Back to Journals » Cancer Management and Research » Volume 13
Prognostic Significance of CDH1, FN1 and VIM for Early Recurrence in Patients with Colorectal Liver Metastasis After Liver Resection
Authors Bogdanovic A , Despotovic J , Galun D, Bidzic N, Nikolic A, Rosic J , Krivokapic Z
Received 31 October 2020
Accepted for publication 3 December 2020
Published 11 January 2021 Volume 2021:13 Pages 163—171
DOI https://doi.org/10.2147/CMAR.S287974
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
Aleksandar Bogdanovic,1,2 Jovana Despotovic,3 Danijel Galun,1,2 Nemanja Bidzic,1,2 Aleksandra Nikolic,3 Jovana Rosic,2 Zoran Krivokapic1,2,4
1HPB Unit, Clinic for Digestive Surgery, Clinical Center of Serbia, Belgrade, 11 000, Serbia; 2School of Medicine, University of Belgrade, Belgrade 11 000, Serbia; 3Laboratory for Molecular Biology, Institute of Molecular Genetics and Genetic Engineering, University of Belgrade, Belgrade, 11 000, Serbia; 4Serbian Academy of Sciences and Arts, Belgrade 11 000, Serbia
Correspondence: Aleksandar Bogdanovic
Clinical Center of Serbia – Clinic for Digestive Surgery, Koste Todorovica 6, Belgrade 11 000, Serbia
Tel +381 64 2461104
Email [email protected]
Purpose: There are limited data on expression of epithelial–mesenchymal transition (EMT) markers in patients with colorectal liver metastases (CRLM). The study aim was to evaluate the expression and prognostic significance of E-cadherin (CDH1), fibronectin (FN1) and vimentin (VIM) in patients with CRLM after curative-intent liver resection.
Patients and Methods: Thirty patients with CRLM managed by curative-intent liver resection were included in this prospective pilot study. Blood samples, colorectal liver metastases and surrounding non-tumor liver tissue were collected. Expression of CDH1, FN1 and VIM was analyzed by quantitative real-time polymerase chain reaction. Expression in CRLM and non-tumor liver tissue was compared, while expression in serum was correlated with CRLM expression. One-year recurrence-free survival was compared between patients with low and high CDH1, FN1 and VIM expression.
Results: The expression of CDH1 was similar in CRLM and non-tumor liver tissues, while FN1 and VIM expression was significantly lower in metastatic tissue (P=0.003 and pP< 0.001, respectively). Serum expression of CDH1 and VIM was detected in 66.7% and 93.3% of patients, respectively, while FN1 was not detected in any of the patients. The correlation of CDH1 and VIM expression between CRLM and serum was not statistically significant. Decreased CDH1 expression in CRLM and decreased VIM expression in serum were associated with early recurrence after surgical treatment of CRLM.
Conclusion: Lower expression of CDH1 in CRLM and lower serum expression of VIM were found to be associated with early recurrence after liver resection for CRLM.
Keywords: epithelial–mesenchymal transition, CDH1, VIM, FN1, colorectal liver metastasis, colorectal cancer
Introduction
Colorectal cancer (CRC) is the third most common cancer and the second leading cause of cancer-related deaths worldwide.1 In 2017, 1.8 million new cases of CRC were diagnosed and 896,000 deaths were registered.2 CRC is characterized by growing incidence and increasing mortality rates in developing countries, mostly in Eastern Europe, Asia and South America.3
Liver is the most common metastatic site of colorectal cancer.4 Approximately 50% of CRC patients develop liver metastases during the course of the disease.5 Currently, liver resection is the only potentially curative treatment option.6 Although long-term survival has been substantially improved, more than half of patients will develop recurrent disease two years after liver resection.7 The majority of patients are managed by perioperative chemotherapy, associated with different side-effects.
Long-term prognosis of patients with colorectal cancer liver metastases (CRLM) is uncertain, while prediction of chemotherapy response is equally difficult. Several predictive scoring systems were developed to stratify prognosis and subsequently allocate the optimal treatment modality.8,9 These scoring systems are clinically applicable in terms of survival but have limited predictive value in terms of patient stratification for clinical management.10 Moreover, patients with similar clinical stages may achieve different survival. This reflects the high degree of biological heterogeneity of CRLM, responsible for differences in prognosis and responsiveness to therapy. In the era of personalized medicine based on genomic analysis and targeted therapies, there is an urgent need for new clinically useful biomarkers. Their use should aid patient stratification and selection of tailored cancer treatment strategies to individual patients.11
Epithelial–mesenchymal transition (EMT) has an important role in promoting tumor invasion and metastasis of cancers with epithelial origin.12 EMT is a process by which epithelial cells lose their cell-cell adhesion and polarity, acquiring a mesenchymal phenotype with increased migratory capacity.13 In recent years, EMT has emerged as well-recognized underlying molecular mechanism in a variety of cancers, including CRC.14–16 The process of EMT is associated with decreased expression of epithelial markers such as E-cadherin (CDH1), and increased expression of mesenchymal markers, such as fibronectin (FN1) and vimentin (VIM).17 E-cadherin is a transmembrane protein and the core component of epithelial adherens junctions, essential for tissue development, differentiation, and maintenance.18 The loss of E-cadherin during the EMT process leads to destroyed cell-cell adhesion, increased cell motility and advanced stages of cancer.19 Fibronectin is an extracellular matrix glycoprotein, having an important role in cell adhesion and migration and affecting cell proliferation.20 Vimentin, as a type III intermediate filament, has a role in maintaining cytoskeleton organization and focal adhesion stability.21
Several studies have investigated expression of individual markers of EMT in primary CRC, and confirmed their potential prognostic role.22,23 However, there are limited data about EMT genes expression in patients with CRLM.24,25 Moreover, relation and correlation between EMT genes expression in serum, metastatic CRC tissue or non-tumor liver tissue have been insufficiently investigated. The prognostic value of EMT markers in prediction of rapid disease progression after curative-intent treatment is still not elucidated.
This study was designed as a pilot study on a limited number of patients with the aim to analyze gene expression status of CDH1, FN1 and VIM in serum, CRLM and non-tumor liver tissue, and to evaluate their eventual prognostic significance for early recurrence after curative-intent liver resection for CRLM.
Patients and Methods
Subjects
Between December 2016 and February 2019, thirty patients with CLRM were recruited for the prospective pilot study. All subjects were managed by curative-intent liver resection at University Clinic for Digestive Surgery, Clinical Center of Serbia, Belgrade. The inclusion criteria were age 18–90 and potentially curative hepatectomy for CRLM, simultaneously or after radical resection of a primary tumor. The exclusion criteria were previous hepatectomy for CRLM and the presence of residual extrahepatic disease. This study was conducted in accordance with the Declaration of Helsinki. The study was approved by the Ethics Committee of Clinical Center of Serbia. Informed consent was obtained from all individual participants included in the study.
Preoperative tumor staging was determined by transabdominal ultrasound, chest radiography, computed tomography (CT) and/or magnetic resonance imaging (MRI). Demographic and clinicopathological features were recorded: age, sex, primary cancer data (localization, neoadjuvant/adjuvant chemo-radiotherapy, TNM and Duke’s classification), number, diameter and lobar distribution of liver metastases, carcinoembryonic antigen (CEA) and carbohydrate antigen (CA 19–9), perioperative chemotherapy, and tumor grading and tumor residual status.
Samples
Tumor tissue and surrounding non-tumor liver tissue samples were obtained from resected liver specimens immediately after surgery. Tissue samples were immersed into RNAlater® RNA Stabilization Solution (ThermoFisher Scientific, Vilnius, Lithuania) and stored at −80°C. Corresponding blood samples were obtained one week prior to surgery. Blood samples were allowed to coagulate for 15 min, and then centrifuged at 3500 rpm for 10 min. Obtained sera were stored at −80°C until extraction of RNA.
RNA Extraction
Collected tumor tissue and non-tumor liver tissue was cut into smaller pieces and homogenized prior to addition of 1 mL of TRIzol Reagent (Thermo Fisher Scientific, Vilnius, Lithuania). Total RNA from tissue was extracted according to the manufacturer’s protocol, while the RNA from serum was extracted using the following protocol. 1 mL of TRIzol Reagent was added to 400 μL of serum together with 1 μL of 1 μg/μL nuclease-free glycogen. The mixture was vortexed for 20 s to dissipate the yellow globules and incubated at room temperature for 10 min. Then, 200 μL of chloroform was added. The tube was shaken vigorously by hand for 20 simmediately after chloroform addition, incubated for 15 min at room temperature and centrifuged at 12,000 x g for 15 min at 4°C. Approximately 800 μL of the upper aqueous phase was transferred to a fresh tube, followed by the addition of 1.2 mL of isopropanol. Mixture was incubated at room temperature for 10 min and centrifuged at 12,000 x g for 8 min at 4°C. The supernatant was carefully aspirated and 1 mL of 75% ethanol was added. The tube was centrifuged at 7500 x g for 5 min at room temperature. The supernatant was removed and the pellet was allowed to dry for 5 min. The RNA pellet was resuspended with 20 μL of nuclease-free water and stored at −80°C. The concentration and purity of total RNA from tissue and serum were determined by measuring the absorbance at 260 nm and 280 nm.
Gene Expression Analysis
Extracted total RNA was treated with Ambion™ DNase I (RNase free) (ThermoFisher Scientific, Vilnius, Lithuania) in the presence of the 10 x DNase Buffer and incubated at 37°C for 30 min. DNase I was inactivated by 2 min incubation with DNase Inactivation Reagent from the DNA-free™ DNA Removal Kit (ThermoFisher Scientific, Vilnius, Lithuania). The mixture was centrifuged at 10,000 x g for 1.5 min and the supernatant containing DNA-free RNA was transferred to a new tube.
The total RNA was reverse-transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Vilnius, Lithuania) according to the manufacturer’s instructions. The reverse-transcription mixture was incubated for 10 min at 25°C, 120 min at 37°C and 5 min at 85°C. Complementary DNA (cDNA) was stored at −20°C until further use.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
qRT-PCR was performed using HOT FIREPol® EvaGreen® qPCR Mix Plus (ROX) (Solis BioDyne, Tartu, Estonia). Primers used for EMT genes were: CDH1 forward 5ʹ-TGCCCAGAAAATGAAAAAGG-3ʹ, CDH1 reverse 5ʹ-GTGTATGTGGCAATGCGTTC-3ʹ, FN1 forward 5ʹ-CAGTGGGAGACCTCGAGAAG-3ʹ, FN1 reverse 5ʹ-TCCCTCGGAACATCAGAAAC-3ʹ, VIM forward 5ʹ-GAGAACTTTGCCGTTGAAGC-3ʹ, VIM reverse 5ʹ-GCTTCCTGTAGGTGGCAATC-3ʹ.26 The expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) housekeeping gene was used as an endogenous control for normalization of CDH1, FN1 and VIM relative gene expression. GAPDH primers were GAPDH forward 5ʹ-GTGAAGGTCGGAGTCAACG-3ʹ and GAPDH reverse 5ʹ-TGAGGTCAATGAAGGGGTC-3ʹ. qRT-PCR was carried out in a 7500 Real-Time PCR System machine (Applied Biosystems, Foster City, California, USA). PCR conditions were as follows: 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 60 s. The qRT-PCR reaction for each sample was done in triplicate. Data were extracted and analyzed using 7500 System Software. Data are presented as average ΔCt values.
Follow Up
Patients were followed up during the first postoperative year after liver resection. Tumor markers CEA and CA 19–9 and transabdominal ultrasonography were performed every three months. Computed tomography and/or magnetic resonance were performed six months and one year after liver resection. Colonoscopy/rectoscopy and positron emission tomography were considered when local or distant recurrence was suspected. Early recurrence was defined by the presence of either local recurrence (at the site of primary cancer resection), appearance of new liver metastases or extrahepatic disease during the first year after surgery. According to the status of recurrent disease, patients were further stratified into recurrence and no recurrence groups.
Statistical Analysis
Statistical analysis was carried out using SPSS version 18.0 (SPSS Inc., Chicago, IL, USA). P value ≤0.05 was considered statistically significant. Continuous variables were expressed as median (range). Categorical variables were expressed as absolute numbers (percentages). Normal distribution of continuous data was tested using Kolmogorov–Smirnov normality test. If data were normally distributed, Student’s t-test was applied, and Pearson bivariate correlation coefficient was determined. If not, Mann–Whitney U-test for two independent samples or Wilcoxon signed ranks test for two related samples were used. Non-parametric Spearman’s rank correlation was determined for non-normal distributions.
Receiver operating characteristic (ROC) curves were created to determine optimal, sample-based cut-off values of CDH1, FN1 and VIM expression in CRLM and serum by Youden index (smaller test results indicate more positive test for CRLM CDH1 and VIM expression and serum VIM expression). Patients were stratified according to cut-off values into the appropriate low- and high- CDH1, FN1 and VIM groups. One-year recurrence-free survival of low- and high- CDH1, FN1 and VIM groups was estimated by the Kaplan-Meier method and compared using Log rank test. Univariate analysis was performed to analyze prognostic factors affecting one year recurrence. Univariate analysis was conducted using Fisher’s exact test for categorical variables or Mann–Whitney U-test for continuous variables. Multivariate analysis was not done since only univariate analysis found significant parameters.
Results
The expression of CDH1, FN1 and VIM was analyzed in CRLM, non-tumor liver tissue and in serum from patients with hepatic metastases of CRC. Demographic and clinicopathological data of the included subjects are summarized in Table 1.
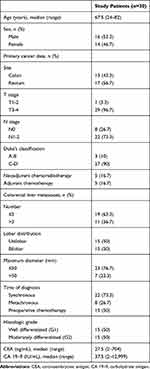 |
Table 1 Patients’ Characteristics |
CDH1
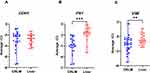
CDH1 expression in CRLM was −2.28 (range, −7.39 to −0.81), in non-tumor liver tissue −2.64 (range, −4.48 to −1.33) and in serum −4.15 (range, −10.48 to −0.56).
FN1
FN1 expression in CRLM was −0.79 (range, −6.06–2.03) and in non-tumor liver tissue 2.75 (range, −6.73–4.75).
VIM
VIM expression in CRLM, non-tumor liver tissue and serum was −1.52 (range, −5.28–2.59), −0.8 (range, −2.24–1.61) and −0.97 (range, −2.64–1.99), respectively.
CDH1 expression in CRLM was similar to non-tumor liver tissue (Figure 1A), while expression of FN1 and VIM in CRLM was significantly lower than in non-tumor liver tissue (P=0.003 and P<0.001, respectively) (Figure 1B and C). Serum expression of CDH1 was detected in 66.7% of patients, while VIM expression was detected in 93.3% of patients. Serum expression of FN1 was not detected in any of the patients.
As shown in Figure 2, there was no significant correlation between expression of CDH1 (Spearman’s rho=−0.255, P=0.322) or VIM (Pearson’s r=0.181, P=0.357) in serum and CRLM (Figure 2A and B).
ROC Curve Analysis and Cut-off Values Estimation
An optimal cut-off value for CRLM expression of CDH1, FN1 and VIM was −3.59 (sensitivity 54.5%, specificity 99.9%), −0.54 (sensitivity 60%, specificity 71.4%) and −1.98 (sensitivity 50%, specificity 60%), respectively. A cut-off value for serum expression of CDH1 and VIM was −3.16 (sensitivity 44.4%, specificity 81.8%) and 0.26 (sensitivity 92.3%, specificity 40%), respectively. According to estimated cut-off values, patients were subsequently divided into corresponding low- and high- groups based on CRLM expression: 6 (24%) in low-CDH1 and 19 (76%) in high-CDH1, 14 (58.3%) in low-FN1 and 10 (41.7%) in high-FN1, and 10 (33.3%) in low-VIM and 19 (63.3%) in high-VIM; and also according to serum expression: 14 (70%) in low-CDH1 and 6 (30%) in high-CDH1 and 21 (75%) in low-VIM and 7 (25%) in high-VIM.
Prognostic Factors of Early Recurrence
Median one-year recurrence-free survival was 12 (range, 4–12) months. Patients in high-CDH1 CRLM group experienced longer one-year recurrence-free survival than patients in low-CDH1 CRLM group (P=0.001), while one-year recurrence-free survival for FN1 and VIM was similar in both groups (P=0.160 and P=0.108, respectively). Based on serum analysis, high-VIM group showed longer recurrence-free survival than low-VIM group (P=0.041), while recurrence-free survival was similar between high- and low-CDH1 groups (P=0.265). Kaplan–Meier curves are shown in detail in Figure 3.
With univariate analysis only CDH1 expression in CRLM was found as a prognostic factor for one-year recurrence prediction (P=0.041). Age more than 65, sex, neoadjuvant chemotherapy, synchronous presentation, bilobar distribution, multiple metastases, number of metastases ≥3, tumor size ≥50 mm, histologic grade G2, residual status R1, FN1 and VIM expression level in CRLM and CDH1 and VIM expression level in serum had no predictive ability for early recurrence (Table 2). Therefore multivariate analysis was not done.
 |
Table 2 Univariate Analysis of Potential Prognostic Factors Related to Early Recurrence After Liver Resection for Colorectal Liver Metastases |
Discussion
The presented pilot study analyzed expression of three genes involved in EMT using different tissue samples and serum of patients with CRLM. A panel of genes was selected based on previous reports that demonstrated clinical relevance of CDH1, FN1 and VIM in CRC patients.27–29 The major findings of this study are: decreased expression of FN1 and VIM in CRLM in comparison to non-tumor liver tissue, and association of decreased CDH1 expression in CRLM and decreased VIM expression in serum with early recurrence after surgical treatment of CRLM.
The majority of published studies focused on identifying potential protein biomarkers using immunohistochemistry analysis of tissue samples or using enzyme-linked immunosorbent assay to measure their plasma concentrations.25,30 Recent attention was given to circulating cell-free nucleic acids (cfNAs), such as DNA, mRNA or microRNA (miRNA), and cellular nucleic acids (cNAs). Currently, the most commonly used methods for cfRNAs and cNAs detection are qRT-PCR, microarray and deep sequencing.31 cfNAs originate from apoptotic and necrotic cancer cells digested by macrophages and then released into the circulation.32 cfNAs from the peripheral blood are easily accessible and may serve as a minimal invasive diagnostic tool – “liquid biopsy”, to replace surgical soft tissue biopsy. Furthermore, cfNAs can be used as prognostic markers, during the follow-up in already treated patients or to evaluate therapeutic response.
Higher concentrations of cfNAs have been registered in cancer patients, although increased concentrations can be found in other conditions, like inflammatory diseases or benign tumors.33 To date, miRNA are extensively investigated in CRC patients and their prognostic role was confirmed in several studies.34,35 Unlike miRNAs, circulating mRNAs are poorly studied as disease biomarkers.
Although various molecules are involved in the process of EMT (i.e. CDH1, CDH2, TCF-8, Claudin-1, Zo-1, β-catenin, Snail, etc.), the presented study is among the first that analyzed cfmRNAs for CDH1, VIM and FN1 as metastatic CRC prognostic biomarkers.
Decreased levels of tumor tissue mRNA for VIM and FN1 were observed in CRLM compared with non-tumor liver tissue. Niknami et al. showed up-regulated VIM and FN1 expression in stromal cells, while VIM was down-regulated in colonic epithelial cells.36 Niknami et al. further demonstrated an association between decreased mRNA expression of VIM and larger tumor size, and increased FN1 levels and higher tumor stages. Chaffer et al. proposed that mesenchymal–epithelial transition (MET) occurred during the invasion of targeted organs by bladder cancer, and that cells start acquiring epithelial phenotype, showing a decreased vimentin expression.37 Other authors have also reported that MET seems to occur after invasion of targeted organs and formation of distant metastases.38,39 Truant et al. and Ionescu et al. showed similar E-cadherin expression in colon cancer and adjacent normal mucosa.40,41 However, Truant et al. reported E-cadherin expression was higher in liver metastases than in non-tumor liver tissue. Although CDH1 expression level tended to increase in CRLM, statistical difference in comparison to non-tumor liver tissue expression was not observed in our study. These findings are consistent with previously described mesenchymal–epithelial and epithelial–mesenchymal cellular plasticity of liver metastases.42 The other possible explanation for this finding is a small sample size.
We were unable to detect serum FN1 mRNA in any of our patients. Inability to detect FN1 mRNA in serum is not related to technical constraints, since GAPDH was successfully detected in all patients in tissue samples and in serum, confirming the sample quality. Also, FN1 mRNA was successfully detected in our tissue samples which eliminates the possibility of poor primer selection. This could mean that FN1 mRNA is either so low that it cannot be detected by qRT-PCR, or is completely absent in serum of mCRC patients, probably because of low stability of FN1 mRNA. Nevertheless, fibronectin exists in serum of CRC patients and it is a useful marker of disease advancement.43
Serum CDH1 and VIM were not found to reflect their expression in CRLM, although this result was expected considering the shedding of tumor cells into the circulation. Despite no correlation being confirmed by the current analysis, the association between serum and malignant tissue expression should be further investigated, since cfmRNAs for CDH1 and VIM might have important roles in CRLM patients.
The presented study confirmed the prognostic significance of CDH1 expression in CRLM tissue for the prediction of early recurrence. The majority of studies have investigated the prognostic impact of EMT markers in primary cancers after curative-intent surgery. Several authors reported a relationship between hepatic metastatic tissue gene expression and long-term prognosis after liver resection.25,44,45 Nanashima et al. analyzed expression of E-cadherin in hepatic metastases of colorectal carcinoma by immunohistochemistry, and found that patients with preserved expression of E-cadherin had better prognosis compared with those with negative expression, although the difference was not significant.44 The same author reported no association between E-cadherin levels and recurrence-free survival period in a larger sample size analysis.45 Also, immunohistochemically analyzed, E-cadherin levels in metastatic liver tissue were not found to be associated with survival.25 We estimated expression of mRNAs for CDH1, VIM and FN1 by qRT-PCR and observed an association between CRLM CDH1 level and early recurrence. This study also reports prolonged recurrence-free survival in patients with high-VIM serum expression, but no difference between the study groups was observed.
The small sample size of this study is a major study limit and should be overcome by conducting larger and multicenter studies. However, the pilot study was designed as the first step aimed to assess prognostic significance of CDH1, VIM and FN1 for early recurrence after curative-intent liver resection. Furthermore, long-term survival was not included in the current analysis, since this study is designed as a pilot project. Thorough evaluation of EMT gene expression as biomarkers will also require long-term follow up of multiple clinical and histopathological parameters.
In summary, the presented pilot study confirmed prognostic significance of CDH1 expression in CRLM tissue for prediction of early recurrence. However, the expression of CDH1 in CRLM and non-tumor liver tissue was similar and did not correlate with its expression in serum, so the clinical application of this biomarker remains unclear. Serum expression of VIM was detected in 93.3% of patients, and it was associated with longer one-year recurrence-free survival, indicating its prognostic potential.
Conclusions
Lower expression of CDH1 in CRLM and lower serum expression of VIM were found to be associated with early recurrence after liver resection for CRLM. Studies with larger samples are needed to confirm these initial results.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Fitzmaurice C, Abate D; Global Burden of Disease Cancer C. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2017: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2019. doi:10.1001/jamaoncol.2019.2996
3. Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66(4):683–691. doi:10.1136/gutjnl-2015-310912
4. Park JH, Kim JH. Pathologic differential diagnosis of metastatic carcinoma in the liver. Clin Mol Hepatol. 2019;25(1):12–20. doi:10.3350/cmh.2018.0067
5. Adam R, Hoti E, Folprecht G, Benson AB. Accomplishments in 2008 in the management of curable metastatic colorectal cancer. Gastrointest Cancer Res. 2009;3(5 Supplement 2):S15–22.
6. Chow FC-L, Chok KS-H. Colorectal liver metastases: an update on multidisciplinary approach. World J Hepatol. 2019;11(2):150–172. doi:10.4254/wjh.v11.i2.150
7. de Jong MC, Pulitano C, Ribero D, et al. Rates and patterns of recurrence following curative intent surgery for colorectal liver metastasis: an international multi-institutional analysis of 1669 patients. Ann Surg. 2009;250(3):440–448. doi:10.1097/SLA.0b013e3181b4539b
8. Sasaki K, Morioka D, Conci S, et al. The Tumor Burden Score: A New “Metro-ticket” Prognostic Tool For Colorectal Liver Metastases Based on Tumor Size and Number of Tumors. Ann Surg. 2018;267(1):132–141. doi:10.1097/SLA.0000000000002064
9. Fong Y, Fortner J, Sun RL, Brennan MF, Blumgart LH. Clinical score for predicting recurrence after hepatic resection for metastatic colorectal cancer: analysis of 1001 consecutive cases. Ann Surg. 1999;230(3):309–318. doi:10.1097/00000658-199909000-00004
10. Gomez D, Cameron IC. Prognostic scores for colorectal liver metastasis: clinically important or an academic exercise? HPB (Oxford). 2010;12(4):227–238. doi:10.1111/j.1477-2574.2010.00158.x
11. Aziz MA, Yousef Z, Saleh AM, Mohammad S, Al Knawy B. Towards personalized medicine of colorectal cancer. Crit Rev Oncol Hematol. 2017;118:70–78. doi:10.1016/j.critrevonc.2017.08.007
12. Cao H, Xu E, Liu H, Wan L, Lai M. Epithelial-mesenchymal transition in colorectal cancer metastasis: A system review. Pathol Res Pract. 2015;211(8):557–569. doi:10.1016/j.prp.2015.05.010
13. Bates RC, Pursell BM, Mercurio AM. Epithelial-mesenchymal transition and colorectal cancer: gaining insights into tumor progression using LIM 1863 cells. Cells Tissues Organs. 2007;185(1–3):29–39. doi:10.1159/000101300
14. Montanari M, Rossetti S, Cavaliere C, et al. Epithelial-mesenchymal transition in prostate cancer: an overview. Oncotarget. 2017;8(21):35376–35389. doi:10.18632/oncotarget.15686
15. Huang J, Li H, Ren G. Epithelial-mesenchymal transition and drug resistance in breast cancer (Review). Int J Oncol. 2015;47(3):840–848. doi:10.3892/ijo.2015.3084
16. Zhu QC, Gao RY, Wu W, Qin HL. Epithelial-mesenchymal transition and its role in the pathogenesis of colorectal cancer. Asian Pac J Cancer Prev. 2013;14(5):2689–2698. doi:10.7314/apjcp.2013.14.5.2689
17. Gonzalez DM, Medici D. Signaling mechanisms of the epithelial-mesenchymal transition. Sci Signal. 2014;7(344):re8. doi:10.1126/scisignal.2005189
18. Daulagala AC, Bridges MC, E-cadherin Beyond KA. Structure: A Signaling Hub in Colon Homeostasis and Disease. Int J Mol Sci. 2019;20(11):11. doi:10.3390/ijms20112756
19. Song Y, Ye M, Zhou J, Wang Z, Targeting ZX. E-cadherin expression with small molecules for digestive cancer treatment. Am J Transl Res. 2019;11(7):3932–3944.
20. Stine JM, Sun Y, Armstrong G, Bowler BE, Briknarova K. Structure and unfolding of the third type III domain from human fibronectin. Biochemistry. 2015;54(44):6724–6733. doi:10.1021/acs.biochem.5b00818
21. Liu CY, Lin HH, Tang MJ, Wang YK. Vimentin contributes to epithelial-mesenchymal transition cancer cell mechanics by mediating cytoskeletal organization and focal adhesion maturation. Oncotarget. 2015;6(18):15966–15983. doi:10.18632/oncotarget.3862
22. Okugawa Y, Toiyama Y, Inoue Y, et al. Clinical significance of serum soluble E-cadherin in colorectal carcinoma. J Surg Res. 2012;175(2):e67–73. doi:10.1016/j.jss.2011.11.009
23. Yi W, Xiao E, Ding R, Luo P, Yang Y. High expression of fibronectin is associated with poor prognosis, cell proliferation and malignancy via the NF-κB/p53-apoptosis signaling pathway in colorectal cancer. Oncol Rep. 2016;36(6):3145–3153. doi:10.3892/or.2016.5177
24. Elzagheid A, et al. E-cadherin expression pattern in primary colorectal carcinomas and their metastases reflects disease outcome. World J Gastroenterol. 2006;12(27):4304–4309. doi:10.3748/wjg.v12.i27.4304
25. Jurcic P, Radulovic P, Balja MP, Milosevic M. E-cadherin and NEDD9 expression in primary colorectal cancer, metastatic lymph nodes and liver metastases. Oncol Lett. 2019;17(3):2881–2889. doi:10.3892/ol.2019.9917
26. Mani SA, Guo W, Liao M-J, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–715. doi:10.1016/j.cell.2008.03.027
27. Repetto O, De Paoli P, De Re V, Canzonieri V, Cannizzaro R. Levels of soluble E-cadherin in breast, gastric, and colorectal cancers. Biomed Res Int. 2014;2014:408047. doi:10.1155/2014/408047
28. Du L, Li J, Lei L, et al. High Vimentin Expression Predicts a Poor Prognosis and Progression in Colorectal Cancer: A Study with Meta-Analysis and TCGA Database. Biomed Res Int. 2018;2018:6387810. doi:10.1155/2018/6387810
29. Kida H, Takano Y, Yamamoto K, et al. A single nucleotide polymorphism in fibronectin 1 determines tumor shape in colorectal cancer. Oncol Rep. 2014;32(2):548–552. doi:10.3892/or.2014.3251
30. Cepowicz D, Zareba K, Pryczynicz A, et al. Blood serum levels of E-cadherin in patients with colorectal cancer. Prz Gastroenterol. 2017;12(3):186–191. doi:10.5114/pg.2017.70471
31. Schwarzenbach H, Nishida N, Calin GA, Pantel K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol. 2014;11(3):145–156. doi:10.1038/nrclinonc.2014.5
32. Schwarzenbach H, Hoon DSB, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11(6):426–437. doi:10.1038/nrc3066
33. Nothnick WB, Al-Hendy A, Lue JR. Circulating Micro-RNAs as Diagnostic Biomarkers for Endometriosis: privation and Promise. Journal of Minimally Invasive Gynecology. 2015;22(5):719–726. doi:10.1016/j.jmig.2015.02.021
34. Kanaan Z, Rai SN, Eichenberger MR, et al. Plasma miR-21: a potential diagnostic marker of colorectal cancer. Ann Surg. 2012;256(3):544–551. doi:10.1097/SLA.0b013e318265bd6f
35. Gao S, Zhao Z-Y, Wu R, Zhang Y, Zhang Z-Y. Prognostic value of microRNAs in colorectal cancer: a meta-analysis. Cancer Manag Res. 2018;10:907–929. doi:10.2147/CMAR.S157493
36. Niknami Z, Eslamifar A, Emamirazavi A, Ebrahimi A, Shirkoohi R. The association of vimentin and fibronectin gene expression with epithelial-mesenchymal transition and tumor malignancy in colorectal carcinoma. EXCLI J. 2017;16:1009–1017. doi:10.17179/excli2017-481
37. Chaffer CL, Brennan JP, Slavin JL. Mesenchymal-to-epithelial transition facilitates bladder cancer metastasis: role of fibroblast growth factor receptor-2. Cancer Res. 2006;66(23):11271–11278. doi:10.1158/0008-5472.CAN-06-2044
38. Thiery JP, Acloque H, Huang RYJ, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–890. doi:10.1016/j.cell.2009.11.007
39. Brabletz T, Jung A, Spaderna S, Hlubek F, Kirchner T. Migrating cancer stem cells — an integrated concept of malignant tumour progression. Nat Rev Cancer. 2005;5(9):744–749. doi:10.1038/nrc1694
40. Truant SC, Gouyer VP, Leteurtre EA, Zerimech F, Huet GM, Pruvot F-R-R. E-Cadherin and β-Catenin mRNA Levels Throughout Colon Cancer Progression. J Surg Res. 2008;150(2):212–218. doi:10.1016/j.jss.2007.12.800
41. Ionescu C, Braicu C, Chiorean R, et al. TIMP-1 expression in human colorectal cancer is associated with SMAD3 gene expression levels: a pilot study. J Gastrointestin Liver Dis. 2014;23(4):413–418. doi:10.15403/jgld.2014.1121.234.smad
42. Ceausu AR, Ciolofan A, Cimpean AM, Magheti A, Mederle O, The Mesenchymal-Epithelial RM. Epithelial-Mesenchymal Cellular Plasticity of Liver Metastases with Digestive Origin. Anticancer Res. 2018;38(2):811–816. doi:10.21873/anticanres.12288
43. Saito N, Nishimura H, Kameoka S. Clinical significance of fibronectin expression in colorectal cancer. Mol Med Rep. 2008;1(1):77–81.
44. Nanashima A, Yamaguchi H, Sawai T, et al. Expression of adhesion molecules in hepatic metastases of colorectal carcinoma: relationship to primary tumours and prognosis after hepatic resection. J Gastroenterol Hepatol. 1999;14(10):1004–1009. doi:10.1046/j.1440-1746.1999.01991.x
45. Nanashima A, Yamaguchi H, Sawai T, et al. Prognostic factors in hepatic metastases of colorectal carcinoma: immunohistochemical analysis of tumor biological factors. Dig Dis Sci. 2001;46(8):1623–1628. doi:10.1023/a:1010680815954
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.