Back to Journals » International Journal of General Medicine » Volume 14
Prognostic Value and Clinical Significance of LIPH in Breast Cancer
Authors Gao P, Liu Q, Ai B, Fang Y , Wang Z, Wang J
Received 2 August 2021
Accepted for publication 26 October 2021
Published 2 November 2021 Volume 2021:14 Pages 7613—7623
DOI https://doi.org/10.2147/IJGM.S332233
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Peng Gao,* Qiang Liu,* Bolun Ai,* Yi Fang, Zhongzhao Wang, Jing Wang
Department of Breast Surgical Oncology, National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, 100021, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Zhongzhao Wang; Jing Wang Email [email protected]; [email protected]
Background: Lipase member H (LIPH), a novel member of the mammalian triglyceride lipase family, is localized on human chromosome 3q27-q28. Exploration of the importance of the new cancer-related gene LIPH in several carcinomas has been reported in previous studies. Our study aims to systematically assess the expression pattern of LIPH in breast cancer.
Methods: Our study explored 2994 breast cancer samples with transcriptome data from the Cancer Genome Atlas (TCGA) and the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) datasets. We systematically evaluated the mRNA expression of LIPH in breast cancer and the overall survival (OS) of patients. The protein expression of LIPH in breast cancer was evaluated with the Human Protein Atlas. We also explored the relationship between LIPH and the immune microenvironment in pan-cancer.
Results: Both mRNA and protein expression LIPH were found to be upregulated in breast cancer tumors. The overall survival rate of patients with high LIPH expression was lower than those of patients with low LIPH expression in both the TCGA dataset (p=0.0067) and METABRIC dataset (p< 0.0001). Outcomes of the multivariate analysis found that the level of LIPH expression was an independent prognostic factor in both TCGA (p=0.001) and METABRIC (p=0.019) databases. The outcomes of the univariate analysis showed that LIPH was an important prognostic factor (p=0.01 in TCGA dataset, p=0.001 in METABRIC dataset). In the TCGA dataset, outcomes showed that LIPH expression was negatively correlated with the AJCC (American Joint Committee on Cancer) stage (p=2.3e-05) and triple-negative breast cancer (TNBC) tissues (p=3.1e-10). High LIPH expression showed lower OS in the TNBC subtype (P=0.011).
Conclusion: Compared to normal tissues, the expression of LIPH was higher in breast cancer tissues in both mRNA and protein levels. This study showed that the high level of LIPH expression might be related to the worse prognosis of breast cancer.
Keywords: breast cancer, lipase member H, LIPH, prognostic factor, immune infiltrates
Introduction
Lipase member H (LIPH), which is localized on human chromosome 3q27-q28, is a novel member of the mammalian triglyceride lipase family.1 LIPH is also called mPA-PLA1, which encodes phospholipase A1, the preferred phospholipase associated with membranes. And phospholipase A1 can decompose the phosphatidic acid into lysophosphatidic acid (LPA). LPA is a lipid mediator with multiple cellular responses including platelet aggregation, smooth muscle contraction, transcellular migration, and growth factor-like biological activities.2–5 Previous studies have shown that high levels of LPA in the ascites of ovarian cancer patients indicate that it has a positive effect on the development of ovarian cancer.6 The transactivation of epidermal growth factor receptor (EGFR) is shown to be induced by the LPA-mediated signal transduction in colorectal cancer cells.7
Breast cancer is the most common malignancy in women, and the latest cancer statistics show that breast cancer alone accounts for 30% of female cancers. An estimated 43,600 Americans will die of breast cancer in 2021. Approximately, 281,550 cases of female breast cancer will be diagnosed in 2021.8
Previous studies mostly focused on the mutation and correlation of LIPH, which is the main reason for autosomal recessive woolly hair/hypotrichosis.9–11 Recent years, studies of LIPH on cancers have been increasingly investigated. Orozco-terWengel et al12 showed that the expression of LIPH was related to immunity in cattle. And Zhuang et al13 further studied the associations between the expression of LIPH and pancreatic cancer immune infiltration. They found that LIPH was a novel unfavorable prognostic biomarker correlated with immunosuppression in pancreatic cancer. Other studies related to the LIPH expression and lung cancer, esophageal adenocarcinoma and papillary thyroid carcinoma were also reported.1,2,14 However, the specific expression of LIPH for patients with breast cancer and their relationships with breast cancer’s biological behavior remain unclear. A few studies further investigated the potential functional effects in large samples of different types and stages of breast cancer.
Therefore, two independent datasets, the Cancer Genome Atlas (TCGA) and the Breast Cancer International Consortium (METABRIC), are utilized in our study. Breast cancer and other human tumors are analyzed using the TCGA Research Network, and TCGA shows the different expression levels of LIPH in different types and stages of breast cancer tissue. Comprehensive molecular portraits of breast cancer in invasive lobular and invasive ductal carcinomas were previously reported in TCGA Research Network.15,16 About 2000 breast cancers’ transcriptomic and genomic architecture were released in the METABRIC. And it is believed that the METABRIC database provides prospects for understanding how LIPH expression affects prognosis and migration, and reveals a new subgroup that might become the target of future breast cancer research.17,18
Our study aims to systematically evaluate the LIPH expression of breast cancer among different clinical parameters. And two independent comprehensive breast cancer datasets (TCGA and METABRIC) are used to further demonstrate the prognostic value of LIPH for patients with breast cancer.
Methods
Data Acquisition
The websites of TCGA (https://cancergenome.nih.gov/), Synapse (https://www.synapse.org/), and cBioPortal (http://www.cbioportal.org/) were used to download the TCGA and METABRIC databases. In 2019, 233 cancer genomics research datasets could be found in cBioPortal, including 14 breast cancer research. This study used the TCGA dataset on breast invasive carcinoma (TCGA, Provisional) and METABRIC dataset on breast cancer (METABRIC, Nature 2012) for further investigation of LIPH expression in breast cancer. Our study was approved by the Ethics Committee of Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College.
Statistical Analyses
In our study, the associations between clinicopathologic characteristics and LIPH expression were analyzed with Chi-square tests. The differences in LIPH expression between clinicopathologic characteristics were assessed with a one-way analysis of variance (ANOVA) or t-test. Kaplan-Meier method was utilized for the estimation of survival analysis. The Log rank test was utilized to analyze any differences in survival. The prognostic role of LIPH expression was evaluated with the methods of univariate and multivariate Cox proportional hazards regression. Gene expression correlations were analyzed using the Spearman correlation. R software version 3.6.0 was used for the whole statistical analysis. And it was deemed to be statistically significant if P<0.05.
Results
Clinicopathologic Characteristics According to the Expression Level of LIPH
The total number of study subjects in the TCGA dataset was 1090, and the total number of subjects in the METABRIC dataset was 1904. Tables 1 and 2 summarize the baseline clinicopathological characteristics of the subjects. In TCGA dataset, patients with age <55 were more common in the LIPH high-expression group (p<0.001). Compared with the low LIPH-expression group, the proportion of estrogen receptor-positive subjects was higher in the high LIPH expression group (p=0.0274). However, a higher proportion of HER2-positive subjects was found in the high LIPH expression group (p=0.012). In METABRIC dataset, no other important difference in clinicopathologic factors was found except tumor grade (p=0.0226). The expression of LIPH in breast cancer among different clinical parameters was also further explored in our study. In the TCGA dataset, LIPH expression was observed to be upregulated in patients with breast cancer (p=0.001) (Figure 1A). In addition, outcomes showed that LIPH expression was negatively correlated with the AJCC (American Joint Committee on Cancer) stage and T stage (Figure 1B and C). Compared with the luminal A subtype, LIPH expression was downregulated in the basal subtype, but differences in the Her2 subtype, and no statistical significance were found in the luminal B subtype and normal tissue subtype (Figure 1D). Significantly upregulation of LIPH expression was found in the ER positive group, PR positive group, and HER2 positive group (Figure 1E–G). LIPH expression was significantly downregulated in triple-negative breast cancer (TNBC) tissues (Figure 1H).
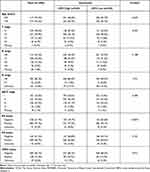 |
Table 1 Clinicopathologic Characteristics According to Expression Level of LIPH in TCGA Databases |
 |
Table 2 Clinicopathologic Characteristics According to Expression Level of LIPH in METABRIC Databases |
 |
Figure 1 LIPH expression pattern in breast cancer in TCGA database (A–H). (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). Abbreviation: ns, no significance. |
Survival Analyses According to Expression Level of LIPH
Compared with the low LIPH expression group, the high LIPH expression group was found to have worse overall survival (OS) in the TCGA dataset (p=0.0067, Figure 2A). In terms of proportion alive, the OS of the group with high LIPH expression was also worse than that of the group with low LIPH expression in the METABRIC dataset (p<0.0001, Figure 2B). In addition, we evaluated the survival curve of LIPH expression at the subtype level. In TCGA dataset, Figure 2C observes that the high LIPH expression group showed lower OS for PAM50, luminal A subtype (p=0.039). High LIPH expression showed lower OS in the TNBC subtype (P=0.011, Figure 2D). And there were no significant statistical differences in other subtypes (Figures 2E–H). Figure 3A finds that high LIPH expression group showed lower OS for basal subtype (p=0.014) and Her2 subtype in METABRIC dataset (p=0.014, Figure 3B). And low LIPH expression group showed higher OS in both the TNBC subtype (P=0.0082) and Non-TNBC subtype (P=0.0018) (Figures 3C and D). Other subtypes expressed no significant statistical differences (Figure 3E and F).
Univariable and Multivariable Analyses
We used the univariate and multivariate analyses to further assess the prognostic value of LIPH in breast cancer on independent TCGA and METABRIC datasets. Both TCGA (p=0.01) and METABRIC (p=0.001) datasets found that the expression level of LIPH played an important role in univariable analysis. Based on the outcomes of TCGA and METABRIC datasets multivariate analysis (p=0.001, p=0.019), the expression of LIPH of breast cancer was also an independent and important prognostic factor (Tables 3 and 4).
 |
Table 3 Univariable Analyses Regarding Proportion Alive Using TCGA Databases |
 |
Table 4 Univariable Analyses Regarding Proportion Alive Using METABRIC Databases |
Immunohistochemistry Staining of LIPH Based on the Human Protein Atlas
The Human Protein Atlas was used in our study to evaluate the immunohistochemistry (IHC) data pertaining to the protein expression of LIPH in breast cancers and normal tissues. And the result showed that the protein expression of LIPH was higher in breast cancer tissues than in normal tissues (Figure 4).
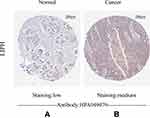 |
Figure 4 Immunohistochemistry staining of LIPH in the normal breast tissue (A) and breast cancer (B). |
Correlation Landscape of LIPH Expression and Immune Cell Infiltration in Pan-Cancer
In order to further evaluate the relationship between LIPH and the immune microenvironment in pan-cancer, 33 cancer types (including a total of 10,201 tumor samples) were enrolled in our study to depict the correlation pattern between LIPH expression and immune cell infiltration (Figure 5). To further investigate the relationship between LIPH and the immune microenvironment in pan-cancer, we depicted the correlation pattern between LIPH expression and immune cell infiltration across 33 cancer types, including a total of 10,201 tumor samples (Figure 5). Our results found that LIPH showed varied correlation patterns with immune cell populations across different cancer types. LIPH was correlated with few immune cell infiltrations in ACC, GBM, SARC, SKCM, STAD, KICH, LGG, OV, PCPG and UCEC, while LIPH was correlated with many types of immune cell infiltration, such as THCA and BRCA, indicating the complex heterogenicity of the different tumor microenvironment. In BRCA, LIPH expression showed a negative correlation with T cell-related infiltration, Macrophage M1 and NK (Natural killer cell) cell activated, while positively correlated with Monocyte, Mast cell activated, Macrophage M2, and B cell naïve.
Discussion
The triglyceride lipase gene family can encode proteins such as pancreatic lipase (PNLIP), lipoprotein lipase (LPL), hepatic lipase (LIPC), endothelial lipase (LIPG) and so on, which can hydrolyze triglycerides and phospholipids to produce fatty acids, thereby promoting intestinal absorption or for energy production or storage.19 LIPH, as a novel member of the mammalian triglyceride lipase family, can encode membrane-associated phosphatidic acid-selective phospholipase A1a. And A1a can produce lysophosphatidic acid (LPA) from phosphatidic acid.20 It is reported that ATX, another enzyme that produces LPA, is expressed in breast cancer, prostate cancer and melanoma. LPA combines with ATX into the receptor-mediated ATX-LPA signaling, which can promote cell proliferation and migration by activating the mitogen-activated protein kinase (MAPK) or PI3K/Akt pathways.21–23 The features of LIPH-induced enhancement of invasiveness might explain our finding that breast cancer patients with high LIPH expression were significantly associated with worse survival regarding OS.
In recent years, studies related to LIPH have mostly focused on the mutations in the LIPH gene, which can cause a rare form of congenital hair loss called autosomal recessive woolly hair/hypotrichosis (ARWH/H).10,24,25 However, the role of the novel cancer-related gene LIPH in several carcinomas has been reported in some studies. The expression of LIPH was correlated with the amount of esophageal tumors, but not with the metastasis, and Ishimine et al14 further confirmed these characteristics using three independent esophageal adenocarcinoma cell lines. Seki et al2 reported that high levels of LIPH in serum were correlated with better survival in patients with early-stage lung cancer after surgery, and LIPH expression was frequently upregulated in the serum and tissues of cancer patients with non-small cell lung carcinoma (NSCLC), especially with adenocarcinoma (AC) and bronchioloalveolar carcinoma (BAC). In addition, the study of Li et al1 showed that LIPH was upregulated in (papillary thyroid carcinoma) PTC and LIPH could positively mediate tumor proliferation and metastasis in vitro. Cui et al26 reported that LIPH had the opposite clinical significance in breast cancer.
Based on univariate and multivariate analysis, the expression of LIPH of breast cancer was also an independent and important prognostic factor. Two well-organized and independent datasets (TCGA and METABRIC datasets) were used for the study. In TCGA dataset, the high LIPH expression group observed higher proportions of patients with age<55, positive estrogen receptor, and positive HER2. Compared with the normal tissue, LIPH expression was significantly upregulated in breast cancer tumors, luminal A subtype, ER positive group, PR positive group, and HER2 positive group in TCGA dataset. And the protein expression of LIPH in the Human Protein Atlas was also higher in breast cancer tissues than in normal tissues. Outcomes of the univariate analysis showed that the expression level of LIPH was an important prognostic factor, and after adjusting for other clinicopathological factors in the TCGA and METABRIC databases, the expression of LIPH was still an independent significant factor. In TCGA database, the LIPH expression was significantly high in patients with age<55 years old and HER2 positive subtype. This might partly explain why young women with HER2 positive subtype had a poor prognosis.
LIPH expression was negatively correlated with the AJCC stage, T stage, N stage and M stage. It demonstrated that LIPH expression was related to the size of breast cancer, lymph node and distant metastasis. And higher LIPH expression predicted a worse overall survival rate along with the tumor progression.
Compared with the non-TNBC tissues, the LIPH expression was significantly downregulated in triple-negative breast cancer (TNBC) tissues. This may be because triple-negative breast cancer accounts for a relatively small percentage of all breast cancers. And the survival curves illustrated that LIPH low-expression group showed a better OS compared with LIPH high-expression group. A recent study reported that the percentage of CD44+/CD24−BCSCs (breast cancer stem cells) in LIPH + TNBC specimens was significantly higher than the percentage in the corresponding LIPH- breast cancer with immunofluorescence, while LIPH silencing reduced the ratio of CD44+/CD24− stem-like cells in vitro and blocked their ability of mammosphere-forming.27 It showed that LIPH expression promoted stemness in TNBC, which demonstrated a poor prognosis in TNBC. This study also verified our outcomes collected from the TCGA and METABRIC databases.
There are several limitations in our study. First, the data in our study were from public databases, and further studies are required to validate our results. Besides, there might be some inherent biases (for example, selection bias). The TCGA and METABRIC datasets have not yet provided certain prognostic factors (for example, the patient’s treatment options and economic status might cause some biases). Moreover, due to lack of information, we could not analyze repetitive patterns based on LIPH expression levels. Other studies are required to further investigate the relationship and mechanisms between LIPH expression and breast cancer.
Conclusion
In our study, higher mRNA and protein expressions of LIPH were found in breast cancer compared to normal tissues. As for overall survival, high LIPH expression was associated with a worse prognosis compared with low LIPH expression in breast cancer. According to univariate and multivariate analysis, the expression of LIPH could be an important prognostic factor for breast cancer overall survival. However, further studies are necessary to validate such possibilities.
Data Sharing Statement
The data generated or analyzed during this study are included in this article, or if absent are available from the corresponding author upon reasonable request.
Ethics Approval and Consent to Participate
The experiments were approved by Ethics Committee of Cancer Hospital of Chinese Academy of Medical Sciences.
Acknowledgments
We would like to acknowledge everyone who contributed to this study.
Funding
This work was supported by the Natural Science Foundation of China (No. 81872160), the Natural Science Foundation of China (No. 82072940), the China National Key R&D (or Research and Development) Program (No. 2020AAA0105000 and 2020AAA0105004), the Beijing Municipal Natural Science Foundation (Key Project) (No. 7191009), the Beijing Municipal Natural Science Foundation (No. 7204293), the Special Research Fund for Central Universities, Peking Union Medical College (No. 3332019053), the Beijing Hope Run Special Fund of Cancer Foundation of China (No. LC2019B03), the Beijing Hope Run Special Fund of Cancer Foundation of China (No. LC2019L07), the Beijing Hope Run Special Fund of Cancer Foundation of China (No. LC2020L01), the Golden Bridge Project Seed Fund of Beijing Association for Science and Technology (No. ZZ20004), the 2021 Chaoyang District Social Development Science and Technology Plan Project (Medical and Health Field) (No. CYSF2115), the Chinese Young Breast Experts Research project (No. CYBER-2021-005), the XianSheng Clinical Research Special Fund of China International Medical Foundation (No. Z-2014-06-2103), and the Beijing Xisike Clinical Oncology Research Foundation (No. Y-Young2021-0017).
Disclosure
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Li Y, Zhou X, Zhang Q, et al. Lipase member H is a downstream molecular target of hypoxia inducible factor-1α and promotes papillary thyroid carcinoma cell migration in BCPAP and KTC-1 cell lines. Cancer Manag Res. 2019;11:931–941. doi:10.2147/CMAR.S183355
2. Seki Y, Yoshida Y, Ishimine H, et al. Lipase member H is a novel secreted protein selectively upregulated in human lung adenocarcinomas and bronchioloalveolar carcinomas. Biochem Biophys Res Commun. 2014;443(4):1141–1147. doi:10.1016/j.bbrc.2013.12.106
3. Aoki J, Inoue A, Okudaira S. Two pathways for lysophosphatidic acid production. Biochim Biophys Acta. 2008;1781(9):513–518. doi:10.1016/j.bbalip.2008.06.005
4. Rivera R, Chun J. Biological effects of lysophospholipids. Rev Physiol Biochem Pharmacol. 2008;160:25–46.
5. Moolenaar W. Bioactive lysophospholipids and their G protein-coupled receptors. Exp Cell Res. 1999;253(1):230–238. doi:10.1006/excr.1999.4702
6. Baker D, Morrison P, Miller B, et al. Plasma lysophosphatidic acid concentration and ovarian cancer. JAMA. 2002;287(23):3081–3082. doi:10.1001/jama.287.23.3081
7. Shida D, Watanabe T, Aoki J, et al. Aberrant expression of lysophosphatidic acid (LPA) receptors in human colorectal cancer. Lab Invest. 2004;84(10):1352–1362. doi:10.1038/labinvest.3700146
8. Siegel R, Miller K, Fuchs H, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33.
9. Taki T, Tanahashi K, Takeichi T, et al. Association of topical minoxidil with autosomal recessive woolly hair/hypotrichosis caused by LIPH pathogenic variants. JAMA Dermatol. 2020;156(10):1030. doi:10.1001/jamadermatol.2020.2195
10. Tahara U, Ono N, Aoki S, et al. Case of autosomal recessive woolly hair/hypotrichosis with a homozygous c.736T>A mutation of LIPH caused by maternal uniparental disomy of chromosome 3. J Dermatol. 2020;47(11):e393–e394. doi:10.1111/1346-8138.15550
11. Akiyama M. Isolated autosomal recessive woolly hair/hypotrichosis: genetics, pathogenesis and therapies. J Eur Acad Dermatol Venereol. 2021;35(9):1788–1796. doi:10.1111/jdv.17350
12. Orozco-terWengel P, Barbato M, Nicolazzi E, et al. Revisiting demographic processes in cattle with genome-wide population genetic analysis. Front Genet. 2015;2(6):191.
13. Zhuang H, Chen X, Wang Y, et al. Identification of LIPH as an unfavorable biomarkers correlated with immune suppression or evasion in pancreatic cancer based on RNA-seq. Cancer Immunol Immunother. 2021. doi:10.1007/s00262-021-03019-x
14. Ishimine H, Zhou R, Sumitomo K, et al. Lipase member H frequently overexpressed in human esophageal adenocarcinomas. Tumour Biol. 2016;37(2):2075–2081. doi:10.1007/s13277-015-3985-y
15. Koboldt DC, Fulton R, McLellan Met al. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi:10.1038/nature11412
16. Ciriello G, Gatza M, Beck A, et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell. 2015;163(2):506–519. doi:10.1016/j.cell.2015.09.033
17. Curtis C, Shah S, Chin S, et al. The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. doi:10.1038/nature10983
18. Pereira B, Chin S, Rueda O, et al. The somatic mutation profiles of 2433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun. 2016;7:11479. doi:10.1038/ncomms11479
19. Carrière F, Withers-Martinez C, van Tilbeurgh H, Roussel A, Cambillau C, Verger R. Structural basis for the substrate selectivity of pancreatic lipases and some related proteins. Biochim Biophys Acta. 1998;1376(3):417–432. doi:10.1016/S0304-4157(98)00016-1
20. Mizukami YA-O, Hayashi R, Tsuruta D, Shimomura Y, Sugawara K. Novel splice site mutation in the LIPH gene in a patient with autosomal recessive woolly hair/hypotrichosis: case report and published work review. J Dermatol. 2018;45(5):613–617. doi:10.1111/1346-8138.14257
21. Mills GB, Moolenaar WH. The emerging role of lysophosphatidic acid in cancer. Nat Rev Cancer. 2003;3(8):582–591. doi:10.1038/nrc1143
22. Houben AJ, Moolenaar WH. Autotaxin and LPA receptor signaling in cancer. Cancer Metastasis Rev. 2011;30(3–4):557–565. doi:10.1007/s10555-011-9319-7
23. Hao F, Tan M, Xu X, et al. Lysophosphatidic acid induces prostate cancer PC3 cell migration via activation of LPA(1), p42 and p38alpha. Biochim Biophys Acta. 2007;1771(7):883–892. doi:10.1016/j.bbalip.2007.04.010
24. Lv H, Li M, Cheng R. Novel small-insertion mutation in the LIPH gene in a patient with autosomal recessive woolly hair/hypotrichosis. J Dermatol. 2020;47(12):1445–1449. doi:10.1111/1346-8138.15581
25. Kinoshita-Ise M, Kubo A, Sasaki T, Umegaki-Arao N, Amagai M, Ohyama M. Identification of factors contributing to phenotypic divergence via quantitative image analyses of autosomal recessive woolly hair/hypotrichosis with homozygous c.736T>A LIPH mutation. Br J Dermatol. 2017;176(1):138–144. doi:10.1111/bjd.14836
26. Cui M, Jin H, Shi X, et al. Lipase member H is a novel secreted protein associated with a poor prognosis for breast cancer patients. Tumour Biol. 2014;35(11):11461–11465. doi:10.1007/s13277-014-2436-5
27. Zhang Y, Zhu X, Qiao X, et al. LIPH promotes metastasis by enriching stem-like cells in triple-negative breast cancer. J Cell Mol Med. 2020;24(16):9125–9134. doi:10.1111/jcmm.15549
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.



