Back to Journals » Cancer Management and Research » Volume 12
Progress in Research on Colorectal Cancer-Related Microorganisms and Metabolites
Authors Han S , Zhuang J, Wu Y, Wu W , Yang X
Received 23 June 2020
Accepted for publication 25 August 2020
Published 21 September 2020 Volume 2020:12 Pages 8703—8720
DOI https://doi.org/10.2147/CMAR.S268943
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Ahmet Emre Eşkazan
Shuwen Han,1 Jing Zhuang,2 Yinhang Wu,3 Wei Wu,4 Xi Yang1
1Department of Oncology, Huzhou Cent Hospital, Affiliated Cent Hospital HuZhou University, Huzhou 313000, People’s Republic of China; 2Graduate School of Nursing, Huzhou University, Huzhou 313000, People’s Republic of China; 3Graduate School of Second Clinical Medicine Faculty, Zhejiang Chinese Medical University, Hangzhou 310053, People’s Republic of China; 4Department of Gastroenterology, Huzhou Cent Hospital, Affiliated Cent Hospital HuZhou University, Huzhou 313000, People’s Republic of China
Correspondence: Xi Yang
Department of Oncology, Huzhou Cent Hospital, Affiliated Cent Hospital HuZhou University, 198 Hongqi Rd, Huzhou, Zhejiang, People’s Republic of China
Tel +8605722555650
Email [email protected]
Wei Wu
Department of Gastroenterology Huzhou Cent Hospital, Affiliated Cent Hospital HuZhou University, 198 Hongqi Road, Huzhou, Zhejiang, People’s Republic of China
Tel +8605722555650
Email [email protected]
Abstract: Intestinal flora is an important component in the human body, which have been reported to be involved in the occurrence and development of colorectal cancer (CRC). Indeed, changes in the intestinal flora in CRC patients compared to those in control subjects have been reported. Several bacterial species have been shown to exhibit the pro-inflammatory and pro-carcinogenic properties, which could consequently have an impact on colorectal carcinogenesis. In this review, we summarize the current knowledge on the potential links between the intestinal microbiota and CRC. We illustrated the mechanisms by which intestinal flora imbalance affects CRC, mainly focusing on inflammation, microbial metabolites, and specific bacteria species. In addition, we discuss how a diet exhibits a strong impact on microbial composition and provides risks for developing CRC. Finally, we describe the potential future directions that are based on intestinal microbiota manipulation for CRC diagnosis and treatment.
Keywords: intestinal flora, colorectal cancer, inflammation, microbial metabolites, diet
Introduction
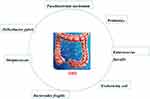
Colorectal cancer (CRC) is a common gastrointestinal cancer. According to the Global Cancer Statistics 2018, in both sexes combined, CRC is the fourth commonly diagnosed cancer based on incidence (6.1%), and the second diagnosed cancer based on mortality (9.2%).1 According to the reports, the incidence of CRCis positively correlated with social economic development.2 It is expected that the number of new cases will increase to 2.2 million and that of death cases will increase to 1.1 million by 2030, especially in developed countries.3 Thus far, the cause of CRC is not very clear. It may be related to genetic factors, inflammatory response, virus infection, and even dietary practices (Figure 1). Some papers have reported that people who were eating red meat had a higher incidence of CRC.4 Determining the pathogenesis of CRC will bring benefits to the treatment of this disease.
With advanced developments in high-throughput sequencing technology, intestinal microecology has received increasing attention in recent years.5 The steady state of intestinal microecology is important for the body to maintain a healthy balance.6 Recent studies have shown that changes in the gut microbiota could be a major factor to trigger CRC.7,8 There are many types and numbers of microorganisms in the intestinal tract. They can participate in the substitution of sugar, protein, starch, vitamins, and other nutrients to determine the body’s immune function and maintain the health of the host. Initial microflora is always in a state of dynamic equilibrium with the intestinal microenvironment. Once the dynamic equilibrium of the group is broken, it may lead to various diseases. Research results suggest that there is a difference in the composition of intestinal microorganisms between CRC patients and healthy individuals.5 The occurrence and development of CRC are in relationship with risk factors such as heredity, immunity, environment, dietary practices, and lifestyle, but the exact pathological mechanism underlying CRC is currently unclear. Animal experiment results9,10 indicate that the incidence of intestinal inflammation and CRC in sterile animals is extremely low. However, once the dysregulated intestinal flora is transplanted into the intestinal tract of sterile animals, the incidence of intestinal inflammation and CRC increases significantly. These results imply that the steady state of intestinal flora is the foundation for maintaining intestinal health. Further, there is also a close connection between the microenvironment of the intestinal flora and CRC. However, the above-mentioned conclusions still lack direct evidence from human subjects, and there is a need for further exploration and in-depth research in the future.
In recent years, studies have shown that the metabolites of intestinal flora are closely related to the occurrence of CRC.11,12 Primary metabolites of intestinal flora, including amino acids, nucleotides, polysaccharides, lipids, and vitamins, are necessary to maintain the growth and reproduction of the intestinal flora.13 The synthesis of primary metabolites is a constant process, and any obstacles in this synthesis will affect the normal activity of the microorganisms. The secondary metabolites of intestinal flora, including alkaloids, phenols, and antibiotics, can determine the function and specificity of the intestinal flora.14 All these findings form studies indicate that the intestinal flora is very valuable in maintaining the intestinal microecological balance. Thus, controlling the metabolites of intestinal flora may be a useful strategy in the treatment of CRC. It is critical to illustrate the interactions between intestinal flora, metabolites and CRC development. In this review, we explore the mechanisms underlying intestinal flora imbalance that affects CRC, mainly including inflammation, microbial metabolites, specific bacteria, and diet. We also elaborate the potential clinical practice implications in this microbiota era.
Overview of the Intestinal Flora
Intestinal flora is a research hotspot in microbiology and medicine in recent years. Intestinal flora refers to the various microorganisms that reside in the gastrointestinal tract of the body, which are combined in a certain ratio. Species are mutually restricted and interdependent to maintain the ecological balance. The human gut has a huge number of microorganisms including bacteria, viruses, and archaea.15–17 Among them, bacteria are present in the largest number, reaching to 10.14 Intestinal flora is mainly divided into three categories: (1) Probiotics such as Bifidobacterium, Lactobacillus, etc., which are beneficial to health; (2) Conditionally pathogenic bacteria such as Escherichia coli, which are not pathogenic under normal conditions but can cause disease when the intestinal microenvironment changes or the balance of the flora is broken; (3) Pathogenic bacteria such as Staphylococcus, PneumoniaCocci, Neisseria, etc., which can cause disease even under normal conditions.
The initial microflora has an important role in maintaining the survival and health of the host organism. Previous research results suggested that the intestinal flora has the following physiological functions at minimum: (1) Preventing the invasion of pathogenic bacteria, adjusting the balance between the human body and the microorganisms state, and maintaining the body’s initial health or physiological state; (2) Immunity function during which the bacterial flora can produce an immune response by stimulating the host to inhibit the propagation of pathogenic bacteria in the intestine; (3) Detoxification function in which the bacterial flora can adjust the peristaltic movement of intermediates and the absorption of water to promote stool and excretion of harmful substances; (4) Nutritional effect, in which the initial bacterial flora can synthesize or promote the absorption of nutrients; (5) The bacterial flora can activate antitumor cytokines by degrading and removing carcinogens to exert its antitumor role; (6) The initial flora can reduce the production of oxygen free radicals and control the arthritis response to delay senescence.
In a healthy gut, the dominant bacteria mainly include Firmucutes, ActinobacteriaProteobacteria, and Bacteroidetes. However, the intestinal flora has a diverse structure at the genus and species levels. Intestinal flora communicates with the host, enhances the epithelial defense against pathogens, and accelerates the maturity of the immune system.18,19 Previous research reported that the intestinal flora has the capacity to defend the body against pathogens by recognizing the conserved antigen of bacteria.20,21 The intestinal flora was reported to protect the local homeostasis. For example, Wang et al reported that gut microbiota could decrease alcohol-associated steatohepatitis.22 Except that, Salmonella typhi was reported as a well-known pathogen that can cause great damage to human health. A variety of factors such as age, diet, drugs, sports, and genotype also have been reported to impact the gut microbial community.23–27
In addition, the relationship between gut microbiota and CRC has become a research hotspot in recent years. Studies have shown that people who begin to take antibiotics from a young age are more likely to develop CRC after 60 years of age. This prompted us to suggest that the disruption of the intestinal microbiota balance may be a main factor of CRC.28 Compared with healthy people, CRC patients have significantly decreased intestinal microbiota diversity and obviously changed microbial abundance.29 Many studies showed that the bacteria in the initial microflora species such as Bacteroides fragilis, Fusobacterium nucleatum, Escherichia coli, and so on, were obviously increased in CRC patients.30,31
Pathogenic Mechanism of Intestinal Flora
Inflammatory Microenvironment and CRC
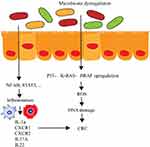
Inflammation is the key factor that promotes CRC progression. Among patients with inflammatory disorders, 10%-15% are more likely to develop CRC compared with that in normal people.32 The dysregulation of intestinal microbiome can make the intestinal tract environment worsen and stimulate intestinal epithelial cells to activate the NF-κB pathway to drive the inflammation (Figure 2). A study on ApcMin/+ mouse reported that colonitis was driven by the high density of microorganisms.33 Microorganisms can accumulate in the polypus and trigger local inflammation. Antibiotics can relieve these symptoms. Previous studies reported that inflammation and gut microbiota are closely linked through a two-way relationship. On the one hand, IL-33 can activate B cell to produce IgA to maintain the homeostasis of the intestinal microbiome. On the other hand, the remodeling of the intestinal microbiota can activate the release of IL-1α to induce tumor formation related to colitis and inflammation.34 Long-term chronic mild inflammation that accompanies the aging process is the main cause of inflammatory-related tumors. Immune depletion may be the main reason. Studies have found that mild chronic inflammation was often accompanied by the reconstruction of intestinal microbiota. Microbial diversity was reduced. The change in gut microbiota can indeed promote the inflammation. According to Wong’s study, they fed the sterile mouse with CRC patients’ fecal samples and found that the treatment group had more polypus, intestinal dysplasia, increased inflammatory factors including CXCR1, CXCR2, IL-17A, IL-22, IL-23A, and enhanced Th1 and Th17 cells than did the control group.35 However, determining the reasons why the inflammation and microbiology reconstruction are the starting factors for CRC development further study is needed.36
Immune cell infiltration has been reported to be closely related to the prognosis of CRC patients. However, the role of chemokines in promoting immune cells to enter colorectal cancer needs to be confirmed by more studies. Some studies found that tumor cells may be an important source of the expression of chemokine CCL5, CXCL9, and CXCL10. Tumor cells that are exposed to gut bacteria can recruit more T cells than the control cells, which suggests that intestinal microbes may play an indispensable role during the chemotaxis process of T cells.37
Bacterial Metabolites and CRC
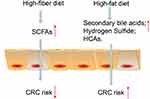
Intestinal microbes play an important role in human health and disease. The metabolic function of intestinal microbes can be considered as a contributing factor for disease development, and their biologically active substances have an important impact on the host’s physiological and pathological processes. After intestinal microflora disorders, intestinal anaerobic bacteria can produce a series of metabolic enzymes to change the metabolic capacity. These metabolic enzymes can act on different substrates (including bile acid, fatty acid, and so on) to produce carcinogens, which, in turn, cause colon cancer.5 According to previous reports, carcinogens produced as a result of bacterial metabolism mainly included hydrogen sulfide, reactive oxygen species (ROS), secondary bile acids, and so on. The type and number of intestinal flora can directly affect the occurrence and development of tumors, and intestinal flora can also indirectly affect CRC cells by regulating body metabolism. At the same time, tumor cells escape the killing effect of these substrates on tumors by downregulating the metabolites of beneficial flora in the process of tumor growth (Figure 3).
Short-Chain Fatty Acids (SCFAs)
The beneficial flora can produce short-chain fatty acids (SCFAs) after fermentation. Common SCFAs include acetate, propionate, and butyrate. Among them, butyrate can enter into the nucleus of tumor cells and function as a histone deacetylase inhibitor to block tumor cell proliferation.38
G-protein-coupled receptor 43 (GPR43) and GPR109A are the main receptors of SCFAs. They are important molecules that mediate the cancer suppressive effect of the bacterial fermentation product SCFA.39 Studies have shown that the expression of GPR109A in CRC can upregulate the expression of apoptotic factors and downregulate the expression of tumor proliferation-related genes, which, in turn, promote tumor cell apoptosis and reduce tumor cell colonization and growth.40 Another study had shown that GRP43 had a low expression level in the primary and metastatic foci of colon cancer.41
The ectopic expression of GPR43 in CRC can lead to cell cycle stagnation and eventually cause apoptosis of tumor cells. Some researchers reported that the ectopic expression of the sodium ion-coupled SCFA transport protein can cause translocation of the apoptosis inhibitor protein from the nucleus to the cell membrane through protein interaction and repress survivin transcription to promote the apoptosis of tumor cells.42 Our previous study also demonstrated that the levels of SCFAs in CRC patients and individuals with a high risk of CRC were higher than those in healthy individuals.8
Bile Acids
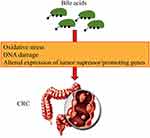
High levels of bile acids have been confirmed to be closely related to human colon cancer (Figure 4). Intestinal bacteria can produce bile acid, especially under a high-fat diet. Clostridiumcan produce secondary bile acids. These secondary bile acids affect the mitotic processes, which induces DNA damage and the production of ROS, thereby leading to the increase in the incidence of colon cancer risk.43
Bile acid is a product of cholesterol catabolism in the liver. It is a general term for a class of bile acids. It can be divided into primary bile acid and secondary bile acid from the source. Primary bile acid is synthesized in the liver cells using cholesterol as a raw material. After the entry of the primary bile acid into the large intestine, it is decomposed and dehydroxylated by bacteria to generate secondary bile acid, deoxycholic acid, and lithocholic acid.44 Secondary bile acids have biological toxic effects such as mutagenesis, cell lysis, and DNA band breakage, which are known inducers of intestinal tumors.45
In vivo studies have shown that the content of secondary cholic acid in the serum and colon of patients with adenomatous polyps and colon cancer was significantly increased.46 In vitro experiments also confirmed that deoxycholic acid and chenodeoxycholic acid in the colon can promote the proliferation of human colon adenoma AA/C1 cells and decrease the apoptosis of cancer cells.46 More importantly, long-term high-fat, high-protein, and low-fiber diets will produce large amounts of secondary bile acids and bile acid fecal enzymes in the colon, which can, in turn, cause colon cancer.47
Hydrogen Sulfide
Hydrogen sulfide is produced by intestinal flora by the reduction of diet-derived sulfate and the metabolism of other compounds including sulfur amino acids and taurine.48 Food residues with a high protein content can stimulate the growth of sulfate-reducing bacteria. Hydrogen sulfide plays a role in promoting inflammation and genotoxic substances, indicating that it is associated with the development of CRC.49 Hydrogen sulfide can cause cell proliferation, differentiation, apoptosis, and inflammation, eventually leading to the malignant transformation of intestinal epithelial cells.50 Thus, hydrogen sulfide levels might be primarily driven by the changes in bacterial activity rather than by bacterial abundance.
Researchers have found that CRC patients had higher hydrogen sulfide levels than healthy people. Furthermore, the detoxification capacity of the colon tissue for hydrogen sulfide in CRC patients was weakened.51 Hydrogen sulfide induces the formation of colon cancer mainly through the induction of DNA damage, the release of free radicals, inflammation of the colonic mucosa, excessive colonic mucosa hyperplasia, and inhibiting cytochrome oxidase, butyrate utilization, mucus synthesis, and DNA methylation.52
Methylamines (TMA/TMAO)
Trimethylamine oxide (TMAO) is one of the important metabolites of intestinal microorganisms. It is first decomposed into trimethylamine (TMA) by the nutrients that are rich in phosphatidylcholine (PC) and L-carnitine under the action of the intestinal microbe TMA lyase. Then it was formed after oxidation by flavin monooxygenase (FMO) 3 in the liver.53 With increasing research on TMAO, it was found that TMAO was related to not only cardiovascular disease, kidney disease, and diabetes but also cancers. Studies have found46 that plasma TMAO levels were lower in patients with stroke and transient ischemic attack (TIA), and it was speculated that stroke or its treatment will reduce plasma TMAO levels. In addition, studies have shown54 that TMAO and Alzheimer’s disease have a strong positive correlation. Clinical data showed55 that plasma TMAO levels were positively correlated with CRC. Experimental studies suggested56 that urine TMAO can be used as a predictor of CRC. On the contrary, a few studies have shown that57,58 TMAO can correct the folding defects in mutant proteins and have a protective effect in the process of CRC cancer. Therefore, the effect of TMAO on cancer needs further study. In addition, some studies have shown that TMAO can cause oxidative damage to the liver59 and can also be used as a factor of poor prognosis of community-acquired pneumonia.60 At the same time, it is also related to inflammatory bowel disease, ulcerative colitis, chronic gastritis, and gastric ulcer.61,62 Some scholars have studied the correlation among diet, microbiome metabolism, and diseases. They found that TMAO-related genes and CRC-related genes share a common genetic pathway in the immune system, cell cycle, cancer pathway, and Wnt signaling pathway. Hence, TMAO may be connected to a high-protein and high-fat diet. Moreover, it may be an important intermediate marker of intestinal microbiome metabolism and CRC risk.63
N-Nitroso Compounds
After dimethylnitrosamine was confirmed to have carcinogenic effects in 1956, more than 200 kinds of nitroso compounds were proved to be carcinogenic. There are two types of nitroso compounds: N-nitrosamine and N-nitrosamide.64 N-nitrosamines are mostly volatile and have no direct mutagenic effect on organs and tissue cells. N-nitrosamide can directly damage DNA and is a direct carcinogen. N-nitroso compounds are ubiquitous in nature and can also be synthesized in the body by nitrates and nitrites in food, and the gastrointestinal tract is the main site for the endogenous synthesis of nitrous compounds.64 Recently, Abu-Ghazaleh et al reported that N-nitroso compounds can modulate CRC progression.65 The content of nitrite in pickled foods such as kimchi and sauerkraut is very high. Therefore, its intake is closely related to the incidence of human CRC.66 N-nitroso compounds have a strong carcinogenic effect,67 especially in inducing human digestive tract cancer such as CRC.68
Ethanol
At present, it is widely recognized that excessive drinking is an important risk factor for carcinogenesis,69,70 while ethanol metabolism by the intestinal flora may further increase its toxicity. Ethanol can be produced by many anaerobic bacteria in vitro when they are grown in pure culture. However, the level of endogenous ethanol production by the colonic microbiota in vivo is unknown. Although ethanol itself is not a substance with obvious carcinogenic effects, its oxidation product acetaldehyde is recognized as a strong carcinogen, which can have a series of effects on the body, including the degradation of vitamin folate and DNA damage.71 Interestingly, studies of the oral microbiota have shown that microorganisms contributed to the production of acetaldehyde from ethanol, which suggested that the gut microbiota might also contribute to this process.71,72
Specific Bacterial Strains Associated with CRC
Current research believes that the composition of intestinal microbes is an important factor affecting tumorigenesis, and some intestinal flora such as Desulfovibrio, Escherichia coli, and those found in fecal intestines (Figure 5) promote the occurrence of CRC.
Fusobacterium nucleatum
Currently, the relationship between Fusobacillus nucleatum (F. nucleatum) and CRC occurrence has become the hotspot. Gur et al73 found that F. nucleumcan protect various tumors from the killing effect of natural killer (NK) cells, and this function was mediated by the ITIM domain (TIGIT). The inhibitory receptor TIGIT exists on human NK cells and T cells, and this inhibitory effect depends on the Fap2 protein in F. Nucleatum. This indicates that the derivative factors of F. Nucleatum can promote tumor immune escape. In addition, proliferation rate, invasion activity, and tumor growth rate in mouse xenograft models can be significantly increased when CRC cells are infected by F. nucleatum.74 F. nucleatum can regulate the tumor immune microenvironment and activate the E-cadherin/β-catenin signaling pathway. Rubinstein et al75 proved that F. nucleatum can bind to E-cadherin by adhering to FadA, thereby activating the β-catenin signaling pathway to induce carcinogenesis and inflammation. Importantly, compared to the CRC tissue, the expression of FadA in healthy tissues is significantly reduced, suggesting that inhibition of this signaling pathway can protect the body from an oncogenic effect. Yu et al76 showed that the relative abundance of F. nucleatum not only was related to the occurrence of CRC but also resulted in resistance to chemotherapy drugs and increased relapse rate by interfering with the signal transmission of TLR4 and MyD88 in CRC patients. The possible mechanism might be that F. nucleatum targets specific miRNAs, resulting in the activation of the autophagy pathway to change the patient’s response to chemotherapy drugs. The enrichment of F. nucleatum in CRC tissues had a positive correlation with a shorter survival rate.77 Therefore, it may serve as a potential prognostic marker for CRC.
Probiotics
In addition to the pathogenic microorganisms in the human intestine, there is also a category of intestinal microorganisms called “probiotic.” They regulate host mucosa and system immune function. They also can produce a beneficial physiological effect on the host by improving the intestinal nutrition and the flora balance. Shadnoush et al78 proved that a stable intestinal environment is the balance between intestine immune and anti-inflammatory reactions, which can be maintained by probiotics. Probiotics can not only relieve lactose intolerance and reduce constipation but also reduce the recurrence of inflammatory bowel disease and prolong the remission period.79 A previous clinical study was conducted on 92 patients with inflammatory bowel disease. Subjective evaluation and clinical remission rate in patients in the treatment group with the addition of probiotics were higher than those in the control group. Sood et al80 also confirmed that probiotics can significantly improve the remission rate in patients with mild-to-moderate active cancer.
Prebiotics are food components that cannot be broken down by enzymes in the intestine. They can promote the growth of probiotics to become the predominant flora. The combination of prebiotics and probiotics is called synbiotics. The application of synbiotics can make fecal flora change significantly. As the number of Bifidobacterium and Lactobacillus increases, the number of Clostridium perfringens decreases, and it can improve the function of the colonic epithelial barrier in patients with polypectomy.81 Probiotic bacteria participate in suppressing allergies, controlling serum cholesterol levels, and regulating immune function to inhibit the growth of potentially harmful bacteria to prevent the occurrence of CRC.82 Based on the anticancer properties of probiotics, it can be used in combination with traditional CRC treatment (including surgery, chemotherapy, etc.).83,84 It can improve the integrity of the intestinal mucosal barrier and reduce the incidence of infectious complications in surgical CRC patients.85 Some studies have analyzed the effects of probiotics and synbiotics and found that both can effectively combat the risk factors of CRC. Yorkshire milk Bacillus (L. johnsonii) can adhere to the surface of the colonic mucosa, reduce the invasion of pathogens into the intestinal tract of patients with CRC during the perioperative period, and adjust its local immune function.86 To counterbalance the adverse complications related to tumor treatment, probiotics can also prevent CRC recurrence and improve the quality of life of patients.87
Enterococcus faecalis
Enterococcus faecalis (E. faecalis) is a gram-positive facultative anaerobic commensal bacterium. Most of them are harmless to humans. However, it has emerged as a human pathogen.88 Balamurugan et al89 reported that significantly higher E. faecalis fecal populations were found in fecal samples collected from CRC patients than in healthy control individuals. E. faecalis has been reported to induce chronic inflammation and to produce extracellular superoxide and hydrogen peroxide.90 Furthermore, production of extra cellular free radicals was shown to induce DNA damage in vitro.91 In addition, E. faecalis was also able to induce DNA damage in colonic cells.91 Reactive oxygen species (ROS) are able to induce chromosomal instability,92 which could be associated with CRC occurrence.93 Moreover, E. faecalis can trigger colitis, dysplasia, and CRC.94 Wang and colleagues also showed that E. faecalis was able to polarize colon macrophages to an M1 phenotype. All of these findings could explain the mechanisms by which E. faecalis exerts effects on colorectal carcinogenesis. Extracellular superoxide produced by E. faecalis is converted to hydrogen peroxide, thereby causing DNA damage to colonic epithelial cells.93,95
Escherichia coli
The content of E. faecalis in CRC patients is higher than that in healthy people. In addition, some studies have found96–98 that some special types of E. coli, Proteobacteria, and enterotoxin-producing Bacteroides fragilis (Enterotoxigenic Bacteroides fragilis, ETBF), Bacteroides/Prevotella spp., Campylobacter, Peptostreptococcus, etc. were found in the colon of CRC patients in much higher quantities than those in normal individuals. Using adenocarcinomas and normal colonic mucosa from CRC patients, mucosa-associated E. coli was found in 50% of adenomas compared to that of 15% of normal mucosal samples.99 Moreover, there is a correlation between poor prognostic factors of CRC and colonization of mucosa by E. coli.100 Some research had reported that E. coli might influence CRC progression by persisting in immune cells and controlling the secretion of pro-tumoral mediators.101
The polyketide synthase (pks) gene of some E. coli strains can encode the colibactin protein. This genotoxic product can induce single-stranded DNA damage.102,103 CRC E. coli causes continuous and low-level colonization in the intestine to induce an asymptomatic and continuous inflammatory response in the colonic mucosa. At the same time, it produces genotoxic substances that can cause DNA damage in colonic epithelial cells, thereby increasing the individual’s vulnerability to CRC.104,105
Bacteroides fragilis
Professor Cynthia recently published a study in Science. She found that enterotoxin-producing Bacteroides fragilis (ETBF) was able to “engulf” a part of the intestinal mucus layer, thus destroying the intestinal barrier and allowing itself and the “genotoxic island” sequence encoded by the pks gene of E. coli (pks+E. coli) invade the inner layer of intestinal mucus and form a biofilm.106 These two common bacteria are important promoters of CRC.
ETBF is an anaerobic bacterium in the colon of healthy people and animals. It is a resident bacterium in the human intestine. ETBF is one of subtypes of the B. fragilis, that can be asymptomatically colonized after the infection and can also cause diseases such as diarrhea. ETBF is also the cause of clinically independent endogenous suppurative infections and is associated with the onset of colorectal tumors. Wu et al107 had focused on the function of ETBF in mice. They found that ETBF and non-toxigenic ETBF can be colonized in mice, but only the ETBF can cause an inflammatory reaction in the colon and significantly increase the incidence of colon tumors in the multiple intestinal neoplasia (Min) mouse model. Goodwin et al108 found that ETBF infection can upregulate spermine oxidase (SMO), which can promote reactive oxygen species, DNA damage etc., thereby promoting tumorigenesis. Treatment with SMO inhibitors can inhibit the inflammatory response produced by ETBF in the Min mouse model and significantly reduce the number of colon tumors. Rhee et al109 found that C57BL/6 mice with intragastric administration of ETBF did not only produce an inflammatory reaction in the colonic mucosa but also caused the proliferation of intestinal mucosa epithelial cells. Toprak et al110 found that the detection rate of fecal ETBF in patients with CRC was 38% and that in normal people was 12%. It was confirmed for the first time that ETBF was highly prevalent in patients with CRC. The pathogenicity of ETBF is related to its products containing sugar capsules, outer membrane proteins, and special enzymes, including enterotoxin fragilysin. Studies have shown that the target of fragilysin is the cell surface protein E-cadherin. ETBF enhances the transcription of proto-oncogenes c-myc and cyclin D1 by activating the Wnt/Wingless signal transduction pathway, leading to tumorigenesis. ETBF directly acts on colonic epithelial cells through Bacteroides fragilis toxins, and at the same time, it can promote the development of CRC by causing an immune inflammation reaction.108,110,111
Streptococcus
Studies have found that Streptococcus bovis was highly enriched in CRC patients.112–117 The role of S. bovis in CRC is currently unclear. Klein et al118 found that most S. bovis can induce colon adenoma or asymptomatic tumors in patients with endocarditis, indicating that S. bovis was involved in the early stages of CRC. In addition, another study found that the level of S. bovis-induced antigen expression of RpL7/L12 was significantly increased in patients with colon polyps and stage I/II CRC but not in patients with advanced lymph nodes or distant metastases.119 These findings suggest that S. bovis may promote the development of CRC at an early stage. At the same time, Streptococcus bovis can also cause specific boarding crypts provided by colorectal tumor lesions, which can cause the release of inflammatory cytokines such as interleukin-8 (IL-8) through direct contact or antigen-stimulated cells, which, in turn, further promotes abnormal colonic recession overproliferation of litters.52,120 In addition, S. bovis also produces inflammatory cytokines such as IL-8 and prostaglandin E2 (PGE2), causing chronic inflammation of the colon. This long-term adverse stimulation will gradually promote normal colonic epithelial cells to become cancerous.121
Helicobacter pylori
Some scholars have found the presence of Helicobacter pylori-DNA in the intestine of patients with CRC.122 In recent years, many domestic and foreign scholars have reported the relationship between Helicobacter pylori infection and CRC.123 At present, the mechanism of colorectal tumors caused by Helicobacter pylori infection is not clear, but hypergastrinemia and the expression of cyclooxygenase-2 (COX-2) are currently considered to be possible mechanisms of CRC caused by Helicobacter pylori infection.124 In animal experiments in mice, it was found that overexpression of gastrin can cause intestinal metaplasia-atypical hyperplasia and eventually gastric cancer in 20 months.125 Therefore, there is a hypothesis that H. pylori infection may indirectly lead to CRC through changes in gastrin levels: H. pylori infection causes atrophic changes in the gastric mucosa and high gastrin levels through the negative feedback mechanism of gastric antrum G cells, thereby promoting colonic mucosa growth.126,127 Studies have shown that high gastrin levels can promote the growth of colon cancer cells cultured in vitro and increase the incidence of CRC in animal models.128–130 In addition, many studies have shown in animal experiments that non-amidated gastrin (including progastrin and glycine extended gastrin) acting as a growth factor for colonic epithelial cells and tumors, may be involved in the development of CRC.131,132 Glycine-extended gastrin promoted the proliferation of the colonic mucosa through Rho/ROCK-dependent pathways, which was mainly manifested by thickening of the intestinal mucosa and increased goblet cells of the gland duct.133,134 Studies have shown that H. pylori infection-induced hypergastrinemia was often accompanied by high expression of COX-2 in the large intestinal mucosa.135,136 Hartwich et al137 further showed that the expression of gastrin, COX-2, and anti-apoptotic mRNA receptors increased in H. pylori-infected colorectal tumor tissues, suggesting that hypergastrinemia and COX-2 may interact and lead to the formation of large intestine tumors. In addition, H. pylori infection may also lead to CRC through immune tolerance mechanisms. Frumento et al138 studies have shown that low tryptophan levels and increase in the concentration of its degradation product kynurenine may directly affect the immune response to antigen-stimulated T cells in tumor patients. However, Engin et al139 found that the H. pylori-positive group of patients with CRC had a significantly higher kynurenine/tryptophan ratio than the H. pylori-negative group. It was speculated that H. pylori infection may lead to cancer development through immune tolerance. At present, hypergastrinemia, high expression of COX-2, and immune tolerance can be considered as one of the mechanisms of the development of colorectal tumors caused by H. pylori infection, but the specific physiological mechanism is still unclear and needs further investigation.
Diet and CRC
High-Fat, High-Protein Diet
In recent years, with continuous improvement in people’s living standards and reduction of dietary fiber content, the proportion of meat and fried grilled food has increased significantly, resulting in an increased risk of CRC. Studies have found that the incidence of CRC was lower in countries with a low-fat diet, while the incidence is higher in countries with a high-fat diet (Figure 6). Studies in Shanghai (China) have also found that the increase in the incidence of CRC was related to the intake of a large number of high-fat diet. The carcinogenic effect may be related to the production of oxides and fatty acids broken down by fat. A high-fat, high-protein diet leads to the onset of CRC.140 It can cause the liver to synthesize and secrete too much of bile acid. Under the influence of anaerobic bacteria in the intestine, the bile acid remaining in the intestine becomes secondary cholic acids such as deoxycholic acid, lithocholic acid, and so on. Secondary cholic acid can directly interfere with DNA metabolism, change DNA synthesis, increase the activity of ornithine decarboxylase in colorectal mucosa cells, reduce immune function, and thereby promote the proliferation of cancer cells. After eight years’ study, Tiemersma et al141 showed that the increase in lean meat intake by men was positively correlated with the incidence of CRC, while the intake of poultry and fish by women could reduce the CRC incidence. An animal experiment in Europe showed that a high-fat diet can induce mutations in the K-ras gene, dysregulate intestinal flora, and promote the development of gastrointestinal tumors.142 While some studies have shown that high-protein diets can produce some potential carcinogenic effects, there is insufficient evidence to prove the relationship between protein diet and CRC, and further proof is needed.
Dietary Fiber
The protective effect of dietary fiber is due to the fact that cellulose can selectively promote the growth of intestinal flora and increase the intestinal motility, increase stool volume, reduce the contact time between stool and carcinogens and intestinal mucosa and dilute the concentration of intestinal carcinogens.143 For example, YEH et al144 found that the intake of fresh vegetables and fruits was negatively correlated with the risk of CRC through a case-control study. Huang Xiuhai et al145 also found that the intake of high fiber, fresh fruits, and vegetables and low saturated fatty acids can reduce the risk of gastrointestinal tumors. Moreover, Bingham et al146 found that dietary fiber intake was negatively correlated with the incidence of CRC through 519,978 case-control studies in 10 European countries. It was not related to the source of dietary fiber and had the greatest protective effect on the left colon. Therefore, the American Cancer Institute of the World Cancer Research Foundation has shown that increasing daily intake of dietary fiber by 10 g can effectively reduce the incidence of CRC by about 10%.147
Trace Elements and Vitamins
A large number of studies have shown that intake of appropriate amounts of vitamins and other nutrients can effectively reduce the incidence of CRC. Important factors for preventing CRC are dietary calcium, folic acid, selenium, and vitamins A, C, D, E, β-carotene, and other micronutrients. Among them, studies have found that long-term use of high-calcium preparations can reduce the risk of distant colon cancer.148 Calcium ions and vitamin D can combine with cholic acid and fatty acids to form an insoluble calcium soap, which can inhibit the growth of intestinal epithelial cells, improve the final differentiation of cells, and play a chemical protective role in intestinal epithelium. In recent years, the observed protective effect of selenium on colon cancer has received increasing attention. Selenomethionine can inhibit tumor cell proliferation by activating the p53 tumor suppressor protein. Studies have also found that the reduction in the risk of colon cancer was related to long-term consumption of foods rich in folic acid.149
Intestinal Flora and Clinical Application in CRC
Immunotherapy can suppress tumor growth by suppressing cell cycle checkpoints, which has a therapeutic effect. However, it is not effective in all populations. Increasing evidence shows that the efficacy of immunotherapy depends on the steady state of the intestinal microbiota, and changes in the intestinal microbiota will have a certain effect on the efficacy of tumor chemotherapy.150,151 Certain intestinal microorganisms such as Bifidobacteria can enhance the activity of dendritic cells, thereby increasing the responsiveness of T cells to checkpoint blockade.152 Currently, anti-CTLA-4 is a commonly used immunotherapy. Sterile mice have no sensitivity to ipilimumab, indicating that the efficacy of ipilimumab, especially the activation of CD4 + T cells, is inseparable from the participation of intestinal microbiota. Further research found that anti-CTLA-4 therapy can change the composition of the gut microbiota in the tumor microenvironment. In mice and patients, the abundance of Bacteroides fragilis increases, which is conducive to killing cancer cells. Fecal transplantation in sterile mice can restore their drug sensitivity. This result gives us hope to control the efficacy of drugs by providing probiotics.153 Understanding microbial-dependent inflammation and immune mechanisms will open up new ways for cancer prevention and improved cancer treatment strategies. Anti-PD-1 immunochemical drugs are currently effective means of treating malignant tumors. There is a significant difference in the sensitivity among different patients to this chemotherapy drug, which may be due to the difference in intestinal microbiota. The tumor growth in mice transplanted with the feces of drug-sensitive patients was slower than that in the control group, and after anti-PD-1 drug treatment, the tumor volume in sensitive mice shrank and the efficacy was better. Matson et al154 found that the abundance of Bifidobacterium longum, Corinella aerogenes and Enterococcus faecium in the feces of patients who were sensitive to chemotherapy drugs was higher. Routy et al155 identified Akkermansia muciniphira (AKK) as a beneficial bacterium with antitumor effects and found that it was highly present in drug-sensitive patients. Gopalakrishnan et al156 identified Faecalibacterium as a beneficial bacterium that can play a positive role in anti-PD-1 immunotherapy. On the contrary, Bacteroides is a harmful bacterium that inhibits the efficacy of anti-PD-1 immunotherapy. The CD8 + T cell activity was higher in the tumor environment of drug-sensitive patients, which suggests that specific intestinal bacteria may enhance the ability of T cells to enter the tumor microenvironment and kill the cancer cells, thereby making the body durable and result in a strong immunotherapy response.
There is a very close relationship between the occurrence and development of malignant tumors and the imbalance in the intestinal flora.157 Studies have found that the concentration of bacteria in tumor tissues was much higher than that in normal tissues. The hypoxic tumor microenvironment provides good conditions for the growth of anaerobic bacteria. At the same time, abnormal blood vessels and high pressure in the tissue gap restrict immune components (granulocytes, antibodies, serum complement, etc.) to enter the bloodstream, protecting bacteria from the body’s immunity. We can see that special microenvironment composition of the tumor tissue will lead to the growth of a large number of bacteria, and excessively proliferating bacteria can compete with tumor cells for nutrition to inhibit tumor growth, and some special metabolites of bacteria can directly inhibit tumors.158 Cell proliferation and induction of tumor cell apoptosis may be achieved through regulation of the intestinal flora to reach the purpose of treating malignant tumors. Colley’s research confirms this point of view. Colley159 found that lumps of tumors in patients with sarcoma infected with acute streptococcus have shrunk; thus, starting the history of using bacteria or bacterial extracts to treat tumors. With improvement in living standards, obesity triggers intestinal flora imbalance, which, in turn, induces a high incidence of colon cancer. Currently, prevention and treatment methods are urgently needed. Therefore, based on the regulation of intestinal flora, exploration of colon cancer treatment methods and mechanisms of action are the main issues that our research group is concerned about. The hope is that through this exploration and research, a certain theoretical reference for the development of drugs based on the intestinal flora can be found, leading to the treatment of colon cancer.
In addition, one emerging translational application of the gut microbiota is its use as a biomarker to indicate the presence of a disease. Nowadays, several studies have reported associations between bacterial markers and treatment efficacies or clinical outcomes. Analysis of the gut microbiota serves as a rich source of potential biomarkers. A higher level of F. nucleatum was found in fecal samples in patients with colorectal adenomas than in healthy individuals.160 Furthermore, the robust association of CRC with S. gallolyticus resulted in the development of a positive test which was used for CRC diagnosis for up to 10 years.161 Moreover, Wang et al found that serum antibodies against F. nucleatum might also serve as a potential biomarker for detecting CRC. Other studies also found the metabolites of intestinal flora, including SCFAs, bile acids, and butyrate,161 were higher in patients with CRC than in healthy controls.163
Current Limitations and Future Directions
With the development of genomics technology, in recent years, intestinal microbiota research on CRC has made rapid progress. However, there are still a series of problems. There are individual differences in the intestinal microbiota. Furthermore, a small sample size in a single experiment makes it difficult to obtain a unified and convincing conclusion. Therefore, it is necessary to establish a multicenter, large sample dataset. The experimental process of similar research lacks standardization. At this moment, it is not clear whether the fecal sampling can completely represent gut microbiota status, and whether there are differences among the different detection platforms. Therefore, the unified collection process and detection methods should be built. Non-cultivated metagenomic big data analysis can only help us understand the state of microbes in the gut from the genetic level. Combining with culture-based culture omics related research will be able to provide a more realistic analysis of the gut microbial composition. We hope to learn more about the gut microbiota to help the clinical detection and prognosis. In addition to routine testing, we hope that microbial markers can be used to detect abnormal states in the early stages of cancer. In addition, we can also use adjuvant probiotic treatment during chemotherapy to achieve better results. As expected, research on the gut microbiota is still in development, and more experimental research is needed. This review summarizes the relationship between intestinal flora and its metabolites and colorectal cancer. It can provide potential microbial targets for screening and diagnosis of colorectal cancer. Moreover, it can provide further research direction for microbial agents as adjuvant therapy for colorectal cancer.
Conclusion
CRC is a disease characterized by complicated causes and pathogenesis. The intestinal microecology is closely related to the occurrence of CRC. Intestinal flora, especially some special bacteria, affect the development of CRC through metabolic and structural changes, which provides research data that can be used to establish prevention strategies of CRC and identify its pathogenic mechanism. We have discussed the important impact of the inflammation of intestinal flora on the development of CRC. The studies that are discussed in this review highlight that progression to CRC is influenced not only by the presence of intestinal flora but also by the metabolic output of the entire microbiota. In the progress of CRC development, the function of metabolites still needs to be further studied. It is expected to provide new ideas and clues. Diet is the most important factor to determine the dynamic changes of intestinal flora composition and function. The intake of dietary fiber and supplementation with beneficial bacteria may maintain intestinal health, which can be very promising for preventing CRC. Moreover, these data have provided an unprecedented opportunity to move microbiota discoveries toward clinical applications. In the future, targeted removal of early-stage carcinogenic members of the gut microbial community, might be a desirable approach to reducing risk factors for CRC.
Methods
We mainly searched for literature paper in PubMed database by using both medical subject heading (MeSH) terminology and relevant keywords. We summarized the correlation between CRC and microorganisms and their metabolites. There are some limitations in the paper, it is inevitable that some literature will be omitted in the process of literature screening. In the process of summarizing relevant literature again, it is possible to misinterpret the author’s original intention.
Author Contributions
All authors participated in the conception and design of the study; wrote the manuscript; designed and draw figures; reviewed and sorted out the literature; read and approved the paper; agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that no potential conflicts of interest exist.
References
1. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Lauby-Secretan B, Vilahur N, Bianchini F, et al. The IARC perspective on colorectal cancer screening. N Engl J Med. 2018;378(18):1734–1740. doi:10.1056/NEJMsr1714643
3. Arnold M, Sierra MS, Laversanne M, et al. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66(4):683–691. doi:10.1136/gutjnl-2015-310912
4. Domingo JL, Nadal M. Carcinogenicity of consumption of red meat and processed meat: a review of scientific news since the IARC decision. Food Chem Toxicol. 2017;105:256–261. doi:10.1016/j.fct.2017.04.028
5. Han S, Gao J, Zhou Q, et al. Role of intestinal flora in colorectal cancer from the metabolite perspective: a systematic review. Cancer Manag Res. 2018;10:199–206. doi:10.2147/CMAR.S153482
6. Charbonneau MR, Blanton LV, DiGiulio DB, et al. A microbial perspective of human developmental biology. Nature. 2016;535(7610):48–55. doi:10.1038/nature18845
7. Gensollen T, Iyer SS, Kasper DL, et al. How colonization by microbiota in early life shapes the immune system. Science. 2016;352(6285):539–544. doi:10.1126/science.aad9378
8. Xi Y, Yuefen P, Wei W, et al. Analysis of prognosis, genome, microbiome, and microbial metabolome in different sites of colorectal cancer. J Transl Med. 2019;17(1):353. doi:10.1186/s12967-019-2102-1
9. Szabo G, Bala S, Petrasek J, et al. Gut-liver axis and sensing microbes. Dig Dis. 2010;28(6):737–744. doi:10.1159/000324281
10. Zhu Q, Jin Z, Wu W, et al. Analysis of the intestinal lumen microbiota in an animal model of colorectal cancer. PLoS One. 2014;9(3):e90849. doi:10.1371/journal.pone.0090849
11. Belcheva A, Irrazabal T, Martin A. Gut microbial metabolism and colon cancer: can manipulations of the microbiota be useful in the management of gastrointestinal health? Bioessays. 2015;37(4):403–412. doi:10.1002/bies.201400204
12. Han S, Wu W, Da M, et al. Adequate lymph node assessments and investigation of gut microorganisms and microbial metabolites in colorectal cancer. Onco Targets Ther. 2020;13:1893–1906. doi:10.2147/OTT.S242017
13. Singh R, Kumar M, Mittal A, et al. Microbial metabolites in nutrition, healthcare and agriculture. 3 Biotech. 2017;7(1):15. doi:10.1007/s13205-016-0586-4
14. Shi Y, Pan C, Wang K, et al. Synthetic multispecies microbial communities reveals shifts in secondary metabolism and facilitates cryptic natural product discovery. Environ Microbiol. 2017;19(9):3606–3618. doi:10.1111/1462-2920.13858
15. Jawad N, Direkze N, Leedham SJ. Inflammatory bowel disease and colon cancer. Recent Results Cancer Res. 2011;185:99–115.
16. Terzic J, Grivennikov S, Karin E, et al. Inflammation and colon cancer. Gastroenterology. 2010;138(6):2101–2114 e5. doi:10.1053/j.gastro.2010.01.058
17. Meyerhardt JA, Sato K, Niedzwiecki D, et al. Dietary glycemic load and cancer recurrence and survival in patients with stage III colon cancer: findings from CALGB 89803. J Natl Cancer Inst. 2012;104(22):1702–1711. doi:10.1093/jnci/djs399
18. Kamada N, Chen GY, Inohara N, et al. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. 2013;14(7):685–690. doi:10.1038/ni.2608
19. Chung H, Pamp S, Hill J, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149(7):1578–1593. doi:10.1016/j.cell.2012.04.037
20. Brenchley JM, Douek DC. Microbial translocation across the GI tract. Annu Rev Immunol. 2012;30(1):149–173. doi:10.1146/annurev-immunol-020711-075001
21. Zeng MY, Cisalpino D, Varadarajan S, et al. Gut microbiota-induced Immunoglobulin G controls systemic infection by symbiotic bacteria and pathogens. Immunity. 2016;44(3):647–658. doi:10.1016/j.immuni.2016.02.006
22. Wang L, Fouts D, Stärkel P, et al. Intestinal REG3 lectins protect against alcoholic steatohepatitis by reducing mucosa-associated microbiota and preventing bacterial translocation. Cell Host Microbe. 2016;19(2):227–239. doi:10.1016/j.chom.2016.01.003
23. Claesson MJ, Jeffery IB, Conde S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488(7410):178–184. doi:10.1038/nature11319
24. Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol. 2011;9(4):279–290. doi:10.1038/nrmicro2540
25. Clarke SF, Murphy EF, O’Sullivan O, et al. Exercise and associated dietary extremes impact on gut microbial diversity. Gut. 2014;63(12):1913–1920. doi:10.1136/gutjnl-2013-306541
26. Marlicz W, Loniewski I. The effect of exercise and diet on gut microbial diversity. Gut. 2015;64(3):519–520. doi:10.1136/gutjnl-2014-307909
27. Salonen A, de Vos WM. Impact of diet on human intestinal microbiota and health. Annu Rev Food Sci Technol. 2014;5(1):239–262. doi:10.1146/annurev-food-030212-182554
28. Cao Y, Wu K, Mehta R, et al. Long-term use of antibiotics and risk of colorectal adenoma. Gut. 2018;67(4):672–678. doi:10.1136/gutjnl-2016-313413
29. Drewes JL, Housseau F, Sears CL. Sporadic colorectal cancer: microbial contributors to disease prevention, development and therapy. Br J Cancer. 2016;115(3):273–280. doi:10.1038/bjc.2016.189
30. Feng Q, Liang S, Jia H, et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun. 2015;6:6528. doi:10.1038/ncomms7528
31. Liang Q, Chiu J, Chen Y, et al. Fecal bacteria act as novel biomarkers for noninvasive diagnosis of colorectal cancer. Clin Cancer Res. 2017;23(8):2061–2070. doi:10.1158/1078-0432.CCR-16-1599
32. Choi C-HR, Bakir IA, Hart AL, et al. Clonal evolution of colorectal cancer in IBD. Nat Rev Gastroenterol Hepatol. 2017;14(4):218–229. doi:10.1038/nrgastro.2017.1
33. Dennis KL, Wang Y, Blatner NR, et al. Adenomatous polyps are driven by microbe-instigated focal inflammation and are controlled by IL-10-producing T cells. Cancer Res. 2013;73(19):5905–5913. doi:10.1158/0008-5472.CAN-13-1511
34. Malik A, Sharma D, Zhu Q, et al. IL-33 regulates the IgA-microbiota axis to restrain IL-1alpha-dependent colitis and tumorigenesis. J Clin Invest. 2016;126(12):4469–4481. doi:10.1172/JCI88625
35. Wong SH, Zhao L, Zhang X, et al. Gavage of Fecal samples from patients with colorectal cancer promotes intestinal carcinogenesis in germ-free and conventional mice. Gastroenterology. 2017;153(6):1621–1633 e6. doi:10.1053/j.gastro.2017.08.022
36. Buford TW. (Dis)Trust your gut: the gut microbiome in age-related inflammation, health, and disease. Microbiome. 2017;5(1):80. doi:10.1186/s40168-017-0296-0
37. Cremonesi E, Governa V, Garzon JFG, et al. Gut microbiota modulate T cell trafficking into human colorectal cancer. Gut. 2018;67(11):1984–1994. doi:10.1136/gutjnl-2016-313498
38. Bultman SJ. Molecular pathways: gene-environment interactions regulating dietary fiber induction of proliferation and apoptosis via butyrate for cancer prevention. Clin Cancer Res. 2014;20(4):799–803. doi:10.1158/1078-0432.CCR-13-2483
39. Ganapathy V, Thangaraju M, Prasad PD, et al. Transporters and receptors for short-chain fatty acids as the molecular link between colonic bacteria and the host. Curr Opin Pharmacol. 2013;13(6):869–874. doi:10.1016/j.coph.2013.08.006
40. Elangovan S, Pathania R, Ramachandran S, et al. The niacin/butyrate receptor GPR109A suppresses mammary tumorigenesis by inhibiting cell survival. Cancer Res. 2014;74(4):1166–1178. doi:10.1158/0008-5472.CAN-13-1451
41. D’Souza WN, Douangpanya J, Mu S, et al. Differing roles for short chain fatty acids and GPR43 agonism in the regulation of intestinal barrier function and immune responses. PLoS One. 2017;12(7):e0180190. doi:10.1371/journal.pone.0180190
42. Coothankandaswamy V, Elangovan S, Singh N, et al. The plasma membrane transporter SLC5A8 suppresses tumour progression through depletion of survivin without involving its transport function. Biochem J. 2013;450(1):169–178. doi:10.1042/BJ20121248
43. Bernstein C, Holubec H, Bhattacharyya AK, et al. Carcinogenicity of deoxycholate, a secondary bile acid. Arch Toxicol. 2011;85(8):863–871. doi:10.1007/s00204-011-0648-7
44. Maillette de Buy Wenniger L, Beuers U. Bile salts and cholestasis. Dig Liver Dis. 2010;42(6):409–418. doi:10.1016/j.dld.2010.03.015
45. Mahmoud NN, et al. Administration of an unconjugated bile acid increases duodenal tumors in a murine model of familial adenomatous polyposis. Carcinogenesis. 1999;20(2):299–303. doi:10.1093/carcin/20.2.299
46. Yin J, Liao S-X, He Y, et al. Dysbiosis of gut microbiota with reduced trimethylamine-N-oxide level in patients with large-artery atherosclerotic stroke or transient ischemic attack. J Am Heart Assoc. 2015;4(11):11. doi:10.1161/JAHA.115.002699
47. Debruyne PR, Bruyneel EA, Li X, et al. The role of bile acids in carcinogenesis. Mutat Res. 2001;480-481:359–369. doi:10.1016/S0027-5107(01)00195-6
48. Magee EA, Richardson CJ, Hughes R, et al. Contribution of dietary protein to sulfide production in the large intestine: an in vitro and a controlled feeding study in humans. Am J Clin Nutr. 2000;72(6):1488–1494. doi:10.1093/ajcn/72.6.1488
49. Greer JB, O’Keefe SJ. Microbial induction of immunity, inflammation, and cancer. Front Physiol. 2011;1:168. doi:10.3389/fphys.2010.00168
50. Yazici C, Wolf PG, Kim H, et al. Race-dependent association of sulfidogenic bacteria with colorectal cancer. Gut. 2017;66(11):1983–1994. doi:10.1136/gutjnl-2016-313321
51. Nguyen LH, Ma W, Wang DD, et al. Association between sulfur-metabolizing bacterial communities in stool and risk of distal colorectal cancer in men. Gastroenterology. 2020;158(5):1313–1325. doi:10.1053/j.gastro.2019.12.029
52. Tjalsma H, Schöller-Guinard M, Lasonder E, et al. Profiling the humoral immune response in colon cancer patients: diagnostic antigens from Streptococcus bovis. Int J Cancer. 2006;119(9):2127–2135. doi:10.1002/ijc.22116
53. Wang Z, Klipfell E, Bennett BJ, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472(7341):57–63. doi:10.1038/nature09922
54. Xu R, Wang Q. Towards understanding brain-gut-microbiome connections in Alzheimer’s disease. BMC Syst Biol. 2016;10(Suppl S3):63. doi:10.1186/s12918-016-0307-y
55. Bae S, Ulrich CM, Neuhouser ML, et al. Plasma choline metabolites and colorectal cancer risk in the women’s health initiative observational study. Cancer Res. 2014;74(24):7442–7452. doi:10.1158/0008-5472.CAN-14-1835
56. Kim K-B, Yang J-Y, Kwack SJ, et al. Toxicometabolomics of urinary biomarkers for human gastric cancer in a mouse model. J Toxicol Environ Health A. 2010;73(21–22):1420–1430. doi:10.1080/15287394.2010.511545
57. Georgescauld F, Mocan I, Lacombe M-L, et al. Rescue of the neuroblastoma mutant of the human nucleoside diphosphate kinase A/nm23-H1 by the natural osmolyte trimethylamine-N -oxide. FEBS Lett. 2009;583(4):820–824. doi:10.1016/j.febslet.2009.01.043
58. Lunn JC, Kuhnle G, Mai V, et al. The effect of haem in red and processed meat on the endogenous formation of N-nitroso compounds in the upper gastrointestinal tract. Carcinogenesis. 2007;28(3):685–690. doi:10.1093/carcin/bgl192
59. Hu Y, Zhao Y, Yuan L, et al. Protective effects of tartary buckwheat flavonoids on high TMAO diet-induced vascular dysfunction and liver injury in mice. Food Funct. 2015;6(10):3359–3372. doi:10.1039/C5FO00581G
60. Ottiger M, Nickler M, Steuer C, et al. Trimethylamine-N-oxide (TMAO) predicts fatal outcomes in community-acquired pneumonia patients without evident coronary artery disease. Eur J Intern Med. 2016;36:67–73. doi:10.1016/j.ejim.2016.08.017
61. Wilson A, Teft WA, Morse BL, et al. Erratum to: trimethylamine-N-oxide: A Novel Biomarker for the Identification of Inflammatory Bowel Disease. Dig Dis Sci. 2016;61(1):325. doi:10.1007/s10620-015-3826-2
62. Wu D, Cao M, Peng J, et al. The effect of trimethylamine N-oxide on Helicobacter pylori-induced changes of immunoinflammatory genes expression in gastric epithelial cells. Int Immunopharmacol. 2017;43:172–178. doi:10.1016/j.intimp.2016.11.032
63. Xu R, Wang Q, Li L. A genome-wide systems analysis reveals strong link between colorectal cancer and trimethylamine N-oxide (TMAO), a gut microbial metabolite of dietary meat and fat. BMC Genomics. 2015;16(Suppl S7):S4. doi:10.1186/1471-2164-16-S7-S4
64. Hebels DGAJ, Jennen DGJ, Kleinjans JCS, et al. Molecular signatures of N-nitroso compounds in Caco-2 cells: implications for colon carcinogenesis. Toxicol Sci. 2009;108(2):290–300. doi:10.1093/toxsci/kfp035
65. Abu-Ghazaleh N, Chua WJ, Gopalan V. Intestinal microbiota and its association with colon cancer and red/processed meat consumption. J Gastroenterol Hepatol. 2020. doi:10.1111/jgh.15042
66. Demeyer D, Honikel K, De Smet S. The World Cancer Research Fund report 2007: a challenge for the meat processing industry. Meat Sci. 2008;80(4):953–959. doi:10.1016/j.meatsci.2008.06.003
67. Robichova S, Slamenova D. Effects of vitamins C and E on cytotoxicity induced by N-nitroso compounds, N-nitrosomorpholine and N-methyl-N’-nitro-N-nitrosoguanidine in Caco-2 and V79 cell lines. Cancer Lett. 2002;182(1):11–18. doi:10.1016/S0304-3835(02)00056-3
68. Lewin MH, Bailey N, Bandaletova T, et al. Red meat enhances the colonic formation of the DNA AdductO6 -carboxymethyl guanine: implications for colorectal cancer risk. Cancer Res. 2006;66(3):1859–1865. doi:10.1158/0008-5472.CAN-05-2237
69. Wiseman M. The second world cancer research fund/american institute for cancer research expert report. Food, nutrition, physical activity and the prevention of cancer: a global perspective. Proc Nutr Soc. 2008;67(3):253–256.
70. McMichael AJ. Food, nutrition, physical activity and cancer prevention. Authoritative report from World Cancer Research Fund provides global update. Public Health Nutr. 2008;11(7):762–763. doi:10.1017/S1368980008002358
71. Homann N. Alcohol and upper gastrointestinal tract cancer: the role of local acetaldehyde production. Addict Biol. 2001;6(4):309–323. doi:10.1080/13556210020077028
72. Hooper SJ, Wilson MJ, Crean SJ, Myers JN. Exploring the link between microorganisms and oral cancer: a systematic review of the literature. Head Neck. 2009;31(9):1228–1239. doi:10.1002/hed.21140
73. Gur C, Ibrahim Y, Isaacson B, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42(2):344–355. doi:10.1016/j.immuni.2015.01.010
74. Yang Y, Weng W, Peng J, et al. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-Like Receptor 4 signaling to nuclear factor-kappaB, and up-regulating expression of MicroRNA-21. Gastroenterology. 2017;152(4):851–866 e24. doi:10.1053/j.gastro.2016.11.018
75. Rubinstein MR, Wang X, Liu W, et al. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14(2):195–206. doi:10.1016/j.chom.2013.07.012
76. Yu T, Guo F, Yu Y, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. 2017;170(3):548–563 e16. doi:10.1016/j.cell.2017.07.008
77. Mima K, Nishihara R, Qian ZR, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016;65(12):1973–1980. doi:10.1136/gutjnl-2015-310101
78. Shadnoush M, Shaker Hosseini R, Mehrabi Y, et al. Probiotic yogurt affects pro- and anti-inflammatory factors in patients with inflammatory bowel disease. Iran J Pharm Res. 2013;12(4):929–936.
79. Ouwehand AC, Salminen S, Isolauri E. Probiotics: an overview of beneficial effects. Antonie Van Leeuwenhoek. 2002;82(1–4):279–289. doi:10.1023/A:1020620607611
80. Sood A, Midha V, Makharia GK, et al. The probiotic preparation, VSL#3 induces remission in patients with mild-to-moderately active ulcerative colitis. Clin Gastroenterol Hepatol. 2009;7(11):1202–9, 1209 e1. doi:10.1016/j.cgh.2009.07.016
81. Rafter J, Bennett M, Caderni G, et al. Dietary synbiotics reduce cancer risk factors in polypectomized and colon cancer patients. Am J Clin Nutr. 2007;85(2):488–496. doi:10.1093/ajcn/85.2.488
82. Commane D, Hughes R, Shortt C, et al. The potential mechanisms involved in the anti-carcinogenic action of probiotics. Mutat Res. 2005;591(1–2):276–289. doi:10.1016/j.mrfmmm.2005.02.027
83. Baldwin C, et al. Probiotic Lactobacillus acidophilus and L casei mix sensitize colorectal tumoral cells to 5-fluorouracil-induced apoptosis . Nutr Cancer. 2010;62(3):371–378.
84. Kahouli I, Tomaro-Duchesneau C, Prakash S. Probiotics in colorectal cancer (CRC) with emphasis on mechanisms of action and current perspectives. J Med Microbiol. 2013;62(Pt 8):1107–1123. doi:10.1099/jmm.0.048975-0
85. Liu Z, Qin H, Yang Z, et al. Randomised clinical trial: the effects of perioperative probiotic treatment on barrier function and post-operative infectious complications in colorectal cancer surgery - a double-blind study. Aliment Pharmacol Ther. 2011;33(1):50–63. doi:10.1111/j.1365-2036.2010.04492.x
86. Gianotti L, et al. A randomized double-blind trial on perioperative administration of probiotics in colorectal cancer patients. World J Gastroenterol. 2010;16(2):167–175. doi:10.3748/wjg.v16.i2.167
87. Xia Y, Yang Z, Chen H-Q, et al. [Effect of bowel preparation with probiotics on intestinal barrier after surgery for colorectal cancer]. Zhonghua Wei Chang Wai Ke Za Zhi. 2010;13(7):528–531. Chinese.
88. Pillar CM, Gilmore MS. Enterococcal virulence–pathogenicity island of E. Faecalis Front Biosci. 2004;9:2335–2346. doi:10.2741/1400
89. Balamurugan R, Rajendiran E, George S, et al. Real-time polymerase chain reaction quantification of specific butyrate-producing bacteria, Desulfovibrio and Enterococcus faecalis in the feces of patients with colorectal cancer. J Gastroenterol Hepatol. 2008;23(8 Pt 1):1298–1303. doi:10.1111/j.1440-1746.2008.05490.x
90. Huycke MM, Joyce W, Wack MF. Augmented production of extracellular superoxide by blood isolates of Enterococcus faecalis. J Infect Dis. 1996;173(3):743–746. doi:10.1093/infdis/173.3.743
91. Huycke MM, Abrams V, Moore DR. Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis. 2002;23(3):529–536. doi:10.1093/carcin/23.3.529
92. Limoli CL, Giedzinski E. Induction of chromosomal instability by chronic oxidative stress. Neoplasia. 2003;5(4):339–346. doi:10.1016/S1476-5586(03)80027-1
93. Wang X, Huycke MM. Extracellular superoxide production by Enterococcus faecalis promotes chromosomal instability in mammalian cells. Gastroenterology. 2007;132(2):551–561. doi:10.1053/j.gastro.2006.11.040
94. Balish E, Warner T. Enterococcus faecalis induces inflammatory bowel disease in interleukin-10 knockout mice. Am J Pathol. 2002;160(6):2253–2257. doi:10.1016/S0002-9440(10)61172-8
95. Wang X, Allen TD, May RJ, et al. Enterococcus faecalis induces aneuploidy and tetraploidy in colonic epithelial cells through a bystander effect. Cancer Res. 2008;68(23):9909–9917. doi:10.1158/0008-5472.CAN-08-1551
96. Sobhani I, Tap J, Roudot-Thoraval F, et al. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS One. 2011;6(1):e16393. doi:10.1371/journal.pone.0016393
97. Yang Y, Xu C, Wu D, et al. γδ T cells: crosstalk between microbiota, chronic inflammation, and colorectal cancer. Front Immunol. 2018;9:1483. doi:10.3389/fimmu.2018.01483
98. Candela M, et al. Inflammation and colorectal cancer, when microbiota-host mutualism breaks. World J Gastroenterol. 2014;20(4):908–922. doi:10.3748/wjg.v20.i4.908
99. Maddocks ODK, Short AJ, Donnenberg MS, et al. Attaching and effacing Escherichia coli downregulate DNA mismatch repair protein in vitro and are associated with colorectal adenocarcinomas in humans. PLoS One. 2009;4(5):e5517. doi:10.1371/journal.pone.0005517
100. Bonnet M, et al. Colonization of the human gut by E coli and colorectal cancer risk. Clin Cancer Res. 2014;20(4):859–867.
101. Raisch J, Rolhion N, Dubois A, et al. Intracellular colon cancer-associated Escherichia coli promote protumoral activities of human macrophages by inducing sustained COX-2 expression. Lab Invest. 2015;95(3):296–307. doi:10.1038/labinvest.2014.161
102. Nougayrede JP, et al. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science. 2006;313(5788):848–851. doi:10.1126/science.1127059
103. Cuevas-Ramos G, Petit CR, Marcq I, et al. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc Natl Acad Sci U S A. 2010;107(25):11537–11542. doi:10.1073/pnas.1001261107
104. Housseau F, Sears CL. Enterotoxigenic Bacteroides fragilis (ETBF)-mediated colitis in Min (Apc±) mice: a human commensal-based murine model of colon carcinogenesis. Cell Cycle. 2010;9(1):3–5. doi:10.4161/cc.9.1.10352
105. Mangan PR, Harrington LE, O’Quinn DB, et al. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441(7090):231–234. doi:10.1038/nature04754
106. Dejea CM, Fathi P, Craig JM, et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science. 2018;359(6375):592–597. doi:10.1126/science.aah3648
107. Wu N, Yang X, Zhang R, et al. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb Ecol. 2013;66(2):462–470. doi:10.1007/s00248-013-0245-9
108. Goodwin AC, Shields CED, Wu S, et al. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc Natl Acad Sci U S A. 2011;108(37):15354–15359. doi:10.1073/pnas.1010203108
109. Rhee K-J, Wu S, Wu X, et al. Induction of persistent colitis by a human commensal, enterotoxigenic Bacteroides fragilis, in wild-type C57BL/6 mice. Infect Immun. 2009;77(4):1708–1718. doi:10.1128/IAI.00814-08
110. Toprak NU, Yagci A, Gulluoglu BM, et al. A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer. Clin Microbiol Infect. 2006;12(8):782–786. doi:10.1111/j.1469-0691.2006.01494.x
111. Wu S, Shin J, Zhang G, et al. The Bacteroides fragilis toxin binds to a specific intestinal epithelial cell receptor. Infect Immun. 2006;74(9):5382–5390. doi:10.1128/IAI.00060-06
112. Corredoira J, Alonso MP, Coira A, et al. Characteristics of Streptococcus bovis endocarditis and its differences with Streptococcus viridans endocarditis. Eur J Clin Microbiol Infect Dis. 2008;27(4):285–291. doi:10.1007/s10096-007-0441-y
113. Lazarovitch T, Shango M, Levine M, et al. The relationship between the new taxonomy of Streptococcus bovis and its clonality to colon cancer, endocarditis, and biliary disease. Infection. 2013;41(2):329–337. doi:10.1007/s15010-012-0314-x
114. Gupta A, Madani R, Mukhtar H. Streptococcus bovis endocarditis, a silent sign for colonic tumour. Colorectal Dis. 2010;12(3):164–171. doi:10.1111/j.1463-1318.2009.01814.x
115. Harrison S, Benziger H, Koerner R. Streptococcus bovis infections, colorectal cancer and liver dysfunction. ANZ J Surg. 2011;81(11):762–763. doi:10.1111/j.1445-2197.2011.05874.x
116. McMahon AJ, Auld CD, Dale BAS, et al. Streptococcus bovis septicaemia associated with uncomplicated colonic carcinoma. Br J Surg. 1991;78(7):883–885. doi:10.1002/bjs.1800780734
117. Wentling GK, Metzger PP, Dozois EJ, et al. Unusual bacterial infections and colorectal carcinoma–Streptococcus bovis and Clostridium septicum: report of three cases. Dis Colon Rectum. 2006;49(8):1223–1227. doi:10.1007/s10350-006-0576-4
118. Klein RS, et al. Streptococcus bovis septicemia and carcinoma of the colon. Ann Intern Med. 1979;91(4):560–562. doi:10.7326/0003-4819-91-4-560
119. Boleij A, Roelofs R, Schaeps RMJ, et al. Increased exposure to bacterial antigen RpL7/L12 in early stage colorectal cancer patients. Cancer. 2010;116(17):4014–4022. doi:10.1002/cncr.25212
120. Ellmerich S, Scholler M, Duranton B, et al. Promotion of intestinal carcinogenesis by Streptococcus bovis. Carcinogenesis. 2000;21(4):753–756. doi:10.1093/carcin/21.4.753
121. Biarc J, et al. Carcinogenic properties of proteins with pro-inflammatory activity from Streptococcus infantarius (formerly. S.bovis). Carcinogenesis. 2004;25(8):1477–1484. doi:10.1093/carcin/bgh091
122. Hong SN, Lee SM, Kim JH, et al. Helicobacter pylori infection increases the risk of colorectal adenomas: cross-sectional study and meta-analysis. Dig Dis Sci. 2012;57(8):2184–2194. doi:10.1007/s10620-012-2245-x
123. Zhang Y, Hoffmeister M, Weck MN, et al. Helicobacter pylori infection and colorectal cancer risk: evidence from a large population-based case-control study in Germany. Am J Epidemiol. 2012;175(5):441–450. doi:10.1093/aje/kwr331
124. Dai YK, Zhang Y-Z, Li D-Y, et al. Interaction of Cyclooxygenase-2 with helicobacter pylori induces gastric chronic nonresolving inflammation and the formation of syndrome of internal block of static blood in helicobacter pylori -related gastric diseases. Evid Based Complement Alternat Med. 2020;2020:7340814. doi:10.1155/2020/7340814
125. Wang TC, Dangler CA, Chen D, et al. Synergistic interaction between hypergastrinemia and Helicobacter infection in a mouse model of gastric cancer. Gastroenterology. 2000;118(1):36–47. doi:10.1016/S0016-5085(00)70412-4
126. Watson SA, Grabowska AM, El-Zaatari M, et al. Gastrin — active participant or bystander in gastric carcinogenesis? Nat Rev Cancer. 2006;6(12):936–946. doi:10.1038/nrc2014
127. Georgopoulos SD, Polymeros D, Triantafyllou K, et al. Hypergastrinemia is associated with increased risk of distal colon adenomas. Digestion. 2006;74(1):42–46. doi:10.1159/000096593
128. Han YM, Park J-M, Kangwan N, et al. Role of proton pump inhibitors in preventing hypergastrinemia-associated carcinogenesis and in antagonizing the trophic effect of gastrin. J Physiol Pharmacol. 2015;66(2):159–167.
129. Chueca E, Lanas A, Piazuelo E. Role of gastrin-peptides in Barrett’s and colorectal carcinogenesis. World J Gastroenterol. 2012;18(45):6560–6570. doi:10.3748/wjg.v18.i45.6560
130. Singh P, Sarkar S, Kantara C, et al. Progastrin peptides increase the risk of developing colonic tumors: impact on colonic stem cells. Curr Colorectal Cancer Rep. 2012;8(4):277–289. doi:10.1007/s11888-012-0144-3
131. Hollande F, Imdahl A, Mantamadiotis T, et al. Glycine-extended gastrin acts as an autocrine growth factor in a nontransformed colon cell line. Gastroenterology. 1997;113(5):1576–1588. doi:10.1053/gast.1997.v113.pm9352860
132. Brown D, Yallampalli U, Owlia A, et al. pp60c-Src Kinase mediates growth effects of the full-length precursor progastrin1-80 peptide on rat intestinal epithelial cells, in vitro. Endocrinology. 2003;144(1):201–211. doi:10.1210/en.2002-220501
133. Koh TJ, Dockray GJ, Varro A, et al. Overexpression of glycine-extended gastrin in transgenic mice results in increased colonic proliferation. J Clin Invest. 1999;103(8):1119–1126. doi:10.1172/JCI4910
134. He H, Pannequin J, Tantiongco J-P, et al. Glycine-extended gastrin stimulates cell proliferation and migration through a Rho- and ROCK-dependent pathway, not a Rac/Cdc42-dependent pathway. Am J Physiol Gastrointest Liver Physiol. 2005;289(3):G478–88. doi:10.1152/ajpgi.00034.2005
135. Konturek SJ, Konturek PC, Hartwich A, et al. Helicobacter pylori infection and gastrin and cyclooxygenase expression in gastric and colorectal malignancies. Regul Pept. 2000;93(1–3):13–19. doi:10.1016/S0167-0115(00)00173-7
136. Hartwich J, Konturek SJ, Pierzchalski P, et al. Molecular basis of colorectal cancer - role of gastrin and cyclooxygenase-2. Med Sci Monit. 2001;7(6):1171–1181.
137. Hartwich A, Konturek SJ, Pierzchalski P, et al. Helicobacter pylori infection, gastrin, cyclooxygenase-2, and apoptosis in colorectal cancer. Int J Colorectal Dis. 2001;16(4):202–210. doi:10.1007/s003840100288
138. Frumento G, Rotondo R, Tonetti M, et al. Tryptophan-derived catabolites are responsible for inhibition of T and natural killer cell proliferation induced by indoleamine 2,3-dioxygenase. J Exp Med. 2002;196(4):459–468. doi:10.1084/jem.20020121
139. Engin AB, et al. Helicobacter pylori and serum kynurenine-tryptophan ratio in patients with colorectal cancer. World J Gastroenterol. 2015;21(12):3636–3643. doi:10.3748/wjg.v21.i12.3636
140. Han S, Pan Y, Yang X, et al. Intestinal microorganisms involved in colorectal cancer complicated with dyslipidosis. Cancer Biol Ther. 2019;20(1):81–89. doi:10.1080/15384047.2018.1507255
141. Tiemersma EW, Kampman E, Bas Bueno de Mesquita H, et al. Meat consumption, cigarette smoking, and genetic susceptibility in the etiology of colorectal cancer: results from a Dutch prospective study. Cancer Causes Control. 2002;13(4):383–393. doi:10.1023/A:1015236701054
142. Schulz MD, Atay Ç, Heringer J, et al. High-fat-diet-mediated dysbiosis promotes intestinal carcinogenesis independently of obesity. Nature. 2014;514(7523):508–512. doi:10.1038/nature13398
143. Lattimer JM, Haub MD. Effects of dietary fiber and its components on metabolic health. Nutrients. 2010;2(12):1266–1289. doi:10.3390/nu2121266
144. Yeh -C-C, Hsieh -L-L, Tang R, et al. Risk factors for colorectal cancer in Taiwan: a hospital-based case-control study. J Formos Med Assoc. 2003;102(5):305–312.
145. Huang XH, Chen L, Gao W, et al. Specific IgG activity of bovine immune milk against diarrhea bacteria and its protective effects on pathogen-infected intestinal damages. Vaccine. 2008;26(47):5973–5980. doi:10.1016/j.vaccine.2008.08.040
146. Bingham SA, Day NE, Luben R, et al. Dietary fibre in food and protection against colorectal cancer in the European Prospective Investigation into Cancer and Nutrition (EPIC): an observational study. Lancet. 2003;361(9368):1496–1501. doi:10.1016/S0140-6736(03)13174-1
147. Glade MJ. Food, nutrition, and the prevention of cancer: a global perspective. American Institute for Cancer Research/World Cancer Research Fund, American Institute for Cancer Research, 1997. Nutrition. 1999;15(6):523–526. doi:10.1016/s0899-9007(99)00021-0
148. Wu K, et al. Calcium intake and risk of colon cancer in women and men. J Natl Cancer Inst. 2002;94(6):437–446. doi:10.1093/jnci/94.6.437
149. Fuchs CS, Willett WC, Colditz GA, et al. The influence of folate and multivitamin use on the familial risk of colon cancer in women. Cancer Epidemiol Biomarkers Prev. 2002;11(3):227–234.
150. J T. Immunotherapy Meets Microbiota. Cell. 2015;163(7):1561.
151. Shuwen H, Xi Y, Quan Q, et al. Relationship between intestinal microorganisms and T lymphocytes in colorectal cancer. Future Oncol. 2019;15(14):1655–1666. doi:10.2217/fon-2018-0595
152. Snyder A, Pamer E, Wolchok J. Immunotherapy. Could microbial therapy boost cancer immunotherapy? Science. 2015;350(6264):1031–1032. doi:10.1126/science.aad7706
153. Vetizou M, Pitt JM, Daillere R, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350(6264):1079–1084. doi:10.1126/science.aad1329
154. Matson V, Fessler J, Bao R, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359(6371):104–108. doi:10.1126/science.aao3290
155. Routy B, Le Chatelier E, Derosa L, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359(6371):91–97. doi:10.1126/science.aan3706
156. Gopalakrishnan V, Spencer CN, Nezi L, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359(6371):97–103. doi:10.1126/science.aan4236
157. Fei Z, Lijuan Y, Xi Y, et al. Gut microbiome associated with chemotherapy-induced diarrhea from the CapeOX regimen as adjuvant chemotherapy in resected stage III colorectal cancer. Gut Pathog. 2019;11(1):18. doi:10.1186/s13099-019-0299-4
158. Stritzker J, Weibel S, Hill P, et al. Tumor-specific colonization, tissue distribution, and gene induction by probiotic Escherichia coli Nissle 1917 in live mice. Int J Med Microbiol. 2007;297(3):151–162. doi:10.1016/j.ijmm.2007.01.008
159. Colley KJ, Kitajima K, Sato C. Polysialic acid: biosynthesis, novel functions and applications. Crit Rev Biochem Mol Biol. 2014;49(6):498–532.
160. Zeller G, Tap J, Voigt AY, et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol Syst Biol. 2014;10(11):766. doi:10.15252/msb.20145645
161. Butt J, Jenab M, Willhauck-Fleckenstein M, et al. Prospective evaluation of antibody response to Streptococcus gallolyticus and risk of colorectal cancer. Int J Cancer. 2018;143(2):245–252. doi:10.1002/ijc.31283
162. Weir TL, Manter DK, Sheflin AM, et al. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS One. 2013;8(8):e70803. doi:10.1371/journal.pone.0070803
163. Wang X, Wang J, Rao B, et al. Gut flora profiling and fecal metabolite composition of colorectal cancer patients and healthy individuals. Exp Ther Med. 2017;13(6):2848–2854. doi:10.3892/etm.2017.4367
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.