Back to Journals » Cancer Management and Research » Volume 13
Prognostic Value of lncRNA DRAIC and miR-3940-3p in Lung Adenocarcinoma and Their Effect on Lung Adenocarcinoma Cell Progression
Authors Liu Z, Yang S, Zhou S, Dong S, Du J
Received 17 May 2021
Accepted for publication 30 August 2021
Published 5 November 2021 Volume 2021:13 Pages 8367—8376
DOI https://doi.org/10.2147/CMAR.S320616
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Beicheng Sun
Zhenghua Liu, Shize Yang, Siyu Zhou, Shiyao Dong, Jiang Du
Department of Thoracic Surgery, The First Affiliated Hospital of China Medical University, Shenyang, 110001, Liaoning Province, People’s Republic of China
Correspondence: Jiang Du
Department of Thoracic Surgery, The First Affiliated Hospital of China Medical University, No. 155 North Nanjing Street, Heping District, Shenyang, Liaoning Province, People’s Republic of China
Tel/Fax +86-24-961200
Email [email protected]
Purpose: Lung adenocarcinoma (LUAD) is a most common malignant tumor, even worse for diseases with relatively poor prognosis. Non-coding RNAs have the potential to be biomarkers for the prognosis of various cancers. LncRNA DRAIC and miR-3940-3p have been screened as dysregulated RNAs in LUAD. The clinical significance and biological function of lncRNA DRAIC and miR-3940-3p in LUAD were assessed in this study.
Patients and Methods: A total of 122 cases of LUAD patients with complete clinical information were enrolled. The expression levels of lncRNA DRAIC and miR-3940-3p were determined via RT-qPCR in LUAD tissues and cells. The relationship between lncRNA DRAIC or miR-3940-3p expression and the clinicopathological features of patients was analyzed based on the Pearson Chi-square test. For the prognostic value, the Kaplan–Meier plot and multi-variate Cox proportional regression analysis were introduced. Finally, the effect of lnc DRAIC and miR-3940-3p on the LUAD cellular function was investigated by CCK-8 and Transwell assay.
Results: lnc DRAIC was upregulated in LUAD tissues and cells, but miR-3940-3p was downregulated. Both of them showed significant associations with and TNM stage, lymph node metastasis, and a poor prognosis. Lnc-DRAIC and miR-3940-3p have the potential as independent prognostic factors for LUAD. Furthermore, the inhibition of lnc DRAIC can inhibit cell proliferation, migration, and invasion of LUAD partly as a ceRNA of miR-3940-3p.
Conclusion: lncRNA DRAIC/miR-3940-3p axis may be involved in the progression of LUAD and can be developed to promising prognostic factors, which may provide new insights into the treatment of LUAD.
Keywords: lung adenocarcinoma, lncRNA DRAIC, miR-3940-3p, progression, prognosis
Introduction
The prognosis outcome of cancer patients is one of the main factors that guide the subsequent treatment, especially for lung cancer with a 19% 5-year relative survival rates.1 Lung cancer, as a heterogeneous disease, comprises several subtypes based on pathologic and clinical relevance, in which non-small cell lung carcinoma (NSCLC) is the main subtype2. Furthermore, the recognition of histologic subtypes of NSCLC, including lung adenocarcinoma (LUAD) as the most frequent subtype, has become important as a determinant of therapy options in this disease.3 In recent years, the identification of abnormalities in the molecular level from a large proportion of patients has allowed the emergence of personalized targeted therapies and has opened new horizons to create new prognosis markers for these patients.4 In turn, utilizing the prognostic predictive biomarkers to identify patients’ survival outcomes has given targeted agents and biotherapies treatment more possibilities and personalization.5 Therefore, it has clinical significance to identify applicable prognostic biomarkers to guide individualized treatment and improve unfavorable prognosis of LUAD patients.
Apart from the protein-coding genes, non-coding RNAs (ncRNAs), comprising long ncRNAs (lncRNAs), microRNAs (miRNAs), and other RNA classes, are an emerging class of transcripts that are coded by the genome but mostly not translated into proteins.6 Among ncRNAs, lncRNAs are transcripts that are longer than 200 nucleotides, while miRNAs are small cellular RNAs with 18 to 24 nucleotides.7 Although not involving in protein translation, ncRNAs act as crucial players in a variety of cellular and physiologic functions.8 The significance of lncRNAs and miRNAs in the carcinogenesis process has been successively underscored, and increasing studies also reported their aberrant expressions in lung cancer samples.9–12 This aberrant expression of lncRNAs and miRNAs has an indicative role in patients’ outcomes in LUAD. For instance, high expression of miR-130b was associated with unfavorable clinical factors and serves as a biomarker for poor prognosis for patients with LUAD.13 Benefited from integrated bioinformatics analysis, five lncRNAs (DIAPH2-AS1, FOXN3-AS2, LINC00652, MEG3, and RHPN1-AS1) have been reported to affect the occurrence and development of LUAD by regulating the expression levels of target genes.14 Similarly, lncRNA DRAIC (lnc DRAIC) and miR-3940-3p have been reported as survival-related lncRNA or miRNA in patients with LUAD from TCGA database.15,16 However, the detailed clinical significance and effects of lnc DRAIC and miR-3940-3p on LUAD have not been investigated and underscored.
So, in this study, the expression level of lnc DRAIC and miR-3940-3p was determined in LUAD tissues and adjacent normal tissues, as well as in cells. Based on the aberrant expressions, the predictive value of lnc DRAIC and miR-3940-3p in LUAD was estimated. Further, their effects on LUAD cell function were investigated.
Materials and Methods
Human LUAD Specimens
Our study comprised 122 patients diagnosed with LUAD at The First Affiliated Hospital of China Medical University with adequate clinical information, and 122 pairs of LUAD tissues and normal adjacent (NA) tissues were collected. All patients underwent surgical resection from 2011 to 2015. Before surgery, no one has received any treatment targeted at LUAD. Tissue samples obtained from the surgery were fresh-frozen and stored at −80°C until analysis. Patients’ overall survival monitoring was performed via telephone follow-up from the day of surgery. The staging of LUAD at diagnosis was classified according to the seventh edition of the International Association for the Study of Lung Cancer (IASLC) tumor, node, metastasis (TNM) classification, seventh edition. All patients provided written informed consent, and the Ethics Committee of The First Affiliated Hospital of China Medical University approved the study protocol. All studies were performed in accordance with the ethical guidelines of the World Medical Association Declaration of Helsinki (revised in 2008).
Cell Lines Culture and Transfection
A set of four human LUAD cells, Calu-3, HCC827, NCI-H441, and NCI-H1975, and a kind of control cells, BEAS-2B cells (an immortalized human bronchial epithelial cell line) were purchased and utilized in this study. All the cells were purchased from the ATCC (Manassas, USA). The RPMI 1640 medium (Corning, USA) containing 10% FBS (Corning, USA) was prepared for the culture of all cells. The cultured condition was in the humidified incubator perfused 5% CO2 and 95 air at 37 °C.
Lnc-DRAIC targeting siRNA (si-DRAIC, 5ʹ-GCUCAACAACGAAAAGCAA-3ʹ), non-specific control siRNA (si-RNA, 5ʹ-UAGCGACUAAACACAUCAA-3ʹ), miR-3940-3p mimic (miR mimic, 5ʹ-CAGCCCGGAUCCCAGCCCACUU-3ʹ), miRNA mimic negative control (mimic NC, 5ʹ-CUCGGGCCAUCCCAGCCCACUU-3ʹ), miR-3940-3p inhibitor (miR inhibitor, 5ʹ- AAGUGGGCUGGGAUCCGGGCUG-3ʹ), and negative control inhibitor (inhibitor NC, 5ʹ-AAGUGGGCUGGGAUGGCCCGAG-3ʹ) were purchased from BioRN Life Science (Nanjing, China). The procedures for transfection of cells were carried out using Lipofectamine™ 3000 Transfection Reagent (ThermoFisher, USA) in line with the manufacturer’s protocol. Shortly, cells were seeded in 6-well plates, incubated with transfection reagent, and indicated sequence. As a negative control, cells were transfected with mimic NC or si-NC or inhibitor NC instead. Mock transfection was prepared as cells incubated solely with RPMI 1640 medium.
Plasmid Constructs and Dual-Luciferase Reporter Assay
Human 3´-UTR sequences containing lnc DRAIC predicted binding sites with miR-3940-3p were amplified by BioRN Life Science and inserted into the pGL3-basic luciferase vector (Promega, USA) formed DRAIC-WT. Constructs containing mutagenized miR-3940-3p binding sites as DRAIC-MUT were obtained from BioRN Life Science. Plasmids were transfected into Calu-3 and HCC827 cells. 7h later, the cells were transfected again with mimic NC or miR-mimic. After 24 h, luciferase activities were quantified using the Dual-Luciferase Reporter Assay System (Promega, USA).
Total RNA Isolation and Quantitative Real-Time Polymerase Chain Reaction (RT-qPCR)
Tissue samples were previously processed in QIAzol Lysis Reagent (Qiagen, Germany) according to the handbook. Total RNA from tissue and cell samples was extracted using the RNeasy Extraction Kit (Qiagen) in line with the manufacturer’s instructions. For the analysis of lnc DRAIC levels, total RNA was reverse-transcribed using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, USA). Lnc DRAIC expression assay was performed using the TaqMan™ Non-coding RNA Assay on a QuantStudio™ 3 Real-Time PCR System (Applied Biosystems), and transcript expression was normalized to the GAPDH levels by using the 2−ΔΔCt formula. The primers for lnc DRAIC were 5ʹ-AATGAGGCGAGGGCTGGGGAACCTA-3ʹ (forward) and 5ʹ-TTGAGGGCGGAGGGTCGGAAATGGC-3ʹ (reverse). The primers for GAPDH were 5ʹ- CCCTTCATTGACCTCAACTACA-3ʹ (forward) and 5ʹ-ATGACAAGCTTCCCGTTCTC-3ʹ (reverse). To measure the expression of miR-3940-3p, the cDNAs were reverse-transcribed by TaqMan miRNA RT kit (Applied Biosystems). The assay was performed using the appropriate TaqMan miRNA assay on a QuantStudio™ 3 Real-Time PCR System (Applied Biosystems). The endogenous control used the ubiquitously expressed U6b snRNA (U6) to normalize miRNA expression by the 2−ΔΔCt formula. The primer for miR-3940-3p was 5ʹ-CTCAAGGACCACCGCATC-3ʹ (forward); The primers for U6 were 5ʹ- CTCGCTTCGGCAGCACA-3ʹ (forward) and 5ʹ- AACGCTTCACGAATTTGCGT-3ʹ (reverse). Each plate was determined in duplicate, and shown as mean ± SD of five independent biological assays.
Cell Proliferation Accessed by Cell Counting Kit-8 (CCK-8) Assay
To estimate growth curves of Calu-3 and HCC827 cells, cells were starved by serum deprivation and then released to growth by reexposure to a complete medium containing serum. In brief, cells at a concentration of 3×103 were seeded into 96-well plates. Before detected, CCK-8 solution (Dojindo, Japan) was added at 0, 24, 48, 72 h and incubate for 2 hours at 37°C and 5% CO2. Cell proliferation curve was based on the corresponding normalized OD values at 450nm by a multifunctional microplate reader (Thermo Fisher Scientific, USA) and each point showed the mean of three independent samples.
Cell Migration and Invasion Determined by Transwell Assay
Cell migration and invasion were detected using HTS Transwell-96 System with 8 μm membrane pore size (Corning, USA). For invasion assay, the upper surface of the chambers was coated with Matrigel (BD Biosciences) preciously, but for migration assay not. Subsequently, cell suspension (containing 1×105 cells) without FBS was placed at the upper chamber, and 500 μL RPMI-1640 medium with 10% FBS was placed at the lower chamber. After incubation at 37°C with 5% CO2 for 24h (migration) or for 48 h (invasion), the cells that did not pass through were wiped off, and the passed cells were fixed, stained, and counted in five random visual fields. Each experiment was performed in triplicate.
Statistical Analysis
Differential expression levels between the LUAD tissues or cells and the normal ones were compared by unpaired t-test. Assessment of difference between groups was carried out with a one-way ANOVA test. Associations between patients’ clinicopathological characteristics and lnc DRAIC or miR-3940-3p were evaluated using the Pearson Chi-Square test. The predictive performances of the biomarker candidates were accessed by multi-Cox regression. The correlations between overall survival and the expression level of lnc DRAIC or miR-3940-3p were estimated via the Kaplan–Meier curves (the Log rank test). P values of less than 0.05 were thought as statistically significant.
Results
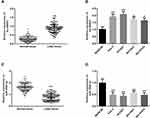
Aberrant Expression of lnc DRAIC and miR-3940-3p Were Detected in LUAD Tissues and Cells
From RT-qPCR assay, lnc DRAIC and miR-3940-3p expression were found with an aberrant level in human LUAD tissue and cells. The expression of lnc DRAIC in LUAD tissues was significantly increased compared to the matched adjacent normal tissues, as well as in LUAD cells and normal lung cells (P < 0.01, Figure 1A and B). The miR-3940-3p expressions were obviously downregulated in LUAD tissues and cells than those in para-cancer normal tissues and cells (P < 0.001, Figure 1C and D).
Expression Levels of lnc DRAIC and miR-3940-3p are Correlated with Clinicopathological Factors of LUAD
Based on the mean value of lnc DRAIC (0.868) and miR-3940-3p (0.759), the patients were classed into low expression and high-expression groups, respectively. The expression levels of lnc DRAIC and miR-3940-3p were included in the Chi-square analysis respectively to access the correlation with various clinicopathological features (Table 1). There was significant relevance between lnc DRAIC and TNM stage (P=0.020), metastasis to lymph nodes (P=0.014), and vascular invasion (P=0.020). miR-3940-3p was significantly correlated to the TNM stage (P=0.020), metastasis to lymph nodes (P=0.039). However, there was no statistically significant correlation between lnc DRAIC or miR-3940-3p with other features.
 |
Table 1 Clinicopathological Parameters and the Expression of Lnc DRAIC and miR-3940-3p Expression in LUAD |
High Expression of lnc DRAIC Predicts Unfavorable Prognosis of LUAD Patients
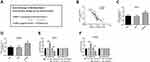
Considering the correlation between lnc DRAIC level and unfavorable clinical factors, the clinical significance of lnc DRAIC in LUAD was worthy to explore further, so Kaplan–Meier curves with the Log rank test and Cox regression analysis were employed. The 5-year survival rate of the LUAD patients whose tumors expressed high levels of lnc DRAIC (45.3%) was significantly lower than that of the patients whose tumors expressed low levels of lnc DRAIC (65.5%). The results of Kaplan–Meier analysis indicated that patients with high expression of lnc DRAIC tended to show shorter overall survival compared with the low-expression patients (P=0.008, Figure 2A). More importantly, among the including features, a high lnc DRAIC level can act as a prognosis biomarker in an independent manner (Table 2).
 |
Table 2 Cox Regression Analysis of Lnc DRAIC Expression and the Clinicopathological Features of LUAD Patients |
Low Expression of miR-3940-3p Was Associated with Poor Prognosis of LUAD Patients
The prognostic value of miR-3940-3p was also explored. When the LUAD patients were stratified into a low (n=64) and a high (n=58) groups with the mean value of the relative miR-3940-3p level, the 5-year survival rate of the LUAD patients with low level of miR-3940-3p was 37.5%, which was significantly lower than that of the patients whose tumors expressed high level of miR-3940-3p (74.1%). Besides, the Kaplan–Meier plot also showed that LUAD patients with a low level of miR-3940-3P expression had a significantly shorter overall survival than those with a high level (P=0.004, Figure 2B). Furthermore, based on the result of Cox regression, a low miR-3940-3p level was significantly correlated with unfavorable survival in an independent manner (Table 3). Taken together, miR-3940-3p can be an independent predictor for prognosis of LUAD patients.
 |
Table 3 Cox Regression Analysis of miR-3940-3p Expression and the Clinicopathological Features of LUAD Patients |
lnc DRAIC Could Bind to miR-3940-3p
LncRNASNP2 database (http://bioinfo.life.hust.edu.cn/lncRNASNP#!/) showed that lnc DRAIC carries putative miR-3940-3p targeting sites (Figure 3A). The expression level of miR-3940-3p was significantly negatively associated with lnc DRAIC expression level in paired tumor tissues and normal tissues (Pearson r=−0.8233, P < 0.0001, Figure 3B). Transfection of si-DRAIC can increase miR-3940-3p level in Calu-3 and HCC827 cells (P < 0.01, Figure 3C and D). Besides, we constructed luciferase reporters containing WT-DRAIC or MUT-DRAIC to co-transfect with miR-3940-3p mimic and found miR-3940-3p mimics can reduce the luciferase activities of the reporter vector containing wild-type WT-DRAIC, but not influence the empty vector or mutant reporter vector in Calu-3 and HCC827 cells (P < 0.001, Figure 3E and F). Based on these results, lnc DRAIC could bind to miR-3940-3p as competing endogenous RNA (ceRNA).
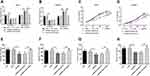
Effect of lnc DRAIC and miR-3940-3p on Cell Proliferation of LUAD Cells
Further, we tried to explore the effects of lnc DRAIC and miR-394-3p on the proliferation of LUAD cells. The si-DRAIC can successfully reduce the expression of lnc DRAIC but increased the expression level of miR-3940-3p, which would be pulled down by miR-3940-3p inhibitor in Calu-3 and HCC827 cells (P < 0.001, Figure 4A and B). CCK-8 assays showed that the inhibition of lnc DRAIC caused by si-DRAIC significantly hindered the proliferation rate of LUAD cells in 72h; however, downregulation of miR-3940-3p can return the cell growth ability (P < 0.01, Figure 4C and D). These results implied inhibition of lnc DRAIC slowed down cell proliferation of LUAD cells, but inhibition of miR-3940-3p can promote it.
Influence of lnc DRAIC and miR-3940-3p on LUAD Cell Migration and Invasion
By transwell assay, the migratory abilities of Calu-3 and HCC827 cells were analyzed after different transfections. Downregulation of lnc DRAIC can attenuate the migratory ability of Calu-3 and HCC827 cells, whereas miR-3940-3p inhibitor can recover that (P < 0.01, Figure 4E and F). Moreover, the invasive ability of Calu-3 and HCC827 cells exhibited a suppression when lnc DRAIC was downregulated by si-DRAIC, nevertheless resumed by miR-3940-3p inhibitor (P < 0.05, Figure 4G and H). These results implied that lnc DRAIC can promote the migration and invasion of LUAD cells but miR-3940-3p suppresses them.
Discussion
Lung cancer remains the most common cause of cancer death worldwide though advances in screening and oncological treatments continued.17 As the most common histological subtype of NSCLC, the 5-year survival rate of LUAD was less than satisfactory showing as 59% for localized adenocarcinoma, 31.7% for regional one, and only 5.8% if distant metastasis.18 The prognosis of LUAD depends on the adenocarcinoma type as well as the stage at diagnosis.19 The TNM staging with other factors such as the site of metastasis, tumor burden, eligibility has a great impact on prognosis.20 However, the performance status and individual variations in the gene would influence the predictive effectiveness of these factors.21 Correlations between gene variations in LUAD and prognosis have been demonstrated by numerous studies.22,23 RNA molecules are another significant group of molecular factors with the prognostic potential in LUAD. NcRNAs have a great impact on the overall survival of LUAD patients and indicate the personal therapy treatment.24,25
Among the ncRNA molecules, lncRNAs are emerging multifunctional regulators in many biological processes.26 In parallel, a rapidly increasing number of studies have unraveled the associations between aberrant lncRNA expression and human cancer prognosis.27,28 Lnc-DRAIC (also known as RP11-279F6.1 or LOC145837), is a long non-coding RNA associated with adverse features of breast cancer and advanced clinical stages of nasopharyngeal carcinoma patients.29,30 This study tried to explore the clinical significance of lnc DRAIC in LUAD. Firstly, the expression of lnc DRAIC was found with a significant increase in LUAD tissues and cells. This abnormal expression implied the underly correlation between lnc DRAIC and prognosis. By the Chi-square test, the higher lnc DRAIC expression level was bound up with TNM III-IV stage, metastasis to lymph nodes, and positive vascular invasion status. With the verification of the Kaplan–Meier plot and Cox regression, patients with high lnc DRAIC levels represented shorter survival time and lower survival rate and lnc DRAIC level was a prognostic predictor in an independent manner. In the latest study, lnc DRAIC also showed the ability of prognosis in a 5 immune-related lncRNA signature established for predicting prognosis of patients with early-stage LUAD.31 All these results indicate that aberrant lnc DRAIC level can be developed to be a mighty biomarker for LUAD prognosis.
Apart from lncRNAs, miRNAs were also increasingly studied with regard to their prognostic value in LUAD.32 miR-3940-5p has been reported to present significant downregulation in NSCLC tissues and correlation with clinicopathological features.33,34 But the role of miR-3940-3p in LUAD has not been clarified. In this study, the expression of miR-3940-3p was determined by RT-qPCR, and a downregulated expression level was obtained in LUAD tissues and cells. Then, the low expression of miR-3940-3p was found to be related to TNM stage and metastasis to lymph nodes. Moreover, LUAD patients with low miR-3940-3p showed a low survival rate and overall survival time. By COX regression, low miR-3940-3p level shows excellent ability in poor prognosis prediction of LUAD. This finding is in line with the study of Xin et al, which constructed a prognostic model with nine miRNAs including hsa-mir-3940 by bioinformatics analysis method.16 So, miR-3940-3p was a potential indicator for the survival of LUAD patients in an independent manner.
Since dysregulation of lncRNAs can play regulatory roles in cancers, increasing emphasis has been placed on their underlying mechanism in tumors to supply the basis of novel molecular targeted therapies.35 Many studies have demonstrated lncRNAs involved in competitive regulatory interactions and can act as microRNA decoys to modulate gene expression, which is known as ceRNA.36,37 In this study, lnc DRAIC has potential binding sites with miR-3940-3p, and the siRNA of lnc DRAIC can significantly increase the expression level of miR-3940-3p. Dual-luciferase reporter assay confirmed the binding between lnc DRAIC and miR-3940-3p. In the cellular function study, downregulation of lnc-DARIC can inhibit the proliferative, migratory, and invasive capacity of LUAD cells, implying the promotor role of lnc DRAIC in LUAD progression. However, the suppression caused by si-DARIC in cell function can be restored by downregulation of miR-3940-3p, which implied the inhibitor role of miR-3940-3p in LUAD progression and further verified the binding relationship between lnc DARIC and miR-3940-3p. Therefore, it is speculated that lnc DRAIC acting as ceRNA of miR-3940-3p promotes the progression of LUAD.
This study brought light to the possibility of prognostic factors in the LUAD and targeted therapy, but further research is needed on a larger number of patients and clinical verification.
Conclusion
In conclusion, lnc DRAIC was found to be significantly upregulated and miR-3940-3p was downregulated in LUAD tissues and cell lines. The dysregulation of lnc DRAIC and miR-3940-3p expression showed a great significance in the prognosis of LUAD. In addition, inhibition of lnc DRAIC significantly suppressed LUAD cell proliferation, migration, and invasion by sponging miR-3940-3p.
Acknowledgments
This study was supported by the China Postdoctoral Science Foundation-Funded Project (Project No. 2018M640266).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34.
2. Nasim F, Sabath BF, Eapen GA. Lung cancer. Med Clin North Am. 2019;103(3):463–473. doi:10.1016/j.mcna.2018.12.006
3. Villalobos P, Wistuba II. Lung cancer biomarkers. Hematol Oncol Clin North Am. 2017;31(1):13–29. doi:10.1016/j.hoc.2016.08.006
4. Jotte RM, Spigel DR. Advances in molecular-based personalized non-small-cell lung cancer therapy: targeting epidermal growth factor receptor and mechanisms of resistance. Cancer Med. 2015;4(11):1621–1632.
5. Naylor EC, Desani JK, Chung PK. Targeted therapy and immunotherapy for lung Cancer. Surg Oncol Clin N Am. 2016;25(3):601–609. doi:10.1016/j.soc.2016.02.011
6. Xue Y, Chen R, Qu L, Cao X. Noncoding RNA: from dark matter to bright star. Sci China Life Sci. 2020;63(4):463–468. doi:10.1007/s11427-020-1676-5
7. Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Rev Cancer. 2021;21(1):22–36. doi:10.1038/s41568-020-00306-0
8. Lekka E, Hall J. Noncoding RNAs in disease. FEBS Lett. 2018;592(17):2884–2900. doi:10.1002/1873-3468.13182
9. Byun Y, Choi YC, Jeong Y, et al. MiR-200c downregulates HIF-1α and inhibits migration of lung cancer cells. Cell Mol Biol Lett. 2019;24(1):28. doi:10.1186/s11658-019-0152-2
10. Chen Z, Chen X, Lu B, et al. Up-regulated LINC01234 promotes non-small-cell lung cancer cell metastasis by activating VAV3 and repressing BTG2 expression. J Hematol Oncol. 2020;13(1):7. doi:10.1186/s13045-019-0842-2
11. Liu C, Yang Z, Deng Z, et al. Upregulated lncRNA ADAMTS9-AS2 suppresses progression of lung cancer through inhibition of miR-223-3p and promotion of TGFBR3. IUBMB Life. 2018;70(6):536–546. doi:10.1002/iub.1752
12. Chen TJ, Gao F, Yang T, et al. LncRNA HOTAIRM1 inhibits the proliferation and invasion of lung adenocarcinoma cells via the miR-498/WWOX axis. Cancer Manag Res. 2020;12:4379–4390. doi:10.2147/CMAR.S244573
13. Kim Y, Kim H, Bang S, Jee S, Jang K. MicroRNA-130b functions as an oncogene and is a predictive marker of poor prognosis in lung adenocarcinoma. Lab Invest. 2021;101(2):155–164. doi:10.1038/s41374-020-00496-z
14. Li J, Yu X, Liu Q, et al. Screening of important lncRNAs associated with the prognosis of lung adenocarcinoma, based on integrated bioinformatics analysis. Mol Med Rep. 2019;19(5):4067–4080.
15. Yu X, Zhang Y. Identification of a long non-coding RNA signature for predicting prognosis and biomarkers in lung adenocarcinoma. Oncol Lett. 2020;19(4):2793–2800.
16. Xin G, Cao X, Zhao W, et al. MicroRNA expression profile and TNM staging system predict survival in patients with lung adenocarcinoma. Math Biosci Eng. 2020;17(6):8074–8083. doi:10.3934/mbe.2020409
17. The Lancet. Lung cancer: some progress, but still a lot more to do. Lancet. 2019;394(10212):1880.
18. Kuhn E, Morbini P, Cancellieri A, Damiani S, Cavazza A, Comin CE. Adenocarcinoma classification: patterns and prognosis. Pathologica. 2018;110(1):5–11.
19. Yu W, Zhao Q, Xia C, et al. Validation of stage groupings in the eighth edition of the tumor node metastasis classification for lung adenocarcinoma. Thorac Cancer. 2019;10(3):483–491. doi:10.1111/1759-7714.12961
20. Travis WD, Asamura H, Bankier AA, et al. The IASLC lung cancer staging project: proposals for coding T categories for subsolid nodules and assessment of tumor size in part-solid tumors in the forthcoming eighth edition of the TNM classification of lung cancer. J Thorac Oncol. 2016;11(8):1204–1223. doi:10.1016/j.jtho.2016.03.025
21. Wang L, Anraku M, Sato M, et al. Impact of the 8th edition of the UICC-TNM classification on clinical stage 0-IA lung adenocarcinoma: does the new classification predict postoperative prognosis more precisely than the previous one? Ann Thorac Cardiovasc Surg. 2018;24(5):223–229. doi:10.5761/atcs.oa.18-00051
22. Qu Y, Cheng B, Shao N, et al. Prognostic value of immune-related genes in the tumor microenvironment of lung adenocarcinoma and lung squamous cell carcinoma. Aging. 2020;12(6):4757–4777. doi:10.18632/aging.102871
23. Jiang H, Xu S, Chen C. A ten-gene signature-based risk assessment model predicts the prognosis of lung adenocarcinoma. BMC Cancer. 2020;20(1):782. doi:10.1186/s12885-020-07235-z
24. Shukla S, Evans JR, Malik R, et al. Development of a RNA-seq based prognostic signature in lung adenocarcinoma. J Natl Cancer Inst. 2017;109(1):djw200.
25. Ghafouri-Fard S, Shoorei H, Branicki W, Taheri M. Non-coding RNA profile in lung cancer. Exp Mol Pathol. 2020;114:104411. doi:10.1016/j.yexmp.2020.104411
26. Chi Y, Wang D, Wang J, Yu W, Yang J. Long non-coding RNA in the pathogenesis of cancers. Cells. 2019;8(9):1015.
27. Yin D, Lu X, Su J, et al. Long noncoding RNA AFAP1-AS1 predicts a poor prognosis and regulates non-small cell lung cancer cell proliferation by epigenetically repressing p21 expression. Mol Cancer. 2018;17(1):92. doi:10.1186/s12943-018-0836-7
28. Zhou H, Zhang H, Chen J, et al. A seven-long noncoding RNA signature predicts relapse in patients with early-stage lung adenocarcinoma. J Cell Biochem. 2019;120(9):15730–15739. doi:10.1002/jcb.28842
29. Liao B, Wang Z, Zhu Y, Wang M, Liu Y. Long noncoding RNA DRAIC acts as a microRNA-122 sponge to facilitate nasopharyngeal carcinoma cell proliferation, migration and invasion via regulating SATB1. Artif Cells Nanomed Biotechnol. 2019;47(1):3585–3597. doi:10.1080/21691401.2019.1656638
30. Zhao D, Dong JT. Upregulation of long non-coding RNA DRAIC correlates with adverse features of breast cancer. Noncoding RNA. 2018;4(4):39.
31. Mu L, Ding K, Tu R, Yang W. Identification of 4 immune cells and a 5-lncRNA risk signature with prognosis for early-stage lung adenocarcinoma. J Transl Med. 2021;19(1):127. doi:10.1186/s12967-021-02800-x
32. Wang C, Tang X, Wang J, Xu Y. Patterns of immune infiltration in lung adenocarcinoma revealed a prognosis-associated microRNA-mast cells network. Hum Cell. 2020;33(1):205–219. doi:10.1007/s13577-019-00300-1
33. Sun Y, Su B, Zhang P, et al. Expression of miR-150 and miR-3940-5p is reduced in non-small cell lung carcinoma and correlates with clinicopathological features. Oncol Rep. 2013;29(2):704–712. doi:10.3892/or.2012.2152
34. Ren K, Li Y, Lu H, Li Z, Han X. miR-3940-5p functions as a tumor suppressor in non-small cell lung cancer cells by targeting cyclin D1 and ubiquitin specific peptidase-28. Transl Oncol. 2017;10(1):80–89. doi:10.1016/j.tranon.2016.11.004
35. Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochim Biophys Acta. 2014;1839(11):1097–1109.
36. Chan JJ, Tay Y. Noncoding RNA:RNA regulatory networks in cancer. Int J Mol Sci. 2018;19(5):1310.
37. Chen R, Wang X, Zhou S, Zeng Z. LncRNA HOXA-AS2 promotes tumor progression by suppressing miR-567 expression in oral squamous cell carcinoma. Cancer Manag Res. 2021;13:5443–5455. doi:10.2147/CMAR.S305946
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.