Back to Journals » OncoTargets and Therapy » Volume 11
Prognostic significance of epidermal growth factor receptor expression in glioma patients
Authors Li JH , Liang RF, Song C, Xiang YF, Liu YH
Received 25 October 2017
Accepted for publication 27 December 2017
Published 7 February 2018 Volume 2018:11 Pages 731—742
DOI https://doi.org/10.2147/OTT.S155160
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Geoffrey Pietersz
Junhong Li,* Ruofei Liang,* Chen Song, Yufan Xiang, Yanhui Liu
Department of Neurosurgery, West China Hospital of Sichuan University, Chengdu, Sichuan Province, People’s Republic of China
*These authors contributed equally to this work
Purpose: There is a great controversy regarding the prognostic significance of epidermal growth factor receptor (EGFR) in glioma patients. The current meta-analysis was conducted to evaluate the effect of abnormal EGFR expression on overall survival in glioma patients.
Materials and methods: A comprehensive literature search of PubMed, EMBASE, Google Scholar, Web of Science, and Cochrane Library was conducted. The combined hazard ratio (HR) and its 95% confidence intervals (CIs) were used to evaluate the association between EGFR expression and survival in glioma.
Results: A total of 476 articles were screened, and 17 articles containing 1,458 patients were selected. The quality assessment of the included studies was performed by the Newcastle–Ottawa Scale. Overexpression of EGFR was found to be an indicator of poor prognosis in overall survival in glioma patients (HR =1.72, 95% CI 1.32–2.25, P=0.000, random effect) and glioblastoma multiforme patients (HR =1.57, 95% CI 1.15–2.14, P=0.004, random effect). Subgroup analysis was conducted to explore the source of high heterogeneity.
Conclusion: This meta-analysis indicated that high expression of EGFR may serve as a biomarker for poor prognosis in glioma patients.
Keywords: epidermal growth factor receptor, glioma, meta-analysis, prognosis
Introduction
Epidermal growth factor receptor (EGFR) is a transmembrane tyrosine kinase on chromosome 7p12, containing an extracellular ligand-binding domain, a transmembrane domain, and an intracellular tyrosine kinase domain, belonging to the HER family. The family includes four members (HER1/EGFR, HER2/neu, HER3, and HER4), with downstream signaling pathways regulating a number of processes, including growth, migration, and survival.1 The pathway is involved in the development and progression of several human malignancies, including non-small-cell lung cancer, gastric cancer, pancreatic cancer, and nasopharyngeal carcinoma.2–6
Glioma is the most common primary malignancy of the central nervous system, accounting for over half of all malignant brain tumors in adults.7 According to the WHO grading (2016), glioma can be divided into four grades. Glioblastoma multiforme (GBM) has the highest grade and the greatest malignancy. The median survival after standard treatment is ~13 months. Median progression-free survival (PFS) is only 7.2 months.8 The EGFR gene is one of the earliest known carcinogenic genes found in GBM. Its increased transcriptional activity directly causes EGFR expression.9 Due to its pro-oncogenic effects, it is not surprising that increased EGFR expression is associated with the malignancy of glioma.10 A study has found that serum levels of EGFR are substantially increased in patients with malignant glioma, suggesting poor survival.11
It has been hypothesized that EGFR may be a potential prognostic biomarker for glioma. However, there remains some controversy as to the prognostic value of EGFR in glioma. Because of the limitations of sample size and research methods, individual studies have not reached consensus on the question. Therefore, this meta-analysis was conducted to evaluate the effect of abnormal EGFR expression on overall survival (OS) in glioma patients.
Materials and methods
Retrieval strategy
Comprehensive literature retrieval was conducted using electronic databases such as PubMed, EMBASE, Google Scholar, Web of Science, and Cochrane Library (up to June 10, 2017). Medical subject headings (MeSH) (Emtree for EMBASE) and free-text words were adopted to balance comprehensiveness and accuracy. The following search terms were used: “Receptor, Epidermal Growth Factor” [Mesh], “Receptor Tyrosine-protein Kinase erbB-1”, “Transforming Growth Factor alpha Receptor”, “TGF-alpha Receptor”, “Epidermal Growth Factor Receptor Kinase”, “erbB-1 Proto-Oncogene Protein”, “erbB-1 Proto-Oncogene Protein”, “Receptor, Transforming-Growth Factor alpha”, “Receptor, TGF-alpha”, “c-ErbB-1 Protein, Proto-oncogene”; “Glioma” [Mesh], “astrocytoma”, “ependymoblastoma”, “ependymoma”, “glioblastoma”, “medulloblastoma”, “oligodendroglioma”, “optic nerve glioma”, “pontine glioma”, and “subependymoma”, and key words related to survival included prognostic, survival, predictive, outcome, and death. The references of retrieved articles were also screened for eligible studies.
Inclusion and exclusion criteria
Inclusion criteria were as follows: 1) studies on pathological diagnosis of glioma in humans and investigation of EGFR expression in glioma; 2) studies containing hazard ratio (HR) or including survival data such as survival curves to estimate HR and its 95% confidence interval (CI); 3) studies published in English; 4) studies in which detection methods for EGFR were restricted to immunohistochemistry (IHC); and 5) randomized controlled trials, retrospective or prospective studies.
Exclusion criteria were as follows: 1) non-human experiments; 2) letters, case reports, and reviews; 3) duplicated publications; and 4) inability to extract HR or necessary information.
Data extraction and quality assessment
All data were extracted independently by two authors (Junhong Li and Ruofei Liang). Disagreements were resolved by consensus with a third investigator (Yanhui Liu). The following data were extracted from the eligible studies: last name of the first author, publication year, region, study design, tumor type, assay, sample size, cutoff point, maximum follow-up time, HR, and its 95% CI.
HR and its 95% CI were extracted directly from the publications. If HRs and 95% CI were not available directly, they were calculated from the available survival data or extracted from the Kaplan–Meier curves by using the methods of Parmar et al12 and Tierney et al.13 If a study contained both univariate and multivariate analyses, both with HR, the HR provided by multivariate analysis was selected preferentially.
The quality of the eligible articles was evaluated on the Newcastle–Ottawa Scale (NOS), including assessments of patient selection, study comparability, follow-up time, and relevant outcome. NOS scores ranged from 0 to 9, with high-quality study defined as a score of >6.
Statistical analysis
HR and 95% CIs were used to evaluate the association between EGFR expression and OS in glioma. An HR of >1 indicated worse survival with elevated EGFR expression, and a 95% CI not overlapping 1 was taken to be statistically significant. Statistical heterogeneity of the included studies was assessed by I2 statistics and chi-square test, and I2 value >50% or Pheterogeneity <0.05 indicated substantial heterogeneity. The random-effects model was used to estimate the pooled HR if heterogeneity appeared. Otherwise, the fixed-effect model was applied.14 All P-values were two-sided, and P<0.05 was considered to be statistically significant. Sensitivity analysis was also performed to evaluate the stability and reliability of the combinative results. The funnel plot with Begg’s test and Egger’s test was applied to evaluate publication bias. STATA 12.0 (StataCorp LP, College Station, TX, USA) was used to perform statistical analysis.
Results
Characteristics of the included studies
A total of 476 articles were screened from the databases, and 17 eligible articles including 1,458 patients were selected (Figure 1 and Table 1).15–31 The publication dates ranged from 1998 to 2015, sample size from 18 to 199, and maximum follow-up time from 25 to 180 months. On NOS scoring, five articles were rated 6 points, one article was rated 7 points, and only one article was rated 8 points. Among all the included studies, only two were prospective studies, and the rest were retrospective studies. In most studies, specimens were fixed with formalin and embedded with paraffin. The treatment of tumor tissues was not clearly defined in two studies. IHC was performed in all studies. Of the 17 articles, seven reported the available HR and 10 provided the Kaplan–Meier curves. Considering glioma subgroups, 10 studies focused on GBM, two studies focused on ependymoma, two focused on low-grade glioma (LGG), one focused on astrocytoma, one included anaplastic astrocytoma (AA) and GBM, and one included glioma of various subgroups and grades. No study reported disease-free survival or PFS.
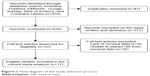  | Figure 1 Flow diagram of the study selection process. |
Prognostic significance of EGFR expression in glioma patients
A total of 1,458 glioma patients were included in the survival analysis. The pooled HR was 1.72 (95% CI 1.32–2.25, P=0.000, random effect; Figure 2). High expression of EGFR had a significant relationship with poor prognosis for glioma patients.
  | Figure 2 Meta-analysis of pooled HRs of OS in gliomas. |
Due to the emergence of high heterogeneity in the study (I2=79.8%, Ph=0.000), a subgroup analysis was conducted to further explore the source of heterogeneity. The subgroups included tumor types, publication years, region, study design, sample size, cutoff value, maximum follow-up time, source of HR, and quality score (Table 2). High EGFR expression was significantly associated with reduced OS in GBM (HR 1.57, 95% CI 1.15–2.14, P=0.004) and mixed gliomas (HR 1.95, 95% CI 1.37–2.77, P=0.000), but not in LGG (HR 1.67, 95% CI 0.037–7.62, P=0.508). There was substantial heterogeneity in GBM (I2=83.8%, Ph=0.000), while heterogeneity was not found in LGG (I2=0%, Ph=0.731) and mixed gliomas (I2=0%, Ph=0.522). In subgroups of publication years, study design, sample size, source of HR, and quality score, the following predicted poor survival: publication year <2010 (HR 1.95, 95% CI 1.37–2.77, P=0.000); publication year ≥2010 (HR 1.52, 95% CI 1.10–2.10, P=0.011); retrospective study (HR 1.75, 95% CI 1.39–2.19, P=0.000); sample size <100 (HR 1.99, 95% CI 1.54–2.57, P=0.000); sample size ≥100 (HR 1.43, 95% CI 1.04–2.97, P=0.027); HRs extracted from the Kaplan–Meier curves (HR 2.10, 95% CI 1.71–2.58, P=0.000); quality score <7 (HR 1.37, 95% CI 1.00–1.87, P=0.048); and quality score ≥7 (HR 1.72, 95% CI 1.32–2.25, P=0.000).
Apparent heterogeneity existed in the subgroup of publication year ≥2010 (I2=80.2%, Ph=0.000), prospective study (I2=81.1%, Ph=0.000), sample size ≥100 (I2=85.5%, Ph=0.000), HRs that were reported (I2=69.5%, Ph=0.003), and quality score ≥7 (I2=84.8%, Ph=0.000). Low heterogeneity existed in the subgroup of publication year <2010 (I2=45.6%, Ph=0.065) and retrospective study (I2=45.8%, Ph=0.024). Sample size <100 (I2=0%, Ph=0.501) and quality score <7 (I2=0%, Ph=0.618) did not show heterogeneity.
The subgroup region included three variables: Europe, America, and Asia, where Europe (HR 1.86, 95% CI 1.32–2.61, P=0.000) and Asia (HR 1.83, 95% CI 1.04–3.23, P=0.037) were significantly related to worse prognosis, but not related to America (HR 1.32, 95% CI 0.98–1.79, P=0.070). The heterogeneity of America (I2=0%, Ph=0.546), Europe (I2=53.3%, Ph=0.036), and Asia (I2=85.7%, Ph=0.000) gradually increased. In the two subgroups of cutoff value and maximum follow-up time, each subitem gave significance for predicting prognosis. The heterogeneity of cutoff value <20% (I2=28.2%, Ph=0.223) and cutoff value ≥20% (I2=43.9%, Ph=0.129) was relatively low, while there was substantial heterogeneity in the subgroup of cutoff value determined by other modes and of unavailable cutoff value (I2=83.3%, Ph=0.000). There was substantial heterogeneity in the subgroup of maximum follow-up time <100 months (I2=81.8%, Ph=0.000), while there was no substantial heterogeneity in the subgroup of maximum follow-up time ≥ 100 months (I2=0%, Ph=0.976) and unavailable maximum follow-up time (I2=0%, Ph=0.944).
Prognostic significance of EGFR expression in GBM patients
GBM is one of the most common gliomas with the highest degree of malignancy. It accounts for a large proportion of the included studies, and therefore, a meta-analysis was conducted to determine the prognostic significance of EGFR expression in GBM patients. Ten studies including 1,074 patients were considered in the survival analysis. The pooled HR was 1.57 (95% CI 1.15–2.14, P=0.004, random effect; Figure 3). This indicated that high expression of EGFR was significantly associated with poor prognosis in GBM patients. Similarly, subgroup analysis was performed because of high heterogeneity (I2=83.8%, Ph=0.000). The grouping and results of subgroup analysis are displayed in Table 3.
Sensitivity analysis and publication bias
All the studies were sequentially removed to determine whether a single study had significant influence on pooled HR and to verify the stability and reliability of HR estimates. It was found that the pooled HRs were not significantly influenced by any individual study (Figures 4 and 5).
  | Figure 4 Sensitivity analysis of the relationship between EGFR expression and overall survival in glioma patients. |
Publication bias was assessed using the funnel plot (Figure 6), Begg’s and Egger’s tests. The funnel plot was asymmetrical. Begg’s test (z =0.54, P > |z| =0.592) revealed no publication bias among the 17 eligible studies, while Egger’s test (t=5.41, P > |t| =0.000) showed clear bias.
  | Figure 6 The funnel plot for all the included studies for overall survival. |
Discussion
EGFR plays an important role in tumor growth, participating in cell motility, adhesion, invasion, and angiogenesis.32 In recent years, EGFR has been a focus of numerous studies on tumor prognosis. Similar meta-analyses involving gastric cancer, non-small-cell lung cancer, nasopharyngeal carcinoma, pancreatic cancer, and esophageal adenocarcinoma have shown poor prognosis associated with EGFR expression.32–36 Comparatively speaking, biomarker studies in glioma have been relatively recent. There have been a large number of inconsistent and even contradictory results regarding the prognostic significance of EGFR in glioma. Among the included studies, 12 suggested that high expression of EGFR indicates poor prognosis, while five others were negative or uncertain on the question. In addition to the 17 studies included in the current meta-analysis, the studies that were excluded according to the inclusion criteria also showed a substantial degree of polarization. High expression of EGFR was not found to predict the poor prognosis of glioma in the studies of Reis-Filho et al,37 Bouvier-Labit et al,38 Smith et al,39 and Dorward et al,40 while other studies suggested that elevated EGFR predicted reduced OS.41–46
Similarly, as the direct source of EGER expression, EFEG amplification was also used as a prognostic marker of glioma. The literature also returned two levels of differentiation. Some studies showed that EGFR amplification was related to worse survival,47–50 while other studies returned the opposite result.51–53 Hobbs et al54 showed that low-to-moderate EGFR amplification was an independent adverse prognostic variable, while high-level EGFR amplification did not play a role in a similar model. In the study of EGFR gene polymorphisms, Costa et al55 found that EGFR variants “-191C/A” and “intron 1 (CA)n repeat” were prognostic markers in GBM patients. Li et al56 conducted a similar gene polymorphism study. In the molecular studies of the Tunisian population, both EGFR amplification and EGFR overexpression predicted significantly poor OS.57 The study on pediatric glioma indicated that no association was apparent between EGFR expression level and either PFS or OS.58,59 All the abovementioned studies have provided rigorous experimental design and accurate and reliable data analysis but contradictory results. A previous meta-analysis of EGFR amplification done by Chen et al60 concluded that there is not enough evidence to suggest that EGFR amplification has prognostic value in GBM patients.
In addition to being a potential prognostic factor, EGFR is also a potential target for the treatment. Some reports claim that 97% of primary GBM show EGFR amplification.61 Because of the important role of EGFR signaling in the pathogenesis of malignant tumor, a growing number of studies have been devoted to developing therapeutic strategies targeting EGFR aberrant activity. As a transmembrane tyrosine kinase, small molecule tyrosine kinase inhibitors (TKIs) may theoretically inhibit EGFR expression and function in malignant tumor. The mechanism would involve displacement of ATP from the catalytic pocket of the enzyme to inhibit kinase activity. Monoclonal antibodies may play a role by combining with the extracellular domain of EGFR.9 Both the methods have been applied in clinical practice with remarkable results in the treatment of non-small-cell lung cancer and colon cancer. However, the use of TKIs and monoclonal antibodies in glioma is challenged. Preusser et al62 concluded in their clinical trial that only a small number of malignant glioma patients benefited from EGFR inhibitor monotherapy. Bevacizumab has a tendency to increase the risk of adverse effects in people older than 65 years, and there is not enough evidence to recommend the use of bevacizumab in pediatric glioma patients.8 Combined with other treatments, such as temozolomide and radiation therapy, stereotactic surgery may be helpful in controlling glioma. Newer and safer targeted therapies have yet to be developed to improve treatment efficacy.
There is a considerable degree of heterogeneity among various studies. Heterogeneity emerged from differences in experimental items, overall experimental design, and analyzed indicators. There was substantial heterogeneity (I2=79.8%, Ph=0.000) in pooled HR in the current meta-analysis. Similar results in pooled HR were observed in the GBM group (I2=83.8%, Ph=0.000). Although the final results suggested poor prognostic role of EGFR, high heterogeneity substantially reduces the reliability of the results. The operations from setting include criteria, subgroup analysis and sensitivity analysis were used to ensure the reliability of the final results, in other words, to provide a strong reference. All the included studies used HR with 95% CI as the index of survival analysis. With I2 and Ph as indicators of heterogeneity, subgroup analysis suggested high heterogeneity in tumor types, publication year, region, sample size, maximum follow-up time, source of HR, and quality score, as follows: GBM (I2=83.8%, Ph=0.000), publication year ≥2010 (I2=80.2%, Ph=0.000), Asia (I2=85.7%, Ph=0.000), sample size ≥100 (I2=85.5%, Ph=0.000), and other methods to divide cutoff value and cutoff value not available (I2=83.3%, Ph=0.000), maximum follow-up time <100 months (I2=81.8%, Ph=0.000), and quality score ≥7 (I2=84.8%, Ph=0.000). There was low heterogeneity in Europe (I2=53.3%, Ph=0.036) when HR was reported (I2=69.5%, Ph=0.003). The abovementioned items were the actual source of heterogeneity; other items showed low or no heterogeneity. Among 22 items in the nine subgroups, only three had HR with no statistical significance, including LGG (HR 1.67, 95% CI 0.37–7.62, P=0.508), America (HR 1.32, 95% CI 0.98–1.79, P=0.070), and HR that were reported (HR 1.28, 95% CI 0.98–1.66, P=0.072). This suggested a stable and reliable final result.
In the GBM subgroup analysis, there was substantial heterogeneity in items such as publication year ≥2010 (I2=82.2%, Ph=0.000), Europe (I2=73.7%, Ph=0.010), Asia (I2=88.1%, Ph=0.000), sample size ≥100 (I2=84.4%, Ph=0.000), other methods to divide cutoff value (I2=80.4%, Ph=0.000), unavailable cutoff value (I2=80.4%, Ph=0.000), maximum follow-up time <100 months (I2=82.0%, Ph=0.000), and quality score ≥7 (I2=86.2%, Ph=0.000). The condition was basically consistent with HR subgroup analysis of the whole set of glioma. Among 19 items in the eight subgroups, pooled HR of eight items showed no statistical significance, and the reliability and stability of the pooled results were lower than that of the entire group of glioma. The greater heterogeneity of the GBM group (83.8% vs 79.8%) is one of the reasons for these results.
In sensitivity analysis, individual studies were sequentially removed, and the bottom limits of 95% CI of pooled HR of other studies were all larger than 1, suggesting that individual studies had limited impact on the entire study. The pooled HR had statistical significance, and the entire study was stable.
Asymmetry of the funnel plot and Egger’s test demonstrated the existence of publication bias (the most common source of bias) affecting the authenticity and reliability of the data to some extent. First, positive results are easier to publish. Of the 17 studies, five were negative, and the inclusion of negative results would theoretically reduce publication bias. Second, repeatedly published theses will also lead to bias, if a single-center or multiple-center results are both published and included. Repeated calculation would lead to larger weight for some studies, affecting the reliability of the clinical research. The research institutions and localities were carefully checked, and overlapping of institutions or experimental data was not found. Next, the small sample size studies return low test efficiencies. In other words, positive results may be caused by opportunities, which may not provide a reliable basis for clinical practice. In the subgroup analysis, HR was merged with a sample size of 100 as the boundary. Our results suggested larger HR and lower P in the subgroup with a sample size of <100, without heterogeneity, while greater heterogeneity was observed in the subgroup with large sample size (≥100). This confirmed that bias was caused by small sample size from the side. There may be many other unknown confounding factors that play a role in publication bias.
As a meta-analysis of prognostic analysis, there are inevitable limitations in several aspects which should be further discussed. First, the areas and population involved in this study are irregular (Africa, the Middle East, South America, and other regions). In addition, China, with a large population, was not considered. Second, there are differences in diagnostic standard and inspection methods in studies on glioma at different times. The accuracy of IHC results is improved by the optimization of the equipment and test methods with the passage of time. The diverse definitions of cutoff values among the studies may also lead to bias. Among experimental designs, there were no randomized controlled trials, which might have improved the accuracy of the analysis. However, there were two prospective studies, which are superior to retrospective studies. Finally, patients in several studies underwent standard therapy with temozolomide plus radiotherapy after surgery15,22,29 and some patients did not undergo uniform treatments, while the remaining studies failed to report postoperative treatments. This too may result in heterogeneity of survival data.
Conclusion
Our meta-analysis suggested that EGFR may serve as a useful biomarker for poor prognosis in glioma patients. In the future, targeted therapy aiming at EGFR signaling pathways may help slow the disease progression of glioma and prolong OS. To verify this result, studies with large sample sizes, covering extensive areas, with more reasonable experimental designs, more standardized diagnostic criteria, and standardized postoperative treatment are required.
Acknowledgments
This study was supported by the Key Research and Development Item from the Department of Science and Technology of Sichuan Province, China (No 2017SZ0006). Junhong Li and Ruofei Liang are the co-first authors.
Disclosure
The authors report no conflicts of interest in this work.
References
Hobbs J, Nikiforova MN, Fardo DW, et al. Paradoxical relationship between the degree of EGFR amplification and outcome in glioblastomas. Am J Surg Pathol. 2012;36(8):1186–1193. | ||
Pan J, Kong L, Lin S, Chen G, Chen Q, Lu JJ. The clinical significance of coexpression of cyclooxygenases-2, vascular endothelial growth factors, and epidermal growth factor receptor in nasopharyngeal carcinoma. Laryngoscope. 2008;118(11):1970–1975. | ||
Matsumura T, Sugimachi K, Takahashi Y, et al. Clinical significance of GAB2, a scaffolding/docking protein acting downstream of EGFR in human colorectal cancer. Ann Surg Oncol. 2014;21(Suppl 4):S743–S749. | ||
Deng W, Gu L, Li X, et al. CD24 associates with EGFR and supports EGF/EGFR signaling via RhoA in gastric cancer cells. J Transl Med. 2016;14:32. | ||
Laurie SA, Goss GD. Role of epidermal growth factor receptor inhibitors in epidermal growth factor receptor wild-type non-small-cell lung cancer. J Clin Oncol. 2013;31(8):1061–1069. | ||
Qin CF, Hao K, Tian XD, Xie XH, Yang YM. Combined effects of EGFR and Hedgehog signaling pathway inhibition on the proliferation and apoptosis of pancreatic cancer cells. Oncol Rep. 2012;28(2):519–526. | ||
Brandes AA, Franceschi E, Tosoni A, Hegi ME, Stupp R. Epidermal growth factor receptor inhibitors in neuro-oncology: hopes and disappointments. Clin Cancer Res. 2008;14(4):957–960. | ||
Ghiaseddin A, Peters KB. Use of bevacizumab in recurrent glioblastoma. CNS Oncol. 2015;4(3):157–169. | ||
Krishnan S, Rao RD, James CD, Sarkaria JN. Combination of epidermal growth factor receptor targeted therapy with radiation therapy for malignant gliomas. Front Biosci. 2003;8:e1–e13. | ||
Horbinski C, Hobbs J, Cieply K, Dacic S, Hamilton RL. EGFR expression stratifies oligodendroglioma behavior. Am J Pathol. 2011;179(4):1638–1644. | ||
Quaranta M, Divella R, Daniele A, et al. Epidermal growth factor receptor serum levels and prognostic value in malignant gliomas. Tumori. 2007;93(3):275–280. | ||
Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med. 1998;17(24):2815–2834. | ||
Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. | ||
Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med. 1997;127(9):820–826. | ||
Srividya MR, Thota B, Arivazhagan A, et al. Age-dependent prognostic effects of EGFR/p53 alterations in glioblastoma: study on a prospective cohort of 140 uniformly treated adult patients. J Clin Pathol. 2010;63(8):687–691. | ||
Viana-Pereira M, Lopes JM, Little S, et al. Analysis of EGFR overexpression, EGFR gene amplification and the EGFRvIII mutation in Portuguese high-grade gliomas. Anticancer Res. 2008;28(2a):913–920. | ||
Umesh S, Tandon A, Santosh V, et al. Clinical and immunohistochemical prognostic factors in adult glioblastoma patients. Clin Neuropathol. 2009;28(5):362–372. | ||
Choi Y, Song YJ, Lee HS, et al. Epidermal growth factor receptor is related to poor survival in glioblastomas: single-institution experience. Yonsei Med J. 2013;54(1):101–107. | ||
Nabika S, Kiya K, Satoh H, et al. Prognostic significance of expression patterns of EGFR family, p21 and p27 in high-grade astrocytoma. Hiroshima J Med Sci. 2010;59(4):65–70. | ||
Korshunov A, Golanov A, Sycheva R, Pronin I. Prognostic value of tumour associated antigen immunoreactivity and apoptosis in cerebral glioblastomas: an analysis of 168 cases. J Clin Pathol. 1999;52(8):574–580. | ||
Senetta R, Miracco C, Lanzafame S, et al. Epidermal growth factor receptor and caveolin-1 coexpression identifies adult supratentorial ependymomas with rapid unfavorable outcomes. Neuro Oncol. 2011;13(2):176–183. | ||
Tini P, Cerase A, Cevenini G, Carbone SF, Miracco C, Pirtoli L. Epidermal growth factor receptor expression may correlate with survival through clinical and radiological features of aggressiveness in glioblastoma treated with radiochemotherapy. Anticancer Res. 2015;35(7):4117–4124. | ||
Varela M, Ranuncolo SM, Morandi A, et al. EGF-R and PDGF-R, but not bcl-2, overexpression predict overall survival in patients with low-grade astrocytomas. J Surg Oncol. 2004;86(1):34–40. | ||
Simmons ML, Lamborn KR, Takahashi M, et al. Analysis of complex relationships between age, p53, epidermal growth factor receptor, and survival in glioblastoma patients. Cancer Res. 2001;61(3):1122–1128. | ||
Saha R, Chatterjee U, Mandal S, Saha K, Chatterjee S, Ghosh S. Expression of phosphatase and tensin homolog, epidermal growth factor receptor, and Ki-67 in astrocytoma: a prospective study in a tertiary care hospital. Indian J Med Paediatr Oncol. 2014;35(2):149–155. | ||
Abdullah KG, Ramayya A, Thawani JP, et al. Factors associated with increased survival after surgical resection of glioblastoma in octogenarians. PLoS One. 2015;10(5):e0127202. | ||
Andersson U, Guo D, Malmer B, et al. Epidermal growth factor receptor family (EGFR, ErbB2–4) in gliomas and meningiomas. Acta Neuropathol. 2004;108(2):135–142. | ||
Etienne MC, Formento JL, Lebrun-Frenay C, et al. Epidermal growth factor receptor and labeling index are independent prognostic factors in glial tumor outcome. Clin Cancer Res. 1998;4(10):2383–2390. | ||
Michaelsen SR, Christensen IJ, Grunnet K, et al. Clinical variables serve as prognostic factors in a model for survival from glioblastoma multiforme: an observational study of a cohort of consecutive non-selected patients from a single institution. BMC Cancer. 2013;13:402. | ||
Mendrzyk F, Korshunov A, Benner A, et al. Identification of gains on 1q and epidermal growth factor receptor overexpression as independent prognostic markers in intracranial ependymoma. Clin Cancer Res. 2006;12(7 pt 1):2070–2079. | ||
Chakravarti A, Seiferheld W, Tu X, et al. Immunohistochemically determined total epidermal growth factor receptor levels not of prognostic value in newly diagnosed glioblastoma multiforme: report from the Radiation Therapy Oncology Group. Int J Radiat Oncol Biol Phys. 2005;62(2):318–327. | ||
Meert AP, Martin B, Delmotte P, et al. The role of EGF-R expression on patient survival in lung cancer: a systematic review with meta-analysis. Eur Respir J. 2002;20(4):975–981. | ||
Ma X, Huang J, Wu X, et al. Epidermal growth factor receptor could play a prognostic role to predict the outcome of nasopharyngeal carcinoma: a meta-analysis. Cancer Biomark. 2014;14(4):267–277. | ||
Luo G, Long J, Qiu L, Liu C, Xu J, Yu X. Role of epidermal growth factor receptor expression on patient survival in pancreatic cancer: a meta-analysis. Pancreatology. 2011;11(6):595–600. | ||
Guo YM, Yu WW, Zhu M, Guo CY. Clinicopathological and prognostic significance of epidermal growth factor receptor overexpression in patients with esophageal adenocarcinoma: a meta-analysis. Dis Esophagus. 2015;28(8):750–756. | ||
Hong L, Han Y, Yang J, et al. Prognostic value of epidermal growth factor receptor in patients with gastric cancer: a meta-analysis. Gene. 2013;529(1):69–72. | ||
Reis-Filho JS, Faoro LN, Carrilho C, Bleggi-Torres LF, Schmitt FC. Evaluation of cell proliferation, epidermal growth factor receptor, and bcl-2 immunoexpression as prognostic factors for patients with World Health Organization grade 2 oligodendroglioma. Cancer. 2000;88(4):862–869. | ||
Bouvier-Labit C, Chinot O, Ochi C, Gambarelli D, Dufour H, Figarella-Branger D. Prognostic significance of Ki67, p53 and epidermal growth factor receptor immunostaining in human glioblastomas. Neuropathol Appl Neurobiol. 1998;24(5):381–388. | ||
Smith JS, Tachibana I, Passe SM, et al. PTEN mutation, EGFR amplification, and outcome in patients with anaplastic astrocytoma and glioblastoma multiforme. J Natl Cancer Inst. 2001;93(16):1246–1256. | ||
Dorward NL, Hawkins RA, Whittle IR. Epidermal growth factor receptor activity and clinical outcome in glioblastoma and meningioma. Br J Neurosurg. 1993;7(2):197–199. | ||
Jiang H, Ren X, Zhang W, et al. A new prognostic scoring scale for patients with primary WHO grade III gliomas based on molecular predictors. J Neurooncol. 2013;111(3):367–375. | ||
Jaros E, Perry RH, Adam L, et al. Prognostic implications of p53 protein, epidermal growth factor receptor, and Ki-67 labelling in brain tumours. Br J Cancer. 1992;66(2):373–385. | ||
Zhu A, Shaeffer J, Leslie S, Kolm P, El-Mahdi AM. Epidermal growth factor receptor: an independent predictor of survival in astrocytic tumors given definitive irradiation. Int J Radiat Oncol Biol Phys. 1996;34(4):809–815. | ||
Saito T, Hama S, Kajiwara Y, et al. Prognosis of cerebellar glioblastomas: correlation between prognosis and immunoreactivity for epidermal growth factor receptor compared with supratentorial glioblastomas. Anticancer Res. 2006;26(2b):1351–1357. | ||
Tini P, Belmonte G, Toscano M, et al. Combined epidermal growth factor receptor and beclin1 autophagic protein expression analysis identifies different clinical presentations, responses to chemo- and radiotherapy, and prognosis in glioblastoma. Biomed Res Int. 2015;2015:208076. | ||
Korshunov A, Golanov A, Sycheva R. Immunohistochemical markers for prognosis of cerebral glioblastomas. J Neurooncol. 2002;58(3):217–236. | ||
Hurtt MR, Moossy J, Donovan-Peluso M, Locker J. Amplification of epidermal growth factor receptor gene in gliomas: histopathology and prognosis. J Neuropathol Exp Neurol. 1992;51(1):84–90. | ||
Zhao LL, Xu KL, Wang SW, Hu BL, Chen LR. Pathological significance of epidermal growth factor receptor expression and amplification in human gliomas. Histopathology. 2012;61(4):726–736. | ||
Dehais C, Laigle-Donadey F, Marie Y, et al. Prognostic stratification of patients with anaplastic gliomas according to genetic profile. Cancer. 2006;107(8):1891–1897. | ||
Korshunov A, Sycheva R, Golanov A. The prognostic relevance of molecular alterations in glioblastomas for patients age <50 years. Cancer. 2005;104(4):825–832. | ||
Houillier C, Lejeune J, Benouaich-Amiel A, et al. Prognostic impact of molecular markers in a series of 220 primary glioblastomas. Cancer. 2006;106(10):2218–2223. | ||
Jain R, Poisson LM, Gutman D, et al. Outcome prediction in patients with glioblastoma by using imaging, clinical, and genomic biomarkers: focus on the nonenhancing component of the tumor. Radiology. 2014;272(2):484–493. | ||
Layfield LJ, Willmore C, Tripp S, Jones C, Jensen RL. Epidermal growth factor receptor gene amplification and protein expression in glioblastoma multiforme: prognostic significance and relationship to other prognostic factors. Appl Immunohistochem Mol Morphol. 2006;14(1):91–96. | ||
Hobbs J, Fardo DW, Cieply K, Dacic S, Hamilton RL, Horbinski C. Glioblastoma survival varies according to degree of EGFR amplification. Neuro Oncol. 2011;13:iii92. | ||
Costa BM, Viana-Pereira M, Fernandes R, et al. Impact of EGFR genetic variants on glioma risk and patient outcome. Cancer Epidemiol Biomarkers Prev. 2011;20(12):2610–2617. | ||
Li B, Zhao W, Li J, et al. Effect of epidermal growth factor receptor gene polymorphisms on prognosis in glioma patients. Oncotarget. 2016;7(39):63054–63064. | ||
Trabelsi S, Chabchoub I, Ksira I, et al. Molecular diagnostic and prognostic subtyping of gliomas in Tunisian population. Mol Neurobiol. 2017;54(4):2381–2394. | ||
Bredel M, Pollack IF, Hamilton RL, James CD. Epidermal growth factor receptor expression and gene amplification in high-grade non-brainstem gliomas of childhood. Clin Cancer Res. 1999;5(7):1786–1792. | ||
Ganigi PM, Santosh V, Anandh B, Chandramouli BA, Sastry Kolluri VR. Expression of p53, EGFR, pRb and bcl-2 proteins in pediatric glioblastoma multiforme: a study of 54 patients. Pediatr Neurosurg. 2005;41(6):292–299. | ||
Chen JR, Xu HZ, Yao Y, Qin ZY. Prognostic value of epidermal growth factor receptor amplification and EGFRvIII in glioblastoma: meta-analysis. Acta Neurol Scand. 2015;132(5):310–322. | ||
Anil R, Colen RR. Imaging genomics in glioblastoma multiforme: a predictive tool for patients prognosis, survival, and outcome. Magn Reson Imaging Clin N Am. 2016;24(4):731–740. | ||
Preusser M, Gelpi E, Rottenfusser A, et al. Epithelial growth factor receptor inhibitors for treatment of recurrent or progressive high grade glioma: an exploratory study. J Neurooncol. 2008;89(2):211–218. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.





