018;9(1):2069. doi:10.1038/s41467-018-04439-7
20. Yang HW, Hu XD, Zhang HM, et al. Roles of CaMKII, PKA, and PKC in the induction and maintenance of LTP of C-fiber-evoked field potentials in rat spinal dorsal horn. J Neurophysiol. 2004;91(3):1122–1133. doi:10.1152/jn.00735.2003
21. Chen Y, Luo F, Yang C, Kirkmire CM, Wang ZJ. Acute inhibition of Ca2+/calmodulin-dependent protein kinase II reverses experimental neuropathic pain in mice. J Pharmacol Exp Ther. 2009;330(2):650–659. doi:10.1124/jpet.109.152165
22. Luo F, Yang C, Chen Y, et al. Reversal of chronic inflammatory pain by acute inhibition of Ca2+/calmodulin-dependent protein kinase II. J Pharmacol Exp Ther. 2008;325(1):267–275. doi:10.1124/jpet.107.132167
23. Kaleli HN, Ozer E, Kaya VO, Kutlu O. Protein kinase C isozymes and autophagy during neurodegenerative disease progression. Cells-Basel. 2020;9(3). doi:10.3390/cells9030553
24. Inoue K, Tsuda M. Microglia in neuropathic pain: cellular and molecular mechanisms and therapeutic potential. Nat Rev Neurosci. 2018;19(3):138–152. doi:10.1038/nrn.2018.2
25. Tsuda M. Microglia in the CNS and neuropathic pain. Adv Exp Med Biol. 2018;1099:77–91. doi:10.1038/nrn.2018.2
26. Khan N, Smith MT. Neurotrophins and neuropathic pain: role in pathobiology. Molecules. 2015;20(6):10657–10688. doi:10.3390/molecules200610657
27. Fernandes V, Sharma D, Vaidya S, et al. Cellular and molecular mechanisms driving neuropathic pain: recent advancements and challenges. Expert Opin Ther Targets. 2018;22(2):131–142. doi:10.1080/14728222.2018.1420781
28. Ikeda K, Hazama K, Itano Y, Ouchida M, Nakatsuka H. Development of a novel analgesic for neuropathic pain targeting brain-derived neurotrophic factor. Biochem Biophys Res Commun. 2020;531(3):390–395. doi:10.1016/j.bbrc.2020.07.109
29. Vanelderen P, Rouwette T, Kozicz T, et al. The role of brain-derived neurotrophic factor in different animal models of neuropathic pain. Eur J Pain. 2010;14(5):
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.