Back to Journals » Cancer Management and Research » Volume 11
Prognostic value of miR17-92 family in patients with digestive system cancers: a systematic review and meta-analysis
Authors Zhang X, Zhang S, He Q, Gao X, Yang L, Xu P, Zhao R, Wang Q, Zhang L, Zhao P
Received 28 September 2018
Accepted for publication 22 December 2018
Published 22 January 2019 Volume 2019:11 Pages 1043—1058
DOI https://doi.org/10.2147/CMAR.S189113
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Harikrishna Nakshatri
Xiaoyun Zhang,1 Shijie Zhang,1 Qin He,1 Xiaojuan Gao,1 Lijun Yang,1 Peipei Xu,2 Rui Zhao,2 Qian Wang,3 Lina Zhang,4 Panpan Zhao5
1Department of Clinical Laboratory, First Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan 450052, China; 2Department of Clinical Laboratory, Third Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan 450052, China; 3Department of Medical Laboratory, Fuwai Central China Cardiovascular Hospital, Zhengzhou, Henan 450052, China; 4Blood Transfusion Room, Fuwai Central China Cardiovascular Hospital, Zhengzhou, Henan 450052, China; 5Department of Clinical Laboratory, People’s Liberation Army 159 Hospital, Zhumadian, Henan 463000, China
Abstract: The miR17-92 family is found to be aberrantly expressed and associated with clinicopathological characteristics in patients with various cancers, including digestive system cancers. However, its prognostic value is not yet established. Therefore, we performed a systematic review and meta-analysis to investigate the association between miR17-92-family expression and clinical outcomes in digestive system cancers. We searched the PubMed, Web of Science, Embase, and CNKI (Chinese) databases to retrieve eligible studies up to June 30, 2018. Prognostic data and clinicopathological features of overall survival (OS), disease-free survival (DFS), and progression-free survival (PFS) were extracted to evaluate correlations of the miR17-92 family with digestive system cancers. We used HRs to assess association between miR17-92-family expression and cancers prognosis. A total of 30 qualifying studies involving 4,056 subjects were included in this meta-analysis. Our results indicated that expression levels of miR17-92 can predict poor OS (HR 1.21, 95% CI 1.03–1.39; P=0). However, there was no relationship between the miR17-92 family and DFS (HR 0.86, 95% CI 0.6–1.11; P=0.170) or PFS (HR 1.37, 95% CI 0.83–1.91; P=0). Moreover, miR17-92 was related to TNM stage (III/IV vs I/II, HR 1.37, 95% CI 1.17–1.570; P=0.012), but there was no relationship between miR17-92 and metastasis (HR 1.64, 95% CI 1.34–1.95; P=0.491) or tumor size (≥5 cm vs <5 cm, HR 1.29, 95% CI 1.09–1.49; P=0.586). Subgroup analysis showed that miR17-92 expression was associated with poor OS among the Chinese subgroup (HR 1.28, 95% CI 1.08–1.48; P=0) and tissue samples (HR 1.12, 95% CI 0.93–1.31; P=0), while there was no association with other characteristics. Our results indicated that miR17-92 expression is significantly associated with poor survival in patients with digestive system cancers, suggesting that miR17-92 may be a romising prognostic marker to monitor prognosis and progression of cancers.
Keywords: miR17-92 family, prognosis, digestive system cancer, meta-analysis
Introduction
Digestive system cancers include esophageal squamous-cell carcinoma (ESCC), gastric cancer (GC), gallbladder carcinoma, hepatocellular carcinoma (HCC), pancreatic cancer (PC), colorectal cancer (CRC), and colon cancer (CC). In spite of advanced development in clinical research in recent years, cancer is still the main factor in death worldwide. It is estimated that ~1.7 million people were diagnosed with cancers and 0.6 million people died of malignancies in the US in 2017.1 High morbidity and mortality rates in digestive system cancers remain an important health problem in developing countries.2 Due to the lack of understanding of the molecular mechanisms of cancer, it is difficult to identify a reliable biomarker to detect cancer and find an effective therapeutic agent for clinical application. Several independent factors can be used to identify and evaluate the clinical outcomes of human cancers, consisting of depth of tumor invasion, histological grade, TNM stage, and metastasis to lymph nodes.3 Pathological biopsy is still the gold standard to diagnose diseases, but is an invasive method and needs high requirements for technology. Blood-based tumor biomarkers are used widely to diagnose cancers and predict the prognosis of neoplasms. However, because of low sensitivity and specificity, this detection method is far from satisfactory.4 Therefore, it is essential to find a less invasive and more accurate marker to apply to clinical medicine urgently.
In recent decades, a number of studies have found that miRNA, which belongs to a class of RNA transcripts 20–22 nucleotides in length without a protein-coding function, is closely related to tumor development and progression.5 According to mRNA degradation and translational repression, miRNA can regulate gene expression posttranscriptionally by binding to the 3’-untranslated region of target mRNAs.6 The earliest proof of miRNA involvement in human cancer was provided by Calin et al from studies attempting to identify tumor suppressors at chromosome 13q14 in B-cell chronic lymphocytic leukemia cells.7 In recent decades, numerous articles have indicated that aberrant expression of miRNA in human cancers is related to many processes of tumorigenesis, including cell proliferation, differentiation, angiogenesis, and metastasis.5–7 Cancer cells with abnormal miRNA-expression levels evolve the property to sustain proliferative signaling, evade growth suppressors, resist cell death, activate invasion and metastasis, and induce angiogenesis.8 Roles of miRNAs in human cancers are examined from the viewpoint of dysregulation. Oncogenic miRNAs are involved in the overexpression of cancers, whereas suppressive miRNAs are involved in the downregulation of cancers.9 Because of these fundamental activities, miRNAs have been proven to act as tumor oncogenes or suppressors.10 In addition, miRNA is stable in circulation (such as whole blood, plasma, serum, sputum) and formalin-fixed paraffin-embedded tissue; therefore, it is regarded as a biomarker for cancer diagnosis and prognosis.11
The miR17-92 family, located at human chromosome 13q31, is one of the most extensively studied miRNA clusters and has been shown to play important roles in the pathogenesis of various cancers, including glioma,12 Burkitt’s lymphoma,13 lung cancer,14 osteosarcoma,15 and digestive system cancers. The miR17-92 family has six members (miR17, miR18a, miR19a, miR20a, miR19b, and miR92a) and two paralogues (miR106a and miR106b).16 High expression of the miR17-92 cluster promotes the metastasis of cancers, indicating its role as an oncogene.15 However, studies have suggested that miR17-5p can inhibit metastasis and invasion of tumors.17 Wang et al18 found that higher expression levels of miR17-5p/20a were significantly correlated with poor overall survival (OS). Xue et al19 found that the OS of patients was negatively associated with high levels of miR20b in GCs, but other studies have demonstrated the contrary role of the miR17-92 cluster in cancer outcomes. Fan et al20 identified that patients with lower expression levels of miR20a had significantly poor recurrence-free survival and OS in HCC patients. Therefore, the role of miR17-92 in cancer development and the exact mechanism are not yet consistent.
Previous discrepant results may due to several factors, including sample size, race, detection method, and tumor metastasis. As such, further studies are needed to evaluate the association between the expression of miR17-92 with the prognosis of cancers. A lot of articles have indicated that a similar sequence of miRNAs may regulate a group of target mRNAs and a set of biomarkers may be a better indicator than a single one. Therefore, we conducted this systematic review and meta-analysis to explore the clinical significance of the miR17-92 family as prognostic markers in human digestive system cancers.
Methods
Search strategy
A comprehensive search was performed on the PubMed, Web of Science, Embase, and CNKI (Chinese) databases for articles published to June 30, 2018. Search terms used were “miR17-3p” OR “miR17-5p” OR “miR18a” OR “miR19a” OR “miR19b” OR “miR20a” OR “miR92a” OR “miR106a” OR “miR106b”, “carcinoma” OR “cancer” OR “tumor” OR “malignancy” OR “neoplasia” OR “sarcoma”, “prognosis” OR “prognostic”, “outcome”, and “survival”. In addition, we attempted to find other potential available studies by searching the references and relevant published articles manually. Because this is a systematic review and meta-analysis, ethical approval and patient written informed consent were not required.
Inclusion and exclusion criteria
All eligible studies were reviewed and evaluated based on PRISMA.21 Inclusion criteria were: expression of miR17-92 detected in digestive system cancers; study based on human research; cohort or case–control study; sufficient data to retrieve HRs for survival and corresponding 95% CIs; and published in Chinese or English. We excluded studies if they were conducted on animals or cell lines, were reviews, letters, case reports, conference meetings, or comments, were duplicate publications, or did not provide or had no available data to calculate HRs and 95% CIs. The quality of the included studies was evaluated and examined by the authors after browsing the abstracts and full texts of manuscripts. The final decision was reached by discussion.
Data extraction
According to the inclusion and exclusion criteria, data were extracted by two investigators (PX and RZ) independently. If an article potentially qualified for the meta-analysis, the full text of the study was required. Any discrepancies were resolved by discussion and consensus. Information extracted was first author’s name, publication year, country, total number of patients, cancer type, specimen source, detection method, follow-up time, cutoff value, TNM stage, metastasis, tumor size, HRs with 95% CIs for OS and cancer progression, including disease-free survival (DFS), progression-free survival (PFS), recurrence-free survival, disease-specific survival, cancer-specific survival, cancer-free survival, and event-free survival. All HRs and 95% CIs were extracted from the original literature. If survival data were provided i only n a Kaplan–Meier curve, HRs with 95% CIs were digitized and extracted using Engauge Digitizer (version 4.1) software, designed by Tierney et al.22 Data were extracted from multivariate analyses if both univariate and multivariate results were provided in the same study.
Quality assessment
The quality of the included studies was systematically evaluated by two investigators based on the Newcastle–Ottawa Scale standard,23 which included three parts: selection (4 points), comparability (2 points), and outcome (3 points). Scores range from 0 to 9 points. A study with a score ≥6 points was regarded as high quality.
Statistical analysis
All extracted data were analyzed using Stata software version 12.0 (StataCorp, College Station, TX, USA). Pooled HRs with 95% CIs for OS, DFS and PFS were calculated to estimate the association between miR17-92 expression and prognosis of digestive system cancers. Cochran’ s Q test and Higgins’s I2 statistic were used to evaluate heterogeneity among the selected studies. If P<0.1 or I2>50%, it indicated that heterogeneity existed, and a random effect-model would be used; otherwise, a fixed-effect model would be applied (P>0.1 or I2<50%). Subgroup analysis was conducted to investigate potential heterogeneity based on country, cancer type, and tumor size. Sensitivity analysis was performed to explore the influence of single studies by omitting one study at a time. Additionally, publication bias was assessed using Begg’s and Egger’s tests. P<0.05 was considered statistically significant.
Results
Data selection and study characteristics
A total of 963 articles were retrieved from the online databases and other sources in accordance with the search strategies. After removal of duplicates, there existed 742 studies. After screening of titles and abstracts, 697 were considered ineligible. A total of 45 potential articles were carefully reviewed via full text, and then 15 studies were excluded due to a lack of sufficient data. Finally, 30 eligible studies18–20,24–50 were included in our meta-analysis. The selection flowchart for this meta-analysis is shown in Figure 1.
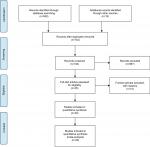  | Figure 1 Flowchart of article-selection process. |
The main characteristics and quality of the eligible studies (2008–2018) assessed with the Newcastle–Ottawa Scale are summarized in Table 1. Among these 30 articles, 21 were from China, four from Japan, two from Spain, one from the US, one from South Korea, and one from Turkey. A total of 4,056 patients were included in our meta-analysis, with a maximum sample size of 735 and a minimum sample size of 22 participants. The type of digestive system cancers included ESCC, GC, HCC, hepatoblastoma, PC, CRC, and CC. HRs with 95% CIs were extracted directly from 26 original studies,18–20,24,26–28,30–38,40–49 while two articles25,50 provided only survival curves, so we indirectly calculated the values from Kaplan–Meier curves based on the method proposed by Tierney.22 Two studies29,39 provided the risk ratios only; therefore, we merged HRs and risk ratios. Quantitative real-time PCR was used to detect miR17-92 expression in 29 studies, while one article used microarrays. Sample types included tissue, serum, and plasma. Because of the variations in cutoff definitions, cutoff values were different in these studies.
Association between miR17-92 expression and clinicopathological features
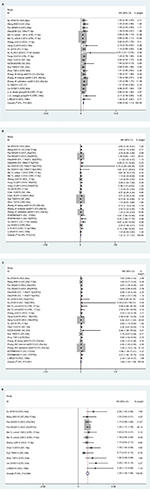
Meta-analysis indicated that there were 23 studies reporting on correlations of miR17-92-expression with sex; however results showed that expression levels were not associated with sex (HR 0.98, 95% CI 0.83–1.13; P=0.940; Figure 2A). There were 27 studies on TNM stage (III/IV vs I/II), and values suggested that miR17-92 expression was linked with advanced TNM stage (HR 1.37, 95% CI 1.17–1.57; P=0.012; Figure 2B). In addition, 24 articles investigated the association between miR17-92 expression and distant metastasis and 12 studies tumor size (≥5 cm vs <5 cm). However, no significance was found for distant metastasis (P=0.491, Figure 2C) or tumor size (P=0.586, Figure 2D). Due to the lack of sufficient data, we did not analyze any association between miR17-92 family expression and other clinicopathological characteristics.
miR17-92 expression and OS
In this meta-analysis, 23 studies reported that expression levels of the miR17-92 family were related to OS. HRs with 95% CIs were retrieved from these articles. We found that miR17-92-expression levels were related to poor OS in digestive system patients (HR 1.21, 95% CI 1.03–1.39; P=0; Figure 3).
  | Figure 3 Forest plot of association between miR17-92 family and overall survival for digestive system cancers. |
To reduce the effect of heterogeneity, we performed a subgroup analysis based on country (Figure 4A), cancer type (Figure 4B), and sample source (Figure 4C). We observed that miR17-92 had a great influence on OS in the HCC subgroup (HR 0.57, 95% CI 0.22–0.91; P=0.020), but no significant effect on GC (P=0.284), CC (P=0.861), CRC (P=0.973), ESCC (P=0.715), or PC (P=0.166). Then, we detected a significant association between miR17-92 expression and poor OS in patients with cancer in the China subgroup (HR 1.28, 95% CI 1.08–1.48; P=0); however, there was no significant effect in the Spain (P=0.273) or Japan (P=0.446) subgroups. Finally, we conducted a subgroup analysis on sample source and found that miR17-92 was strongly related to tissue samples (HR 1.12, 95% CI 0.93–1.31; P=0), while there was no influence on plasma (P=0.697) or serum (P=0.724) samples.
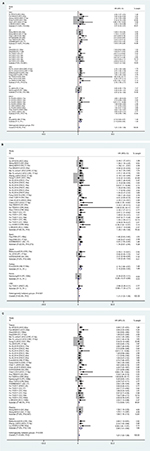  | Figure 4 Forest plot of subgroup analysis according to different group types Note: (A) Country; (B) cancer type; (C) sample source. |
miR17-92 expression and DFS
Eight studies focused on DFS analysis. Due to the relatively high heterogeneity value (I2=74.2%), we used a random-effect model to calculate HRs and 95%CIs for DFS. The results showed that there was no statistical association between miR17-92 expression and cancer DFS (HR 0.86, 95% CI 0.60–1.11; P=0; Figure 5).
  | Figure 5 Forest plot of association between miR17-92 family and disease-free survival for digestive system cancers. |
miR17-92 expression and PFS
PFS analysis was done in five studies. No association was found between miR17-92 expression and cancer PFS (HR 1.37, 95% CI 0.83–1.91; P=0.170; Figure 6). Because only five articles evaluated PFS, this was too small a sample to conduct subgroup analysis for PFS.
  | Figure 6 Forest plot of association between miR17-92 family and progression-free survival for digestive system cancers. |
Sensitivity analysis and publication bias
Sensitivity analysis was conducted with Stata 12.0 to evaluate whether any individual study influenced the overall consequences. The results showed that any single article had little effect on final values (Figure 7). We used Begg’s and Egger’s funnel plots to evaluate the publication bias of the studies included in our meta-analysis, and results indicated that there was no significant publication bias in the pooled analysis of cancer prognosis (Figure 8).
  | Figure 7 Sensitivity analysis of included studies for association between miR17-92 family and overall survival for digestive system cancers. |
  | Figure 8 Begg’s funnel plot of relationship between miR17-92 family and overall survival for digestive system cancers. |
Discussion
The occurrence and development of cancers are multifactorial, multistep, and complicated processes.1 Due to the lack of early diagnostic and prognostic indices, lots of patients are diagnosed in the advanced stage. Recently, many studies have demonstrated that miRNAs play important roles in the tumorigenesis of various cancers, including angiogenesis, cell proliferation, differentiation, invasion, apoptosis, and metastasis.5,6 As such, miRNAs have been considered oncogenes and cancer suppressors. It is hypothesized that miRNAs can regulate more than a third of eukaryotic genes.9 Exploring the functions of miRNAs and their target genes involved in biogenesis may improve understanding of the potential mechanisms of tumor procession and offer significant insights into the diagnosis, prognosis, and treatment of cancers.12,13 Published results have indicated that miRNAs are stable in circulation and tissue and can be promising noninvasive biomarkers for diagnosis and prognosis of cancers.17
Previous research has concluded that aberrant expression of miR17-92 is relevant in cancer development, including Burkitt’s lymphoma,13 breast cancer,17 and GC.18 Many studies have demonstrated that high miR17-92 expression indicates poor clinical characteristics and worse prognosis in digestive system cancers; however, results have not reached agreement as yet. Therefore, we performed the first meta-analysis of eligible studies to evaluate systematically the prognostic significance of miR17-92 in digestive system cancers. In our meta-analysis, the results suggested that high expression levels of miR17-92 represented a risk factor for poor OS (HR 1.21, 95% CI 1.03–1.30; P=0) in digestive system cancers. This demonstrates that the miR17-92 family could be indicators for the prognosis of cancers. Unfortunately, there was no association between miR17-92 expression and DFS (HR 0.86, 95% CI 0.60–1.11; P=0.000) or PFS (HR 1.37, 95% CI 0.83–1.91; P=0.170) in this meta-analysis. Moreover, investigating the effect of pathological features on OS, we found that high expression levels of miR17-92 were significantly associated with TNM stage (III/IV vs I/II, HR 1.37, 95% CI 1.17–1.57; P=0.012), but there was no correlation with metastasis (P=0.491) or tumor size (P=0.586).
In addition, we conducted subgroup analyses to explore the prognostic value of miR17-92 in OS and have successfully acquired some valuable conclusions for clinical application. Results showed that miR17-92 levels predicted poor prognosis in the China subgroup (HR 1.28, 95% CI 1.08–1.48; P=0.000), but not in Spain (P=0.273) or Japan (P=0.446). This diversity may be due to geographical locations, ethnicity, climate, and different lifestyles. Meanwhile, to evaluate the relationship between miR17-92 expression and prognosis based on sample sources, we performed subgroup analyses and found that the tissue subgroup had poor OS (HR 1.12, 95% CI 0.93–1.31; P=0), but no such relationship was found with serum (P=0.724) or plasma (P=0.697). Furthermore, we also noticed that miR17-92 expression was associated with a favorable prognosis in HCC (HR 0.57, 95% CI 0.22–0.91; P=0.02). Then, sensitivity analyses were carried out to assess whether the heterogeneity of data had an effect on results. After removal one study at a time, there was no significant influence on the final outcome. This demonstrated that our results were relatively stable and credible. Also, publication bias did not reach statistical significance.
The miR17-92 cluster was the first miRNA gene implicated in human cancers. However, the potential mechanisms of the miR17-92 cluster in cancer prognosis have not been fully elucidated. Some researchers have demonstrated that miR17-92 functions may be connected with changes in cancer-related proteins and pathways (Figure 9).51,54 Jung et al52 globally investigated Ago2-bound mRNAs and found that miR17-92 obviously repressed numerous targets involved in the instability of mRNA, while the miRNAs repressed expression of their targets, enhanced stability, and lengthened the poly-A tails of nontarget mRNAs. Additionally, the expression of miR17-92 was negatively associated with expression of BTG3, TOB1, CSNK1A1, and ANKRD52 in cancer cell lines. Yang et al53 demonstrated that up-regulation of miR17-92 contributed to the downregulation of QKI2 expression, and then, by decreasing the expression of β-catenin, they inhibited the proliferation, migration and invasion of tumor cells. All these results suggest that miR17-92 can promote tumorigenesis not only by posttranscriptionally increasing global gene expression but also by repressing downstream molecules.
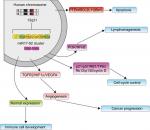  | Figure 9 Underlying biological function of miR17-92 family cluster. |
Strength and limitations
It should be stressed that there are limitations in our meta-analysis. First, most of the included studies were from Asia, while European articles were few in number, which may have been an important source of the heterogeneity and inconsistent results found in our meta-analyses. This emphasizes the need for future studies to test the association of miR17-92 with prognosis in digestive system cancers in Western countries. Second, not all digestive system cancers were included in this meta-analysis. Third, some data25,50 using HRs with 95% CIs were extracted from Kaplan–Meier survival curves, which inevitably brought tiny errors. Fourth, there was no uniform cutoff value to estimate expression levels of miR17-92, and actual values may have been in disagreement due to different algorithms and resulted in some heterogeneity. Finally, there were insufficient data to completely investigate the association between miR17-92 and clinicopathological characteristics of cancers, which needs more studies. Although with some limitations, our study is a comprehensive update, review, and meta-analysis focusing on the correlation of aberrant miR17-92 expression with the development and prognosis of digestive system cancers, providing new insight into the pathogenesis of digestive system cancers.
Conclusion
This systematic review and meta-analysis primarily investigated the expression of miR17-92 and clinical outcomes of patients with digestive system cancers. miR17-92 expression was associated with TNM stage in cancers. Our results demonstrated that the miR17-92 family might be promising prognostic biomarkers for digestive system cancers. Considering the limitations of this meta-analysis, further large-scale and high-quality prospective studies should be performed to validate these findings before clinical guidance using miR17-92 in the prognosis of cancers.
Author contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. | ||
Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. | ||
Svoronos AA, Engelman DM, Slack FJ. OncomiR or tumor suppressor? The Duplicity of microRNAs in cancer. Cancer Res. 2016;76(13):3666–3670. | ||
Perez-Gracia JL, Sanmamed MF, Bosch A, et al. Strategies to design clinical studies to identify predictive biomarkers in cancer research. Cancer Treat Rev. 2017;53:79–97. | ||
Leichter AL, Sullivan MJ, Eccles MR, Chatterjee A. MicroRNA expression patterns and signalling pathways in the development and progression of childhood solid tumours. Mol Cancer. 2017;16(1):15. | ||
Chang CC, Yang YJ, Li YJ, et al. Corrigendum to “MicroRNA-17/20a functions to inhibit cell migration and can be used a prognostic marker in oral squamous cell carcinoma” [Oral Oncol. 49(9) (2013) 923-931]. Oral Oncol. 2017;72:202–203. | ||
Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99(24):15524–15529. | ||
Peng Y, Croce CM. The role of microRNAs in human cancer. Signal Transduct Target Ther. 2016;1(1):15004. | ||
Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20(5):515–524. | ||
Takasaki S. Roles of microRNAs in cancers and development. Methods Mol Biol. 2015;1218:375–413. | ||
Bobbili MR, Mader RM, Grillari J, Dellago H. OncomiR-17-5p: alarm signal in cancer? Oncotarget. 2017;8(41):71206–71222. | ||
Lu S, Wang S, Geng S, Ma S, Liang Z, Jiao B. Increased expression of microRNA-17 predicts poor prognosis in human glioma. J Biomed Biotechnol. 2012;2012(6):1–6. | ||
Robaina MC, Faccion RS, Mazzoccoli L, et al. MiR-17-92 cluster components analysis in Burkitt lymphoma: overexpression of miR-17 is associated with poor prognosis. Ann Hematol. 2016;95(6):881–891. | ||
Chen Q, Si Q, Xiao S, et al. Prognostic significance of serum miR-17-5p in lung cancer. Med Oncol. 2013;30(1):353. | ||
Li X, Yang H, Tian Q, Liu Y, Weng Y. Upregulation of microRNA-17-92 cluster associates with tumor progression and prognosis in osteosarcoma. Neoplasma. 2014;61(04):453–460. | ||
Fedeli M, Riba M, Garcia Manteiga JM, et al. miR-17~92 family clusters control iNKT cell ontogenesis via modulation of TGF-β signaling. Proc Natl Acad Sci USA. 2016;113(51):E8286–E8295. | ||
Li J, Lai Y, Ma J, et al. miR-17-5p suppresses cell proliferation and invasion by targeting ETV1 in triple-negative breast cancer. BMC Cancer. 2017;17(1):745. | ||
Wang M, Gu H, Wang S, et al. Circulating miR-17-5p and miR-20a: molecular markers for gastric cancer. Mol Med Rep. 2012;5(6):1514. | ||
Xue TM, Tao LD, Zhang M, Xu GC, Zhang J, Zhang PJ. miR-20b overexpression is predictive of poor prognosis in gastric cancer. Onco Targets Ther. 2015;8:1871–1876. | ||
Fan MQ, Huang CB, Gu Y, Xiao Y, Sheng JX, Zhong L. Decrease expression of microRNA-20a promotes cancer cell proliferation and predicts poor survival of hepatocellular carcinoma. J Exp Clin Cancer Res. 2013;32(1):21. | ||
Knobloch K, Yoon U, Vogt PM. Preferred reporting items for systematic reviews and meta-analyses (PRISMA) statement and publication bias. J Craniomaxillofac Surg. 2011;39(2):91–92. | ||
Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8(1):16. | ||
Wells G. The Newcastle-Ottawa scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. Appl Eng Agric. 2014;18(6):727–734. | ||
Su X, Wang H, Ge W, et al. An in vivo method to identify microRNA targets not predicted by computation algorithms: p21 targeting by miR-92a in cancer. Cancer Res. 2015;75(14):2875–2885. | ||
Ç E, Aktaş S, Tosun Yildirim H. MicroRNA-17, microRNA-19b, microRNA-146a, MicroRNA-302d expressions in hepatoblastoma and clinical importance. J Pediatr Hematol Oncol. 2018;41(1):7–12. | ||
Díaz R, Silva J, García JM, et al. Deregulated expression of miR-106a predicts survival in human colon cancer patients. Genes Chromosom Cancer. 2010;47(9):794–802. | ||
Su ZX, Zhao J, Rong ZH, Wu YG, Geng WM, Qin CK. Diagnostic and prognostic value of circulating miR-18a in the plasma of patients with gastric cancer. Tumour Biol. 2014;35(12):12119–12125. | ||
Ma Y, Zhang P, Wang F, et al. Elevated oncofoetal miR-17-5p expression regulates colorectal cancer progression by repressing its target gene p130. Nat Commun. 2012;3(1):1291. | ||
Matsumura T, Sugimachi K, Iinuma H, et al. Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br J Cancer. 2015;113(2):275–281. | ||
Zheng J, Dong P, Gao S, Wang N, Yu F. High expression of serum miR-17-5p associated with poor prognosis in patients with hepatocellular carcinoma. Hepatogastroenterology. 2013;60(123):549–552. | ||
Hung CL, Yen CS, Tsai HW, Su YC, Yen CJ. Upregulation of microRNA-19b predicts good prognosis in patients with hepatocellular carcinoma presenting with vascular invasion or multifocal disease. BMC Cancer. 2015;15(1):665. | ||
Yu J, Ohuchida K, Mizumoto K, Fujita H, Nakata K, Tanaka M. MicroRNA miR-17-5p is overexpressed in pancreatic cancer, associated with a poor prognosis, and involved in cancer cell proliferation and invasion. Cancer Biol Ther. 2010;10(8):748–757. | ||
Xu XL, Jiang YH, Feng JG, Su D, Chen PC, Mao WM. MicroRNA-17, microRNA-18a, and microRNA-19a are prognostic indicators in esophageal squamous cell carcinoma. Ann Thorac Surg. 2014;97(3):1037–1045. | ||
Fang L, Li H, Wang L, et al. MicroRNA-17-5p promotes chemotherapeutic drug resistance and tumour metastasis of colorectal cancer by repressing PTEN expression. Oncotarget. 2014;5(10):2974–2987. | ||
Wu CW, Dong YJ, Liang QY, et al. MicroRNA-18a attenuates DNA damage repair through suppressing the expression of ataxia telangiectasia mutated in colorectal cancer. PLoS One. 2013;8(2):e57036. | ||
Chen YJ, Wu H, Zhu JM, et al. MicroRNA-18a modulates p53 expression by targeting IRF2 in gastric cancer patients. J Gastroenterol Hepatol. 2016;31(1):155–163. | ||
Chen ZL, Zhao XH, Wang JW, et al. microRNA-92a promotes lymph node metastasis of human esophageal squamous cell carcinoma via E-cadherin. J Biol Chem. 2011;286(12):10725–10734. | ||
Katada T, Ishiguro H, Kuwabara Y, et al. microRNA expression profile in undifferentiated gastric cancer. Int J Oncol. 2009;34(2):537–542. | ||
Chen L, Jiang M, Yuan W, Tang H. miR-17-5p as a novel prognostic marker for hepatocellular carcinoma. J Invest Surg. 2012;25(3):156–161. | ||
Ke TW, Wei PL, Yeh KT, Chen WT, Cheng YW. MiR-92a promotes cell metastasis of colorectal cancer through PTEN-mediated PI3K/Akt pathway. Ann Surg Oncol. 2015;22(8):2649–2655. | ||
Namkung J, Kwon W, Choi Y, et al. Molecular subtypes of pancreatic cancer based on miRNA expression profiles have independent prognostic value. J Gastroenterol Hepatol. 2016;31(6):1160–1167. | ||
Zhou T, Zhang G, Liu Z, Xia S, Tian H. Overexpression of miR-92a correlates with tumor metastasis and poor prognosis in patients with colorectal cancer. Int J Colorectal Dis. 2013;28(1):19–24. | ||
Zhang JX, Song W, Chen ZH, et al. Prognostic and predictive value of a microRNA signature in stage II colon cancer: a microRNA expression analysis. Lancet Oncol. 2013;14(13):1295–1306. | ||
Valladares-Ayerbes M, Blanco M, Haz M, et al. Prognostic impact of disseminated tumor cells and microRNA-17-92 cluster deregulation in gastrointestinal cancer. Int J Oncol. 2011;39(5):1253–1264. | ||
Hu Y, Correa AM, Hoque A, et al. Prognostic significance of differentially expressed miRNAs in esophageal cancer. Int J Cancer. 2011;128(1):132–143. | ||
Yu G, Tang JQ, Tian ML, et al. Prognostic values of the miR-17-92 cluster and its paralogs in colon cancer. J Surg Oncol. 2012;106(3):232–237. | ||
Liu GH, Zhou ZG, Chen R, et al. Serum miR-21 and miR-92a as biomarkers in the diagnosis and prognosis of colorectal cancer. Tumour Biol. 2013;34(4):2175–2181. | ||
Li J, Liu Y, Wang C, et al. Serum miRNA expression profile as a prognostic biomarker of stage II/III colorectal adenocarcinoma. Sci Rep. 2015;5(1):12921. | ||
Li BK, Huang PZ, Qiu JL, Liao YD, Hong J, Yuan YF. Upregulation of microRNA-106b is associated with poor prognosis in hepatocellular carcinoma. Diagn Pathol. 2014;9:226. | ||
Komatsu S, Ichikawa D, Tsujiura M, et al. Prognostic impact of circulating miR-21 in the plasma of patients with gastric carcinoma. Anticancer Res. 2013;33(1):271. | ||
Jin HY, Lai M, Xiao C. microRNA-17~92 is a powerful cancer driver and a therapeutic target. Cell Cycle. 2014;13(4):495–496. | ||
Jung E, Seong Y, Jeon B, Kwon YS, Song H. MicroRNAs of miR-17-92 cluster increase gene expression by targeting mRNA-destabilization pathways. Biochim Biophys Acta Gene Regul Mech. 2018;1861(7):603–612. | ||
Yang H, Peng Z, Liang M, et al. The miR-17-92 cluster/QKI2/β-catenin axis promotes osteosarcoma progression. Oncotarget. 2018;9(38):25285–25293. | ||
George K, Chao-Yi W, Huang-Yu Y. MiR-17-92 cluster and immunity. J Formos Med Assoc. 2018;pii: S0929-6646(18):30016–30020. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.