Back to Journals » Cancer Management and Research » Volume 11
Prognostic significance of serum lactate dehydrogenase in patients with breast cancer: a meta-analysis
Authors Liu D, Wang D, Wu C, Zhang L, Mei Q, Hu G, Long G, Sun W
Received 22 December 2018
Accepted for publication 21 March 2019
Published 26 April 2019 Volume 2019:11 Pages 3611—3619
DOI https://doi.org/10.2147/CMAR.S199260
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Dongbo Liu,1 Dingkun Wang,2 Cheng Wu,1 Linli Zhang,1 Qi Mei,1 Guangyuan Hu,1 Guoxian Long,1 Wei Sun1
1Department of Oncology, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, Hubei 430030, People’s Republic of China; 2Department of Integrated Traditional Chinese and Western Medicine, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, Hubei 430030, People’s Republic of China
Background: Multiple studies have assessed the prognostic significance of serum lactate dehydrogenase (LDH) in patients with breast cancer, but their results remain controversial. This study aimed to evaluate the prognostic value of LDH in breast cancer by meta-analysis.
Methods: Electronic searches for relevant articles were conducted in PubMed, Embase and Web of Science databases. The HR and their 95% CI were used to assess the prognostic value of serum LDH. Stata Statistical Software 12.0 was applied for statistical analysis.
Results: A total of 11 studies involving 6,102 patients were subjected to final analysis. Our results showed that higher serum LDH had significant effect on poor overall survival (HR, 1.88; 95% CI, 1.68–2.11) and progression-free survival (HR, 1.98; 95% CI, 1.46–2.68). Moreover, the results of subgroup analyses were consistent with that of overall outcomes. No significant heterogeneity and publication bias were found in this study.
Conclusion: Serum LDH could act as a prognostic factor for patients with breast cancer. Future data are needed to validate and update our results.
Keywords: breast cancer, lactate dehydrogenase, prognosis, biomarker, meta-analysis
Introduction
Breast cancer is the most common cancer in women and the second leading cause of cancer-related deaths in females, with an estimated 1.67 million new cases and 0.52 million cancer deaths occurred in 2012 worldwide.1 Although significant achievements have been made in therapeutic strategies, the clinical outcome of breast cancer patients remains unsatisfactory due to recurrence, metastasis or treatment-resistant. Identifying factors related to aggressive phenotypes and prognosis of breast cancer is essential for disease surveillance and precise therapy.
Breast cancer is a heterogeneous disease and presents with different clinical and biological characteristics. Nowadays, TNM staging system is widely used for cancer prognosis. However, survival of breast cancer patients after chemotherapy varies greatly, even within the same stages.2 Several clinicopathological factors such as age of the patient, menopause status, tumor size, lymph node status, tumor grade, estrogen receptor, progesterone receptor, epidermal growth factor receptor 2 (HER-2) and Ki-67 expression parameters have been developed for making treatment plan.3 Recently, more and more serum biomarkers were identified as prognostic factors for breast cancer, including circulating microRNAs, inflammatory factors and stem cell markers.4–6 These easily detected and objective markers may provide new prognostic information for patients with breast cancer.
It is now generally accepted that the metabolism of cancer cells differs from that of normal cells. Under normal oxygen concentrations, tumor tissues, but not adjacent normal tissues, exhibit a high rate of glucose consumption. This phenomenon has been widely exploited for the diagnosis and staging of human solid cancers. 18F-fluorodeoxyglucose positron emission tomography (18F-FDG-PET) utilizes the differences between glucose uptake of cancer cells and normal cells to make an imaging technique for detecting tumors. Besides, cancer cells preferentially metabolize glucose by glycolysis to generate energy even in the presence of adequate oxygen.7 Lactate dehydrogenase (LDH) is the most important metabolic enzyme involved in glycolysis as it can convert pyruvate to lactate at the end of glycolysis.8 LDH is a 140-kDa tetramer molecule that exists in five major isoenzymes, numbered LDH-1 through LDH-5, formed by the association of two different types of 35 kDa subunits, M (Muscle) and H (Heart), encoded by LDHA and LDHB genes, respectively. Increased levels of this protein are in fact released in blood as a consequence of massive cell death and are associated with acute diseases. High levels of LDH in serum are also measured during neoplastic diseases, as a consequence of tissue destruction caused by the cancerous growth. The prognostic value of serum LDH level has been identified in different types of cancers.9–11 The international prognostic index, a common clinical tool used to predict outcomes in patients with non-Hodgkin’s lymphoma, contains five risk factors including serum LDH level.12 In addition, serum LDH level is included in TNM staging system for melanoma.13 Thus, serum LDH which can be detected in clinical practices may allow widespread clinical use and contribute to prognosis estimation.
The prognostic value of serum LDH level in breast cancer patients has been investigated in several studies.5,9,14–22 However, it is still difficult to confirm the prognostic role of serum LDH in breast cancer. First, the sample sizes in some published studies are small.5,18 Second, the existing studies are conflicting in their results. Some studies suggested that serum LDH was associated with poor prognosis in breast cancer,5,9,14 whereas other studies failed to demonstrate such correlations.16,19 Therefore, we performed a meta-analysis to evaluate the prognostic role of serum LDH in patients with breast cancer.
Methods
Study design
We developed a protocol that defined search strategy, inclusion criteria, information extraction, and analysis plan based on the Cochrane Handbook for Systematic Reviews of Interventions.23 The checklist requirements of this meta-analysis adhere to the PRISMA statements.24
Literature search
Electronic searches for relevant articles in PubMed, Embase, and Web of Science databases published up to November 30, 2018 were conducted. The search strategy was generated by combining key words related to LDH (“lactate dehydrogenase” or “LDH”), breast cancer (“breast cancer” or “breast carcinoma”), and prognosis (“prognosis” or “prognostic”). Moreover, we manually searched the reference lists of relevant articles for additional publications. We did not limit the search through the use of any restrictions.
Quality assessment
We assessed study quality according to the method developed by Hayden.25 It was based on the identification of six sources of potential study bias that should be assessed in a review of prognostic studies: study participation, study attrition, prognostic factor measurement, outcome measurement, confounding measurement and account, and analysis. The quality assessment score of each section was on a scale of 0 to −2, and 12 was the maximum total score in each every study. Studies scoring 10 or more were considered high quality.
Inclusion criteria
Studies were included in this meta-analysis if they met the following criteria: 1) all patients recruited in the study were diagnosed with breast cancer; 2) the serum level of LDH was measured; 3) the prognostic value of LDH was tested in the article; 4) only English-language studies were included; 5) the HR and their 95% CI were described or could be statistically extracted from the study; 6) when several articles were from the same patient population, the newest or most informative single article was included.
Information extraction
The following information was extracted from each study: first author’s last name, publication year, country of origin, number of patients, tumor types, cutoff level of LDH, and survival data (HR and its 95% CIs for overall survival (OS) and progression-free survival (PFS)). When HRs and their 95% CIs were not given explicitly in an article, these statistical variables were calculated from available numerical data using methods reported by Parmar26 and Tierney.27
Statistical analysis
HRs and their 95% CIs were adopted to estimate the prognostic value of serum LDH. The individual HR estimates were combined into an overall HR, and the results were presented graphically in the form of a forest plot. Pooled effect sizes were considered to be significantly different if their 95% CIs did not include 1 (P<0.05). HR >1 implied a poor survival for the higher LDH group. The Cochran Q test and I2 test were performed to assess the heterogeneity between studies.28 When the Cochran Q test P-value was ≤0.10 and I2 test I2 value was ≥50%, statistically significant heterogeneity was considered to be present. When heterogeneity was absent, fixed effects models were employed; otherwise, random effects models were adopted. Subgroup analyses were conducted according to clinical stages of breast cancer. Funnel plots, Begg’s test and Egger’s test were performed to test publication bias. An asymmetry of the funnel plot with a P-value of <0.05 was regarded as a significant publication bias. All analyses were carried out by using Stata Statistical Software, version 12.0 (Stata Corporation, College Station, TX, USA).
Results
Search results
Literature search and eligibility assessment were performed independently by two reviewers and disagreements among them were resolved by consensus. Figure 1 illustrates the process of study selection; 724 studies were initially found by our search strategy. After the article titles and abstracts were checked, 19 articles were reviewed in detail.5,9,14–22,29–36 Eight papers were excluded from the meta-analysis,29–36 leaving 11 studies which fulfilled the eligibility criteria (Table 1).5,9,14–22 Among these excluded studies, seven could not provide sufficient survival data for extracting HR29–35 and the other one was excluded because an identical patient cohort had been used in other selected study.36 The total number of patients was 6,102, ranging from 70 to 2,425 cases per study. Seven studies focused on metastatic breast cancer5,17–22 and three studies dealt with non-metastatic breast cancer.14–16 There were two studies reported triple-negative breast cancer.9,14 The majority of these studies tested serum LDH before treatment. Ten studies assessed OS5,9,14–16,18–22 and five studies evaluated PFS9,14,16,17,19 in patients with breast cancer. The quality assessment scores of included studies ranged from 7 to 11. No one obtained a perfect score of 12, and six scored less than 10.
 | Table 1 Characteristics of studies included in this meta-analysis |
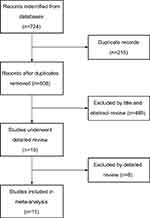 | Figure 1 Flow diagram of study selection. |
Survival analysis
We used OS and PFS as clinical outcomes in this meta-analysis. The combined HR for OS was 1.88 (95% CI, 1.68–2.11; P=0.000), indicating that higher serum LDH had a significant effect on poor survival in patients with breast cancer (Figure 2). Similar to the results of OS, our study revealed that serum LDH was a predictor for short PFS time (HR, 1.98; 95% CI, 1.46–2.68; P=0.000; Figure 3). The heterogeneity test showed no significant heterogeneity in analysis dealing with OS (I2=17.9%, Q test P=0.279) but a slight heterogeneity in analysis regarding PFS (I2=50.2%, Q test P=0.09).
 | Figure 2 Forest plot of HR for the association between serum LDH and OS in breast cancer. Abbreviations: LDH, lactate dehydrogenase; OS, overall survival. |
 | Figure 3 Forest plot of HR for the association between serum LDH and PFS in breast cancer.Abbreviations: LDH, lactate dehydrogenase; PFS, progression-free survival. |
Subgroup analysis
We also performed the subgroup analyses to evaluate the prognostic role of serum LDH in patients with different clinical stages. When we limited the analysis to the six studies dealing with metastatic breast cancer, the combined HR was 1.91 (95% CI 1.71–2.13; P=0.000) without inter-study heterogeneity (I2=2.3%, Q test P=0.402; Figure 4). In the non-metastatic breast cancer subgroup, higher serum LDH indicated a poor OS with a pooled HR of 1.76 (95% CI 1.18–2.62; P=0.006) and an acceptable heterogeneity (I2=47.8%, Q test P=0.147; Figure 5). All subgroup analyses showed statistically significant association between serum LDH and patients’ survival, indicating that serum LDH was able to suggest poor survival of patients with breast cancer.
 | Figure 4 Forest plot of HR for the association between serum LDH and OS in breast cancer in metastatic breast cancer. Abbreviations: LDH, lactate dehydrogenase; OS, overall survival. |
 | Figure 5 Forest plot of HR for the association between serum LDH and OS in nonmetastatic breast cancer. Abbreviations: LDH, lactate dehydrogenase; PFS, progression free survival. |
Publication bias
We assessed the publication bias by visually assessing a funnel plot for asymmetry and by quantitatively performing Begg’s test and Egger’s test. As shown in Figure 6, there was no clear evidence of funnel plot asymmetry by visual assessment. Moreover, publication bias statistics displayed in Table 2 indicated that no publication bias was detected either from Begg’s test or Egger’s test.
 | Table 2 Results of Begg’s test and Egger’s test for publication bias |
Discussion
The prognostic value of serum LDH has been investigated extensively in various tumors.37–39 These studies suggested that serum LDH could predict poor survival of cancer patients. In breast cancer, the association between serum LDH and clinical outcome has also been widely studied. However, it is still difficult to confirm the prognostic value of serum LDH in breast cancer as the existing studies are often small and conflicting in their results. In this study, we provided the evidence that serum LDH could act as an available prognostic factor for OS and PFS in patients with breast cancer.
To the best of our knowledge, this is the first comprehensive meta-analysis to confirm the prognostic significance of serum LDH in breast cancer. A total of 11 studies were enrolled to investigate the effect of serum LDH on survival of breast cancer patients. We got the combined HR value of 1.88 (95% CI: 1.68–2.11; P=0.000) for OS, which suggested that patients with higher serum LDH had a shorter OS time. In addition, a significant association was also found between serum LDH and poor PFS (HR, 1.98; 95% CI, 1.46–2.68; P=0.000). These results hinted that serum LDH could work as a prognostic factor for patients with breast cancer.
Total serum LDH level elevation is a predictive marker of tissue damage and inflammation. Its prognostic value on the follow up of patients with malignant hematologic diseases and solid tumors is known. Serum LDH levels are used as a prognostic factor in chronic lymphocytic leukemia and metastatic melanoma.12,13 High-serum LDH levels have been suggested as a marker of relapse in asymptomatic non-Hodgkin lymphoma patients and are also known to be an important factor in determining an appropriate treatment strategy. However, the underlying mechanism linking LDH to poor survival remains unknown. It has been hypothesized that serum LDH level may reflect the extent of hypoxia in tumor cells. Tumor cells are often starved of oxygen due to their rapid proliferation. In producing energy, cancer cells can use anaerobic glycolysis which enables them to be independent of oxygen supply. This phenomenon is known as the Warburg effect and is one of the predominant metabolic processes that occur during malignant transformation.40 LDH, as the key kinase of this process, ensures the efficiency of this process and can be detected in the serum. Besides, elevated serum LDH level was suggested to be a marker of immune suppression in cancer patients. Ding et al found that LDH allows tumor cells to suppress and evade the immune system by altering the tumor microenvironment.41 Taken together, serum LDH level may reflect the hypoxia in tumor cells and immune suppression in patients which lead to poor prognosis. What’s more, LDH is emerging as one of the most interesting molecular targets for the development of glycolytic inhibitors to possibly use in cancer therapy.42 Further deeper investigations toward LDH may promote its clinical utility in cancers. Thus, serum LDH correlates with poor survival and targeting LDH may be an effective way to control breast cancer.
Given that histological types and clinical stages of breast cancer are correlated with patient’s prognosis, we conducted subgroup analyses in this study. In terms of histological types in each study, two researched patients with triple-negative breast cancer9,14 and others considered patients with all pathological types. Thus, this type of subgroup analysis could not be performed. In terms of clinical stages in each study, seven studies focused on metastatic breast cancer5,17–22 and three dealt with non-metastatic breast cancer.14–16 Consequently, we combined studies considering patients with different clinical stages for our subgroup analysis. When analysis was limited to metastatic breast cancer, the combined HR for OS was 1.91 (95% CI 1.71–2.13; P=0.000). In the non-metastatic breast cancer subgroup, higher serum LDH indicated a poor OS with a pooled HR of 1.76 (95% CI 1.18–2.62; P=0.006). The elevation of LDH usually indicates a heavy tumor burden and tumor metastasis. Mishra et al performed a study to evaluate the blood levels of biochemical parameters in breast cancer with and without metastasis.43 They found that breast cancer patients with metastasis had higher pre-treatment serum LDH levels than those without metastasis (mean LDH level was 730 and 433 U/L, respectively). This phenomenon may be interpreted by medical conditions in patients with advanced or metastatic stages that are more likely to be influenced by systemic effects from the cancers. The results of our subgroup analyses were consistent with that of pooled analyses and demonstrated the prognostic significance of serum LDH once again. This further proves that LDH is a reliable prognostic factor for breast cancer patients.
Estimating heterogeneity and publication bias is an essential part of a meta-analysis. In this study, heterogeneity test revealed no significant heterogeneity when analyzing the associations between serum LDH and OS. However, a slight heterogeneity appeared when assessing the value of serum LDH in predicting PFS. The heterogeneity may partly come from the variations in assessing serum LDH. The cutoff values for judging high blood levels of LDH varied in the studies, ranging from 165 U/L14 to 480 U/L.5,16 Some studies applied receiver operating curve analysis to calculate optimal cutoff values for serum LDH level,14 while other studies adopted the normal value18,19,22 or upper limit of normal value17 as cutoff values. The majority of these studies tested serum LDH before treatment, but they did not provide the detailed methods for LDH testing. The standardization of LDH testing may resolve this problem in the future, including the cutoff value, test time and test methods. Factors related to the clinical variables could also contribute to heterogeneity. In subgroup analysis, heterogeneity reduced when we combined studies considering with metastatic breast cancer, indicating that the difference in clinical stages could be another source of heterogeneity. With regard to publication bias, no clear evidence of funnel plot asymmetry was found by visual assessment. Moreover, no publication bias was detected according to both Begg’s test and Egger’s test. These findings suggested that our results were robust and not far from the actual situation.
However, the present meta-analysis still has some limitations. First, it is a literature-based meta-analysis, making our results less reliable than individual patient data-based analysis. Second, studies that cannot provide sufficient data to extract HR were excluded. The exclusion of these studies may make our pooled effect sizes differ from their true value on some level. Third, we only conduct subgroup analysis in terms of clinical stages. Considering these issues, our results need to be validated and updated in the near future.
Conclusion
In view of this study, our findings showed that high serum LDH correlated with poor OS and PFS in patients with breast cancer. Thus, serum LDH can act as a prognostic factor for breast cancer, which might help to define high-risk patients and guide clinical decision-making. However, future large randomized trials are needed to improve and update our results due to the limitation of meta-analysis.
Acknowledgments
This work was funded by the Tongji Hospital Foundation (number 2018A08).
Disclosure
The authors declare that they have no conflicts of interest in this work.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi:10.3322/caac.21262
2. Lai J, Wang H, Peng J, Chen P, Pan Z. Establishment and external validation of a prognostic model for predicting disease-free survival and risk stratification in breast cancer patients treated with neoadjuvant chemotherapy. Cancer Manag Res. 2018;10:2347–2356. doi:10.2147/CMAR.S171129
3. Esbah O, Oksuzoglu B. Prognostic & predictive factors for planning adjuvant chemotherapy of early-stage breast cancer. Indian J Med Res. 2017;146(5):563–571. doi:10.4103/ijmr.IJMR_1354_17
4. Li P, Dong J, Zhou X, et al. Expression patterns of microRNA-329 and its clinical performance in diagnosis and prognosis of breast cancer. Onco Targets Ther. 2017;10:5711–5718. doi:10.2147/OTT.S147974
5. Petekkaya I, Unlu O, Roach EC, et al. Prognostic role of inflammatory biomarkers in metastatic breast cancer. J BUON. 2017;22(3):614–622.
6. Kong Y, Lyu N, Wu J, et al. Breast cancer stem cell markers CD44 and ALDH1A1 in serum: distribution and prognostic value in patients with primary breast cancer. J Cancer. 2018;9(20):3728–3735. doi:10.7150/jca.28032
7. Hsu PP, Sabatini DM. Cancer cell metabolism: warburg and beyond. Cell. 2008;134(5):703–707. doi:10.1016/j.cell.2008.08.021
8. Gallo M, Sapio L, Spina A, Naviglio D, Calogero A, Naviglio S. Lactic dehydrogenase and cancer: an overview. Front Biosci (Landmark Ed). 2015;20:1234–1249.
9. Jia Z, Zhang J, Wang Z, et al. An explorative analysis of the prognostic value of lactate dehydrogenase for survival and the chemotherapeutic response in patients with advanced triple-negative breast cancer. Oncotarget. 2018;9(12):10714–10722. doi:10.18632/oncotarget.24246
10. Namikawa T, Ishida N, Tsuda S, et al. Prognostic significance of serum alkaline phosphatase and lactate dehydrogenase levels in patients with unresectable advanced gastric cancer. Gastric Cancer. 2018. doi:10.1007/s10120-018-0897-8
11. Ma L, Qiu J, Zhang Y, et al. Prognostic factors for operable biliary tract cancer: serum levels of lactate dehydrogenase, a strong association with survival. Onco Targets Ther. 2018;11:2533–2543. doi:10.2147/OTT.S150502
12.
13. Balch CM, Gershenwald JE, Soong SJ, et al. Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol. 2009;27(36):6199–6206. doi:10.1200/JCO.2009.23.4799
14. Chen B, Dai D, Tang H, et al. Pre-treatment serum alkaline phosphatase and lactate dehydrogenase as prognostic factors in triple negative breast cancer. J Cancer. 2016;7(15):2309–2316. doi:10.7150/jca.16622
15. Liu X, Meng QH, Ye Y, Hildebrandt MA, Gu J, Wu X. Prognostic significance of pretreatment serum levels of albumin, LDH and total bilirubin in patients with non-metastatic breast cancer. Carcinogenesis. 2015;36(2):243–248. doi:10.1093/carcin/bgu247
16. Petekkaya I, Aksoy S, Roach EC, et al. Impact of inflammatory markers on the prognosis of patients with operable breast cancer. J BUON. 2014;19(3):673–680.
17. Bidard FC, Hajage D, Bachelot T, et al. Assessment of circulating tumor cells and serum markers for progression-free survival prediction in metastatic breast cancer: a prospective observational study. Breast Cancer Res. 2012;14(1):R29. doi:10.1186/bcr3169
18. Brunetto AT, Sarker D, Papadatos-Pastos D, et al. A retrospective analysis of clinical outcome of patients with chemo-refractory metastatic breast cancer treated in a single institution phase I unit. Br J Cancer. 2010;103(5):607–612. doi:10.1038/sj.bjc.6605812
19. Er O, Frye DK, Kau SW, et al. Clinical course of breast cancer patients with metastases limited to the liver treated with chemotherapy. Cancer J. 2008;14(1):62–68. doi:10.1097/PPO.0b013e3181629a7b
20. Pierga JY, Asselain B, Jouve M, et al. Effect of adjuvant chemotherapy on outcome in patients with metastatic breast carcinoma treated with first-line doxorubicin-containing chemotherapy. Cancer. 2001;91(6):1079–1089.
21. Ryberg M, Nielsen D, Osterlind K, Skovsgaard T, Dombernowsky P. Prognostic factors and long-term survival in 585 patients with metastatic breast cancer treated with epirubicin-based chemotherapy. Ann Oncol. 2001;12(1):81–87.
22. Yamamoto N, Watanabe T, Katsumata N, et al. Construction and validation of a practical prognostic index for patients with metastatic breast cancer. J Clin Oncol. 1998;16(7):2401–2408. doi:10.1200/JCO.1998.16.7.2401
23. Higgins JP, Green S Cochrane handbook for systematic reviews of interventions. John Wiley & Sons; 2011. Available from
24. Moher D, Liberati A, Tetzlaff J, Altman DG;
25. Hayden JA, Côté P, Bombardier C. Evaluation of the quality of prognosis studies in systematic reviews. Ann Intern Med. 2006;144(6):427–437.
26. Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med. 1998;17(24):2815–2834.
27. Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi:10.1186/1745-6215-8-16
28. Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. doi:10.1136/bmj.327.7414.557
29. Nieder C, Dalhaug A, Pawinski A, Mannsaker B, Haukland E. Survival after palliative radiotherapy in patients with breast cancer and bone-only metastases. In Vivo. 2016;30(6):879–883.
30. Agrawal A, Gandhe MB, Gupta D, Reddy MV. Preliminary study on serum lactate dehydrogenase (LDH)-prognostic biomarker in carcinoma breast. J Clin Diagn Res. 2016;10(3):BC06–BC08. doi:10.7860/JCDR/2016/17111.7364
31. Madic J, Kiialainen A, Bidard FC, et al. Circulating tumor DNA and circulating tumor cells in metastatic triple negative breast cancer patients. Int J Cancer. 2015;136(9):2158–2165. doi:10.1002/ijc.29265
32. Brown JE, Cook RJ, Lipton A, Coleman RE. Serum lactate dehydrogenase is prognostic for survival in patients with bone metastases from breast cancer: a retrospective analysis in bisphosphonate-treated patients. Clin Cancer Res. 2012;18(22):6348–6355. doi:10.1158/1078-0432.CCR-12-1397
33. Nisman B, Barak V, Hubert A, et al. Prognostic factors for survival in metastatic breast cancer during first-line paclitaxel chemotherapy. Anticancer Res. 2003;23(2C):1939–1942.
34. Baur M, Schlappack O, Havelec L, Wrba F, Dittrich C. Prognostic significance of liver metastases as first site of generalisation in patients with breast cancer-a retrospective analysis. Acta Med Austriaca. 2001;28(5):135–140.
35. Kamby C, Sengelov L. Pattern of dissemination and survival following isolated locoregional recurrence of breast cancer. A prospective study with more than 10 years of follow up. Breast Cancer Res Treat. 1997;45(2):181–192.
36. Pierga JY, Hajage D, Bachelot T, et al. High independent prognostic and predictive value of circulating tumor cells compared with serum tumor markers in a large prospective trial in first-line chemotherapy for metastatic breast cancer patients. Ann Oncol. 2012;23(3):618–624. doi:10.1093/annonc/mdr263
37. Li B, Li C, Guo M, et al. Predictive value of LDH kinetics in bevacizumab treatment and survival of patients with advanced NSCLC. Onco Targets Ther. 2018;11:6287–6294. doi:10.2147/OTT.S171566
38. Chen Z, Guo Q, Lu T, et al. Pretreatment serum lactate dehydrogenase level as an independent prognostic factor of nasopharyngeal carcinoma in the intensity-modulated radiation therapy era. Med Sci Monit. 2017;23:437–445.
39. Zhu L, Li X, Shen Y, et al. A new prognostic score based on the systemic inflammatory response in patients with inoperable non-small-cell lung cancer. Onco Targets Ther. 2016;9:4879–4886. doi:10.2147/OTT.S107279
40. Biswas S, Lunec J, Bartlett K. Non-glucose metabolism in cancer cells – is it all in the fat? Cancer Metastasis Rev. 2012;31(3–4):689–698. doi:10.1007/s10555-012-9384-6
41. Ding J, Karp JE, Emadi A. Elevated lactate dehydrogenase (LDH) can be a marker of immune suppression in cancer: interplay between hematologic and solid neoplastic clones and their microenvironments. Cancer Biomark. 2017;19(4):353–363. doi:10.3233/CBM-160336
42. Di Stefano G, Manerba M, Di Ianni L, Fiume L. Lactate dehydrogenase inhibition: exploring possible applications beyond cancer treatment. Future Med Chem. 2016;8(6):713–725. doi:10.4155/fmc.16.10
43. Mishra S, Sharma DC, Sharma P. Studies of biochemical parameters in breast cancer with and without metastasis. Indian J Clin Biochem. 2004;19(1):71–75. doi:10.1007/BF02872394
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

