Back to Journals » OncoTargets and Therapy » Volume 11
Promoter methylation analysis of CDH1 and p14ARF genes in patients with urothelial bladder cancer
Authors Bayramov B, Gunes S , Buyukalpelli R, Aydın O, Henkel R
Received 28 November 2017
Accepted for publication 1 April 2018
Published 19 July 2018 Volume 2018:11 Pages 4189—4196
DOI https://doi.org/10.2147/OTT.S158259
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jianmin Xu
Bayram Bayramov,1 Sezgin Gunes,1,2 Recep Buyukalpelli,2,3 Oğuz Aydın,4 Ralf Henkel5
1Medical Biology, Faculty of Medicine, Ondokuz Mayis University, Samsun, Turkey; 2Multidisciplinary Molecular Medicine, Health Sciences Institute, Ondokuz Mayis University, Samsun, Turkey; 3Urology, Faculty of Medicine, Ondokuz Mayis University, Samsun; 4Pathology, Faculty of Medicine, Ondokuz Mayis University, Samsun, Turkey; 5Department of Medical Bioscience, University of the Western Cape, Bellville, South Africa
Background/aim: Urothelial bladder cancer arises from the accumulation of multiple epigenetic and genetic changes. We aimed to investigate the specificity and sensitivity of gene-specific promoter methylation of CDH1 and p14ARF genes in the early diagnosis of bladder cancer and compare those with other diagnostic tests in our population.
Patients and methods: In the current study, 65 patients with urothelial bladder cancer and 35 controls without any history of cancer were recruited. Methylation profiles of CDH1 and p14ARF genes from tumor and urine samples were determined by methylation-specific polymerase chain reaction method.
Results: Methylation of CDH1 and p14ARF genes in tumor samples was 95.4% and 78.5%, respectively. The methylation frequencies were found to be 68.8% for CDH1 gene and 72.9% for p14ARF gene in urine samples. Sensitivities of CDH1, p14ARF and urine cytology were found to be 67.4%, 72.1% and 34.9%, respectively, while their specificities were 93.9%, 63.6% and 93.9%, respectively.
Conclusion: Aberrant promoter methylation of CDH1 and p14ARF genes can be used to detect urothelial bladder cancer. In low-grade tumors, when compared with urine cytology, combined methylation analysis of CDH1 and p14ARF genes may not increase the sensitivity to identify malignant cells in urine samples.
Keywords: biomarker, CDH1, urine cytology, urothelial bladder cancer, p14ARF, urine
Introduction
Urothelial bladder cancer (UBC) is the most prevalent type of urinary tract malignancy, particularly in men.1,2 Numerous genetic and epigenetic factors play a significant role in progression, recurrence and metastasis of UBC.3 Certain genetic factors such as activation of proto-oncogenes (EGFR, FGFR, HER/neu [c-erb-B2], c-myc, MDM2 and others), inactivation of tumor suppressor genes (p53 mutation, Rb homozygous deletion and others) and alteration of cell cycle regulators (p21, p27, Ki-67, cyclin D1, cyclin E and others) and cell adhesion molecules (MMP-2, E-cadherin, etc) have been observed to be related to bladder cancer.4–7 Aberrant promoter methylation of tumor suppressor genes might change normal cellular growth properties by causing decrease in gene expression. Methylation occurs in the early stages of carcinogenesis and can be determined in body fluids, indicating that a noninvasive and early cancer detection method can be developed. Bladder cancer studies of hypermethylation in urine DNA samples are in progress from day to day and refer to the potential of epigenetics in cancer diagnosis.8
Cadherin 1 (CDH1, E-cadherin, epithelial cadherin) is a Ca2+-dependent transmembrane glycoprotein that mediates cell–cell adhesion and is highly expressed in normal epithelial tissues.9,10 Cadherins (CDHs) make cell–cell connection with extracellular, transmembrane and intracellular domains.11 Loss of CDH1 expression is one of the characteristics of epithelial–mesenchymal transition (EMT) frequently observed in carcinogenesis. Somatic mutations, transcriptional repressors (SNAI1 and SNAI2, TWIST, ZEB1/ZEB2), loss of heterozygosity and promoter hypermethylation of CDH1 are involved in transcriptional repression and reduced CDH1 expression.12
The INK4a/ARF locus is localized on chromosome 9p21 and encodes tumor suppressor proteins such as p16INK4a and p14ARF, which are the negative regulators of the cell cycle via 16INK4a-Rb and p14ARF-p53 pathways.13–16 p14ARF proteins prevent abnormal cell growth and division in response to oncogene (Ras, c-myc, E1A and E2F1) activation. Overexpression of E2F1, radiations, genotoxic drugs and DNA damage induce the expression of p14ARF and prevent the aberrant cell cycle progression.17,18 Activation of p14ARF inhibits E3 ubiquitin–protein ligase, mouse double minute 2 (MDM2) protein, and prevents degradation of p53 by ligase activity of MDM2. Activated p53 arrests abnormal cell cycle at G1 and G2 phases.17,19,20
The current study aimed to investigate and validate the methylation patterns of CDH1 and p14ARF genes in urothelial bladder carcinoma tissues and voided urine samples as epigenetic diagnostic biomarkers and compared those with other diagnostic methods including urine cytology. In addition, we investigated whether aberrant methylation of CDH1 and p14ARF genes can replace urine cytology in bladder cancer.
Patients and methods
Study subjects
The study protocol was approved by the institutional review board of the Ondokuz Mayis University (OMU), and all participants signed an informed consent form stating their full consent and their own free will to participate in the study after receiving detailed information about the study. A total of 65 histologically confirmed UBC patients diagnosed at the urology clinic of OMU and 35 healthy control individuals were enrolled in this study. All patients were investigated by standard urological evaluation including urine cytology and flexible cystoscopy. Tumors were examined and classified according to the World Health Organization/International Society of Urological Pathology (WHO/ISUP) (2010) and were staged according to TNM (2010). Paired preoperative voided urine samples were collected from 48 patients. The exclusion criteria for controls included urinary infections, a history of cancer and benign conditions. A total of 35 healthy voided urine samples were selected from gender- and age-matched (±5) volunteers. The healthy control individuals with suspicious results for urine cytology were examined in the OMU Urology Clinic, and inflammatory, benign or malignant conditions were not observed.
DNA isolation
The tumor samples were immediately frozen and stored at −80°C until the DNA isolation procedure was performed. Fresh urine samples were used for DNA extraction. DNA isolation from tumor and urine samples was performed with ZR Genomic DNA™ Tissue MiniPrep Kits (Zymo Research, Irvine, CA, USA) and ZR Urine DNA Isolation KitTM (Zymo Research), respectively, according to the manufacturer’s instructions. DNA samples were stored at −20°C until bisulfite modification.
Bisulfite modification and methylation-specific polymerase chain reaction (MS-PCR)
Bisulfite modification of extracted DNA samples was carried out by EZ DNA Methylation™ Kit (Zymo Research). Approximately 200 ng DNA/20 μL and 500 ng DNA/20 μL were used for bisulphite conversion of urine and tumor samples, respectively.
MS-PCR was run for the promoter regions of both CDH1 and p14ARF genes using methylated and unmethylated primer pairs. The reaction was carried out in a final volume of 50 μL containing 2 μL of bisulfite-treated DNA, 0.25 mM of each deoxynucleotide triphosphate (dNTP) (Zymo Research), 0.5 μM of each of the primers and 2 U of Taq polymerase (Zymo Research). After initial denaturation at 95°C for 10 minutes, the subsequent steps of denaturation at 95°C for 30 seconds, annealing at 57°C (CDH1, for both methylated [M] and unmethylated [U]) and 60°C (p14ARF, for both M and U) for 30 seconds, and extension at 72°C for 60 seconds were repeated for 40 cycles.21,22 Half of the PCR products were electrophoresed on a 2.5% agarose gel. Methylated and unmethylated products of CDH1 were 116 bp and 97 bp, respectively, while methylated and unmethylated products of p14ARF were 122 bp and 132 bp, respectively. The gel images of methylation status of the CDH1 and p14ARF promoters in tumor and urine samples are shown in Figure 1. Universal methylated human DNA standard (in vitro-methylated DNA [IVD]; Zymo Research) was used as a positive control for methylation. Fifty base pair DNA ladder (New England BioLabs, Ipswich, MA, USA) was used as a marker.
The gene methylation status was indicated as methylated when amplification products were detected in the reactions performed with primers M or both M and U.
Statistical analyses
For statistical analyses, MedCalc Statistical Software, version 17.2 (MedCalc Software bvba, Ostend, Belgium) was used. After testing for normal data distribution using the Kolmogorov–Smirnov test, the nonparametric Spearman’s rank correlation and Mann–Whitney U-test were applied as needed. k test was performed to compare the methylation status of CDH1 and p14ARF genes in tumor and paired urine samples. P-value of <0.05 was considered significant.
Results
In the current study, we analyzed the methylation status of CDH1 and p14ARF promoters by MS-PCR in 65 bladder tumor samples, 48 paired urine samples and 35 urine samples of healthy individuals. The clinical and pathological data of patients and controls are summarized in Table 1.
  | Table 1 Clinicopathological characteristics of UBC patients and controls |
Frequency of methylation in tumor
Methylation analyses of CDH1 and p14ARF genes were performed on 65 tumor samples from bladder cancer patients. Overall, 95.4% (62/65) and 78.5% (51/65) of tumor samples had methylation in CDH1 and p14ARF promoter, respectively (Table 2).
  | Table 2 Promoter methylation frequencies of CDH1 and p14ARF genes in the patient and control groups |
Frequency of methylation in urine samples
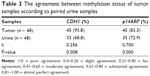
Methylation pattern of CDH1 and p14ARF genes in paired preoperative voided urine samples was analyzed in 48 patients and 35 controls. The methylation frequencies of urine samples of patients were 68.8% (33/48) for CDH1 and 72.9% (35/48) for p14ARF gene. Moreover, methylation frequencies for CDH1 and p14ARF genes were 5.7% (2/35) and 37.1% (13/35; Table 2), respectively, in urine samples of controls without cancer history. The paired urine samples of the patient and control groups have shown a significant difference (P < 0.001) regarding the promoter methylation profile of CDH1 and p14ARF genes. k test was performed to compare the promoter methylation status of CDH1 and p14ARF in 48 tumor and paired urine samples. The results of k test demonstrated the methylation status of two genes in both samples showed reliability (Table 3).
The methylation pattern of paired urine samples was identical to tumor samples in almost in all cases; however, the methylation pattern of tumor samples was not identical in a few urine samples. The agarose gel images of the methylation analysis of CDH1 and p14ARF genes are shown in Figure 1.
We evaluated the positive predictive value (PPV) and negative predictive value (NPV) for CDH1 and p14ARF genes. The PPV and NPV for CDH1 methylation analysis were 96.9% and 91.7%, respectively, with 95% accuracy. These values were lower for p14ARF gene, 79.7% and 61.1%, respectively, with 73% accuracy (Table 4).
Matching of MS-PCR and urine cytology results
A urine cytology test was performed in urine samples of 48 patients and 35 healthy individuals. However, five of the 48 patients voided urine samples and two of the 35 urine samples of the controls could not be diagnosed cytologically. Urine cytology and MS-PCR results were compared in 43 patient samples with tests being positive in 34.9% (15/43) and negative in 23.3% (10/43) of urine samples. The remaining patients’ urine samples were undiagnosed. Urine cytology test results of low-grade bladder tumors demonstrated 41.7% (5/12) suspicious results, and there was no positive result for 12 low-grade patient samples. In low-grade tumor specimens, the sensitivity of the test was 66.7% (8/12) for CDH1 gene and 75% (9/12) for p14ARF gene with the methylation of one of the two genes being 83.3% (10/12) of the samples. In addition, the results of the urine cytology test of the control group were suspicious and negative in 6.1% (2/33) and 93.9% (31/33) of the samples, respectively.
Sensitivity and specificity of urine cytology and MS-PCR results for CDH1 and p14ARF genes are summarized in Table 5. The test was significant for CDH1 gene with a sensitivity of 67.4% and a specificity of 93.9% (P < 0.0001). In addition, sensitivity and specificity for p14ARF were calculated to be 72.1% and 63.6%, respectively. On the other hand, the sensitivity and specificity in the control group for CDH1 were 34.9% (15/33) and 93.9% (31/33), respectively.
The PPV and NPV for the urine cytology test were 100% and 54.1%, respectively (Table 4). Our results have demonstrated a significant relation between the smoking status in patients and controls with regard to the methylation of CDH1 (P = 0.01) and p14ARF (P = 0.0015). In addition, we have found a significant positive association between age and promoter methylation of CDH1 and p14ARF genes (P < 0.05).
We have not observed any correlation among tumor grade, stage, recurrence, gender factor, intravesical therapy and routine second transurethral resection (Re-TURB) with methylation of CDH1 and p14ARF genes (Table 6).
  | Table 6 Correlations among the methylation status of CDH1 and p14ARF genes and tumor stage, recurrence, intravesical therapy and Re-TURB |
Discussion
UBCs are the mixture of heterogeneous cell populations; therefore, tumors with similar pathological characteristics may behave differently.2 Numerous diagnostic methods are used to diagnose and follow up the recurrence and progression of the UBC.5 Flexible cystoscopy is the current standard for the early detection of UBC; however, this method is expensive and invasive under the local anesthesia in bladder cancer patients for histopathological diagnosis.23,24 Urine cytology is a useful noninvasive diagnostic test in the detection of high-grade tumors in urine samples with high specificity, whereas the sensitivity is low in low-grade bladder tumors.5,25 These limitations of the currently available diagnostic techniques have increased the focus on identifying other clinically useful, reliable and noninvasive early diagnostic markers with high specificity for the diagnosis of bladder cancer.
We have found a significant positive association between urothelial bladder carcinoma and methylation of CDH1 and p14ARF genes. The methylation level in our study has been found higher than in previous studies using the same methods.9,22,26 However, the sample size of our study was larger than those of previous studies. Lin et al22 studied the methylation status of four genes (CDH1, p14, CDKN2A and RASSF1A) in 57 bladder tumor specimens and associated preoperative urine samples with the urine samples of 20 healthy control individuals using MS-PCR. Hypermethylation of CDH1, p14 and RASSF1A genes has been suggested as a valuable diagnostic biomarker to urine cytology for low-grade bladder cancer.
Lin et al22 calculated the overall sensitivity of E-cadherin and p14ARF genes to be 35% and 33%, respectively, in detecting bladder carcinoma. In contrast, these researchers found 7.7% positive results for low-grade tumors by urine cytology. In this study, the sensitivity of CDH1 and p14ARF genes separately and combined was higher than the sensitivity of urine cytology in urine samples. In addition, the specificity of urine cytology was the same as the specificity of CDH1 methylation, but higher than the specificity of p14ARF methylation in urine samples and slightly lower than the combined specificity based on two genes. Therefore, our results indicate that combined methylation analysis of these genes may be a useful marker compared with the urine cytology to increase the sensitivity for detecting bladder cancer.
Although the specificity of MS-PCR analysis of urine samples of patients was significantly higher than the results of urine cytology, methylation could not be detected in paired urine samples of six patients despite positive methylation in their tumor samples. This situation can be explained with the low number of malignant cells in urine samples from tumor tissues or a low level of methylated alleles in cancer cells within the urine samples, which could therefore not be enough to be detected by MS-PCR.27 Indeed, methylation results of tumor and urine samples demonstrated a fair and substantial agreement for CDH1 and p14ARF, respectively. Hoque et al28 investigated multiple tumor suppressor genes (APC, ARF, CDH1, GSTP1, MGMT, CDKN2A, RARβ2, RASSF1A and TIMP3) as potential biomarkers for the diagnosis of bladder cancer. Similarly, they observed promoter methylation in tumor samples but not in all paired urine samples.
Numerous studies demonstrated a correlation between promoter methylation of certain genes with tumor grade, stage, recurrence, intravesical therapy and Re-TURB.8,29,30 The methylation of 10 tumor suppressor genes in bladder cancer patients (N = 98) was examined. Methylation of CDH1 was found to be significantly associated with poor survival (P = 0.003) and an independent predictor of survival in a multivariate analysis (P = 0.02).31 In a recent study, Xiong et al32 conducted a large trial involving 687 patients with bladder cancer using MS-PCR. The promoter methylation level of CDH1 was found to be significantly associated with a higher tumor stage compared with lower tumor stage. However, there was no association between the aberrant methylation of the CDH1 and p14ARF genes and tumor stage, grade and recurrence in the current study. Interestingly, several studies showed similar findings in the different types of malignancies.33–35
In our study, the control group consisted of age- and gender-matched individuals, and a significant relationship was observed among the increased methylation of CDH1 and p14ARF genes, aging and malignity. Similarly, Yates et al36 reported an increase in the frequency and extent of methylation with age and malignity in bladder cancer patients. Nevertheless, two studies demonstrated an association between promoter methylation of CDH1 gene and aging independent of cancer.26,37 These findings might indicate that aging may be a risk factor for bladder cancer. On the other hand, Chan et al38 analyzed the methylation of RARβ, DAPK, E-cadherin, p16, p15, GSTP1 and MGMT genes in bladder transitional cell carcinoma and found no association between age and methylation status of these. Two studies observed no methylation in tumor suppressor gene panels including E-cadherin, p16, p14ARF, APC and RASSF1A in urine DNAs from normal healthy individuals.22,36 We have found a significant relation between smoking status of patients and controls regarding the methylation of CDH1 and p14ARF. Indeed, numerous studies indicate that smoking is a risk factor for bladder cancer in both men and women.39,40
Finally, it is important to note that our study is limited by the nonquantitative nature of the detection method of the methylation status of CDH1 and p14ARF genes as well as the sample size. These results must be verified in large cohorts using different approaches.
Conclusion
Our preliminary results have demonstrated that the combined methylation analysis of CDH1 and p14ARF genes as biomarkers may be a sensitive method to detect malignant cells in urine samples. However, larger cohorts and well-defined subgroups of bladder cancer patients are required to reveal the potential role of CDH1 and p14ARF genes as novel biomarkers.
Acknowledgment
This study was supported by OMU Research Foundation (project no PYO.TIP.1904.14.017).
Disclosure
The authors report no conflicts of interest in this work.
References
Ibragimova I, Dulaimi E, Slifker MJ, Chen DY, Uzzo RG, Cairns P. A global profile of gene promoter methylation in treatment-naive urothelial cancer. Epigenetics. 2014;9(5):760–773. | ||
Kim YJ, Kim WJ. Can we use methylation markers as diagnostic and prognostic indicators for bladder cancer? Investig Clin Urol. 2016;57(suppl 1):S77–S88. | ||
Liu L, Qiu M, Tan G, et al. miR-200c inhibits invasion, migration and proliferation of bladder cancer cells through down-regulation of BMI-1 and E2F3. J Transl Med. 2014;12:305. | ||
Kang Z, Li Y, Yu Y, Guo Z. Research progress on bladder cancer molecular genetics. J Cancer Res Ther. 2014;10(suppl):C89–C94. | ||
Kim YK, Kim WJ. Epigenetic markers as promising prognosticators for bladder cancer. Int J Urol. 2009;16(1):17–22. | ||
Knowles MA, Hurst CD. Molecular biology of bladder cancer: new insights into pathogenesis and clinical diversity. Nat Rev Cancer. 2015;15(1):25–41. | ||
Schulz WA, Goering W. DNA methylation in urothelial carcinoma. Epigenomics. 2016;8(10):1415–1428. | ||
Kandimalla R, van Tilborg AA, Zwarthoff EC. DNA methylation-based biomarkers in bladder cancer. Nat Rev Urol. 2013;10(6):327–335. | ||
Ribeiro-Filho LA, Franks J, Sasaki M, et al. CpG hypermethylation of promoter region and inactivation of E-cadherin gene in human bladder cancer. Mol Carcinog. 2002;34(4):187–198. | ||
Li G, Liu Y, Yin H, et al. E-cadherin gene promoter hypermethylation may contribute to the risk of bladder cancer among Asian populations. Gene. 2013;S0378–S1119(13)01422–01424. | ||
Rieger-Christ KM, Cain JW, Braasch JW, et al. Expression of classic cadherins type I in urothelial neoplastic progression. Hum Pathol. 2001;32(1):18–23. | ||
Schneider MR, Kolligs FT. E-cadherin’s role in development, tissue homeostasis and disease: insights from mouse models: tissue-specific inactivation of the adhesion protein E-cadherin in mice reveals its functions in health and disease. Bioessays. 2015;37(3):294–304. | ||
Dominguez G, Silva J, Garcia JM, et al. Prevalence of aberrant methylation of p14ARF over p16INK4a in some human primary tumors. Mutat Res. 2003;530(1–2):9–17. | ||
Lefrerebelda M. Profiles of the 2 INK4a gene products, p16 and p14ARF, in human reference urothelium and bladder carcinomas, according to pRb and p53 protein status*1. Hum Pathol. 2004;35(7):817–824. | ||
Kim WY, Sharpless NE. The regulation of INK4/ARF in cancer and aging. Cell. 2006;127(2):265–275. | ||
Sharpless NE. INK4a/ARF: a multifunctional tumor suppressor locus. Mutat Res. 2005;576(1–2):22–38. | ||
Ozenne P, Eymin B, Brambilla E, Gazzeri S. The ARF tumor suppressor: structure, functions and status in cancer. Int J Cancer. 2010;127(10):2239–2247. | ||
Gallagher SJ, Kefford RF, Rizos H. The ARF tumour suppressor. Int J Biochem Cell Biol. 2006;38(10):1637–1641. | ||
Kawamoto K, Enokida H, Gotanda T, et al. p16INK4a and p14ARF methylation as a potential biomarker for human bladder cancer. Biochem Biophys Res Commun. 2006;339(3):790–796. | ||
Sherr CJ, Weber JD. The ARF/p53 pathway. Curr Opin Genet Dev. 2000;10(1):94–99. | ||
Yegin Z, Gunes S, Buyukalpelli R. Hypermethylation of TWIST1 and NID2 in tumor tissues and voided urine in urinary bladder cancer patients. DNA Cell Biol. 2013;32(7):386–392. | ||
Lin HH, Ke HL, Huang SP, Wu WJ, Chen YK, Chang LL. Increase sensitivity in detecting superficial, low grade bladder cancer by combination analysis of hypermethylation of E-cadherin, p16, p14, RASSF1A genes in urine. Urol Oncol. 2010;28(6):597–602. | ||
Urquidi V, Rosser C, Goodison S. Molecular diagnostic trends in urological cancer: biomarkers for non-invasive diagnosis. Curr Med Chem. 2012;19(22):3653. | ||
Dahmcke CM, Steven KE, Larsen LK, et al. A prospective blinded evaluation of urine-DNA testing for detection of urothelial bladder carcinoma in patients with gross hematuria. Eur Urol. 2016;70(6):916–919. | ||
Ye F, Wang L, Castillo-Martin M, et al. Biomarkers for bladder cancer management: present and future. Am J Clin Exp Urol. 2013;2(1):1–14. | ||
Bornman DM, Mathew S, Alsruhe J, Herman JG, Gabrielson E. Methylation of the E-cadherin gene in bladder neoplasia and in normal urothelial epithelium from elderly individuals. Am J Pathol. 2001;159(3):831–835. | ||
Dulaimi E, Uzzo RG, Greenberg RE, Al-Saleem T, Cairns P. Detection of bladder cancer in urine by a tumor suppressor gene hypermethylation panel. Clin Cancer Res. 2004;10(6):1887–1893. | ||
Hoque MO, Begum S, Topaloglu O, et al. Quantitation of promoter methylation of multiple genes in urine DNA and bladder cancer detection. J Natl Cancer Inst. 2006;98(14):996–1004. | ||
Naghitorabi M, Mohammadi-Asl J, Sadeghi HMM, Rabbani M, Jafarian-Dehkordi A, Javanmard SH. Quantitation of CDH1 promoter methylation in formalin-fixed paraffin-embedded tissues of breast cancer patients using differential high resolution melting analysis. Adv Biomed Res. 2016;5(1):91. | ||
Chen P-C, Tsai M-H, Yip SK, et al. Distinct DNA methylation epigenotypes in bladder cancer from different Chinese sub-populations and its implication in cancer detection using voided urine. BMC Med Genomics. 2011;4(1):45. | ||
Maruyama R, Toyooka S, Toyooka KO, et al. Aberrant promoter methylation profile of bladder cancer and its relationship to clinicopathological features. Cancer Res. 2001;61(24):8659–8663. | ||
Xiong G, Liu J, Tang Q, et al. Prognostic and predictive value of epigenetic biomarkers and clinical factors in upper tract urothelial carcinoma. Epigenomics. 2015;7(5):733–744. | ||
Wang Q, Wang B, Zhang Y-M, Wang W. The association between CDH1 promoter methylation and patients with ovarian cancer: a systematic meta-analysis. J Ovarian Res. 2016;9(1):23. | ||
Liu J, Sun X, Qin S, et al. CDH1 promoter methylation correlates with decreased gene expression and poor prognosis in patients with breast cancer. Oncol Lett. 2016;11(4):2635–2643. | ||
Lin H-H, Ke H-L, Wu W-J, Lee Y-H, Chang L-L. Hypermethylation of E-cadherin, p16, p14, and RASSF1A genes in pathologically normal urothelium predict bladder recurrence of bladder cancer after transurethral resection. Urol Oncol. 2012;30(2):177–181. | ||
Yates DR, Rehman I, Meuth M, Cross SS, Hamdy FC, Catto JW. Methylational urinalysis: a prospective study of bladder cancer patients and age stratified benign controls. Oncogene. 2006;25(13):1984–1988. | ||
Fraga MF, Esteller M. Epigenetics and aging: the targets and the marks. Trends Genet. 2007;23(8):413–418. | ||
Chan MW, Chan LW, Tang NL, et al. Hypermethylation of multiple genes in tumor tissues and voided urine in urinary bladder cancer patients. Clin Cancer Res. 2002;8(2):464–470. | ||
Battista Di Pierro G, Gulia C, Cristini C, et al. Bladder cancer: a simple model becomes complex. Curr Genomics. 2012;13(5):395–415. | ||
Castelao JE, Yuan J-M, Skipper PL, et al. Gender-and smoking-related bladder cancer risk. J Natl Cancer Inst. 2001;93(7):538–545. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.