Back to Journals » OncoTargets and Therapy » Volume 11
Prognostic role of long non-coding RNA HNF1A-AS1 in Chinese cancer patients: a meta-analysis
Authors Zhuang C, Zheng L, Wang P
Received 25 January 2018
Accepted for publication 27 June 2018
Published 31 August 2018 Volume 2018:11 Pages 5325—5332
DOI https://doi.org/10.2147/OTT.S163575
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jianmin Xu
Chunbo Zhuang,1 Lei Zheng,1 Pei Wang2
1Department of Clinical Laboratory Medicine, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan, People’s Republic of China; 2Department of Gastroenterology, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan, People’s Republic of China
Background: Long non-coding RNAs (LncRNAs) play important roles in tumorigenesis and progression. Recent studies have demonstrated that LncRNA HNF1A antisense RNA 1 (HNF1A-AS1) is aberrantly expressed in several types of cancers and is associated with poor outcomes. This meta-analysis was conducted to investigate the relationship between HNF1A-AS1 expression and clinical outcomes in cancer patients.
Methods: We searched PubMed, Embase, Web of Science, China National Knowledge Infrastructure (CNKI), and Wan Fang databases (updated until December 31, 2017) for literature. A total of eight studies with 789 cancer patients were finally included in the present meta-analysis.
Results: The results showed that high expression of HNF1A-AS1 significantly predicted poor overall survival (HR=3.10, 95% CI: 1.58–6.11, P=0.001), which was further validated using The Cancer Genome Atlas (TCGA) dataset. Moreover, high HNF1A-AS1 expression was also associated with advanced TNM stage (OR=3.32, 95% CI: 2.28–4.83, P<0.001), lymph node metastasis (OR=3.08, 95% CI: 1.95–4.85, P<0.001), and distant metastasis (OR=5.53, 95% CI: 1.94–15.77, P=0.001).
Conclusion: Our results suggested that elevated HNF1A-AS1 was associated with poor clinical outcomes and might serve as a potential prognostic biomarker of cancer.
Keywords: cancer, overall survival, TCGA, long non-coding RNA, HNF1A-AS1, meta-analysis
Introduction
Cancer is a major public health problem with increasing incidence and mortality.1 In 2017, 1,688,780 new cancer cases and 600,920 cancer deaths are projected to occur in the United States.2 Although the evolving treatment strategies such as targeted therapy and biotherapy improved the outcomes of cancer patients, the applicable patients were limited.3 In addition, the 5-year survival rates for most cancers are still very low, which is mainly because of the late-stage diagnosis and insufficient understanding of the molecular mechanisms underlying cancer development. Therefore, it is critical to identify useful prognostic biomarkers and novel potential therapeutic targets for cancer therapy.
Long non-coding RNAs (LncRNAs) are a group of RNAs that are larger than 200 nucleotides in length and lack the protein-coding capability.4 In the last decade, numerous studies have reported the critical roles of these kinds of transcripts in biological processes, such as cell differentiation, proliferation, and apoptosis.5 Moreover, accumulating evidence have shown the dysregulated expression of numerous LncRNAs in human cancers and that LncRNAs may act as oncogenes or tumor suppressors participating in tumorigenesis and metastasis.6 A number of LncRNAs have been shown to be novel promising prognostic biomarkers in human cancers.7
HNF1A antisense RNA 1 (HNF1A-AS1), a natural antisense transcript of HNF1A, is a newly identified LncRNA located at chromosomal band 12q24.31 with 2,455 nucleotides in length.8 HNF1A-AS1 has been reported to be upregulated and act as an oncogene in many types of cancers.8–19 Moreover, the expression of HNF1A-AS1 is associated with poor prognosis and high risk of metastasis in several cancers, such as osteosarcoma, colon cancer, lung cancer, and hepatocellular carcinoma (HCC).9,10,12,17,20 However, individual study may be inaccurate or insufficient due to the limitations of sample sizes or research programs. Therefore, we collected all relevant publications and conducted this meta-analysis to explore the association of HNF1A-AS1 with clinical outcomes and to investigate whether HNF1A-AS1 could serve as a potential biomarker for prognosis in cancer patients.
Materials and methods
Literature search
We conducted a comprehensive literature search in the following databases: PubMed, Embase, Web of Science, China National Knowledge Infrastructure (CNKI), and Wan Fang (updated until December 31, 2017). The key words for search included the following: “HNF1A antisense RNA 1”, “HNF1A-AS1”, “HNF1AAS1”, “Long non-coding RNA HNF1A-AS1”, “LncRNA HNF1A-AS1”, “tumor”, “cancer”, “carcinoma”, “neoplas*”, and “malignan*”. A manual review of the reference lists of relevant articles was also performed to identify potentially eligible papers.
Inclusion and exclusion criteria
The inclusion criteria of this meta-analysis were the studies in which: 1) the expression of HNF1A-AS1 in tumor tissues was measured, 2) patients were divided into two groups based on high and low expression levels of HNF1A-AS1, 3) the associations of HNF1A-AS1 expression levels with prognosis or clinicopathological features were described, and 4) HRs or ORs with 95% CIs were reported or sufficient data were available for the computation.
Exclusion criteria were as follows: 1) duplicated articles, 2) editorials, letters, expert opinions, case reports, and reviews, and 3) studies without available data. Two authors screened the studies independently, and the disagreement was resolved by consensus.
Data collection
Two investigators (Chunbo Zhuang and Lei Zheng) extracted the data independently from the eligible studies, and disagreements were resolved by a third investigator (Pei Wang). The following information were extracted: the name of first author, year of publication, country, cancer type, sample size, detection method of HNF1A-AS1, internal control, cutoff values, tumor stage, outcome, follow-up time, HR and its corresponding 95% CI, and the clinicopathological parameters from each eligible study. If both the univariate and multivariate analyses were provided by the eligible studies, the multivariate values were selected as they had higher precision on interpreting confounding factors. For the studies only reporting Kaplan–Meier curves, the Engauge Digitizer (Version 4.1) software was used to extract the survival data, as previously described.21
Public data and tools
The Gene Expression Profiling Interactive Analysis (GEPIA) is an online database for analyzing the RNA sequencing expression data of tumors and normal samples from The Cancer Genome Atlas (TCGA) and the Genotype-Tissue Expression (GTEx) projects, using a standard processing pipeline (http://gepia.cancer-pku.cn/). In this study, we used the GEPIA database to analyze the tumor/normal differential expression of HNF1A-AS1 and its correlation with overall survival (OS) in TCGA dataset. One-way ANOVA was used for differential expression analysis, and the Kaplan–Meier method was used for survival analysis. The HR and 95% CI were also shown in the survival plot.
Statistical analyses
The Stata SE12.0 and Revman 5.3 software were used to perform all statistical analyses. HRs and 95% CIs were calculated to assess the association between HNF1A-AS1 and OS. An observed HR of >1 indicated poor prognosis in patients with high HNF1A-AS1 expression. In addition, ORs and 95% CIs were used to assess the correlation between HNF1A-AS1 expression and the clinicopathological features. The heterogeneity between the included studies was determined by I2 value derived from the Q test and a P-value from the Chi square test. The fixed-effects model was used if there was no obvious heterogeneity (I2≤50% or P≥0.05); otherwise, a random-effects model was applied. Begg’s test with a funnel plot and Egger’s test were used to estimate the publication bias. The sensitivity analysis was also performed to assess the stability of the results. P<0.05 was considered statistically significant.
Results
Study characteristics
The details of literature screening procedure are presented in Figure 1. Our initial search retrieved 94 papers from PubMed, Embase, Web of Science, CNKI, and Chinese Wan Fang databases. After the removal of duplicates, 59 articles remained. After carefully screening the titles and abstracts, we excluded 41 articles including letters, reviews, conference abstracts, and unrelated studies. The remaining 18 full-text articles were further reviewed and assessed, and 10 of them were excluded because of incomplete data. Ultimately, eight articles were included in this meta-analysis.
  | Figure 1 Flow diagram of the literature search and selection. |
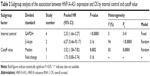
All the eight studies were from China and related to six types of cancers, including osteosarcoma, colon cancer, non-small cell lung cancer, HCC, bladder cancer, and lung adenocarcinoma. A total number of 789 patients were included in this study. The expression of HNF1A-AS1 was detected by quantitative reverse transcription PCR (qRT-PCR) in all the included studies, while the cutoff values were different. HRs and 95% CIs were directly extracted from four studies and were calculated by survival curve in two studies. The characteristics of the included studies are summarized in Table 1.
Association between HNF1A-AS1 expression and OS
Six studies reported the association between HNF1A-AS1 expression and OS. A random-effects model was applied to calculate the pooled HR and 95% CI due to the existence of significant heterogeneity across these six studies (P<0.001, I2=80%). The result suggested that high level of HNF1A-AS1 expression was associated with poor OS (HR=3.10, 95% CI: 1.58–6.11, P=0.001) (Figure 2).
Subsequently, we performed subgroup analyses according to internal control and cutoff values to evaluate the prognostic role of HNF1A-AS1 in cancers. As shown in Table 2, high expression of HNF1A-AS1 was significantly associated with poor OS when detected using GAPDH as an internal control or using median value as a cutoff.
Association between HNF1A-AS1 expression and clinicopathological parameters
In order to comprehensively analyze the potential role of HNF1A-AS1 in various cancers as a prognostic biomarker, we investigated the association between HNF1A-AS1 expression and clinicopathological characteristics. As shown in Table 3, high HNF1A-AS1 expression was significantly associated with advanced TNM stage (OR=3.32, 95% CI: 2.28–4.83, P<0.001; Figure 3A), lymph node metastasis (OR=3.08, 95% CI: 1.95–4.85, P<0.001; Figure 3B), and distant metastasis (OR=5.53, 95% CI: 1.94–15.77, P=0.001; Figure 3C). However, no significant association was observed between HNF1A-AS1 expression and age, gender, tumor size, or tumor differentiation.
Sensitivity analysis and publication bias
Given that significant heterogeneity was observed in the meta-analysis of OS (P<0.001, I2=80%), we performed sensitivity analysis to access the influence of individual study on the synthetic results. As shown in Figure 4, the pooled HR was not significantly affected after removing any single study. However, the result also showed that the study by Fang et al10 was responsible for the heterogeneity.
  | Figure 4 Sensitivity analysis on the association between HNF1A-AS1 expression and OS. |
We performed Begg’s funnel plot and Egger’s test to evaluate the publication bias of the included studies for OS. The Begg’s funnel plot is shown in Figure 5, and the P-value of Egger’s test was 0.399. These results suggested that there was no significant publication bias in this meta-analysis.
  | Figure 5 Begg’s funnel plot of publication bias for OS. |
Validation of the results in TCGA dataset
To validate the abovementioned results, we first evaluated the expression of HNF1A-AS1 in five kinds of cancers using the data from TCGA. As shown in Figure 6A, HNF1A-AS1 was overexpressed in four of them, including colon adenocarcinoma (COAD), esophageal carcinoma (ESCA), liver hepatocellular carcinoma (LIHC), and rectum adenocarcinoma (READ) (|Log2FC| cutoff >0.6 and P<0.01). Then, we accessed the association of HNF1A-AS1 expression with OS in all the cancers from TCGA dataset. A total of 8,787 patients were divided into high or low expression group based on median expression of HNF1A-AS1. As shown in Figure 6B, patients in the high expression group showed a worse OS than those in the low expression group. These results confirmed that HNF1A-AS1 was upregulated and significantly associated with poor prognosis in various cancers.
Discussion
Recently, a growing number of studies have reported the association between LncRNAs and cancer, and the functional roles of these tumor-associated LncRNAs in tumorigenesis and progression were gradually characterized.6 In addition, LncRNAs exhibit genome-wide expression patterns in various tissues and tissue-specific expression characteristics, making them novel promising biomarkers and therapeutic targets for cancer. Accumulating studies have suggested that LncRNAs could serve as potential prognostic biomarkers of different cancers.22–25 HNF1A-AS1 is one of these hot LncRNAs.
HNFA-AS1 has been reported to be upregulated in various types of cancers. Recent studies demonstrated that HNF1A-AS1 promoted tumorigenesis and progression by regulating various cellular processes, including proliferation, migration, invasion, apoptosis, and autophagy.18 However, the underlying mechanisms have not been fully elucidated. It was reported that knockdown of HNF1A-AS1 could inhibit the expression of H19, a well-known cancer-related LncRNA.8 Knockdown of HNF1A-AS1 suppressed tumor cell invasion and migration by reducing the epithelial–mesenchymal transition program in osteosarcoma, lung adenocarcinoma, and nasopharyngeal carcinoma.9,15,19 In line with these findings, Wang et al13 demonstrated that HNF1A-AS1 promoted HCC cell proliferation through repressing NKD1 and p21 expression by interacting with EZH2. Moreover, as an oncogene in osteosarcoma and colorectal cancer, LncRNA HNF1A-AS1 promoted tumor cell proliferation and metastasis by activating the Wnt/β-catenin signaling pathway.17,26 In addition, HNF1A-AS1 has been demonstrated to function as a competing endogenous RNA (ceRNA) that binds to and inhibits the expression of miRNAs such as miR-30b and miR-34a. In HCC, HNF1A-AS1 inhibited apoptosis by the miR-30b/Bcl-2 pathway and promoted autophagy by the miR-30b/AGT5 pathway.11 In another study by Fang et al,10 HNF1A-AS1 facilitated colon cancer cell proliferation and metastasis in an miR-34a/p53-dependent manner. These studies suggested that HNF1A-AS1 might serve as an oncogene and predict poor outcomes in cancer patients.
In the present meta-analysis, a total of eight studies including 789 patients were included, and the result suggested that enforced HNF1A-AS1 expression was significantly associated with poor prognosis in patients with various types of cancers. The increased HNF1A-AS1 expression was associated with shorter OS. The pooled HR for OS was 3.10 (95% CI: 1.58–6.11, P=0.001). The association between HNF1A-AS1 expression and clinicopathological characteristics was also analyzed in this meta-analysis. Our results showed that increased HNF1A-AS1 expression was associated with advanced TNM stage, lymph node metastasis, and distant metastasis. The pooled ORs were 3.32 (95% CI: 2.28–4.83, P<0.001), 3.08 (95% CI: 1.95–4.85, P<0.001), and 5.53 (95% CI: 1.94–15.77, P=0.001), respectively.
However, there are still some limitations that should be noted in this meta-analysis. First, the total sample size was relatively small, and all the included patients were from China. Second, statistical heterogeneity was observed, which may be due to the differences in cancer types, internal control, cutoff value, clinical characteristics, and sample sizes. Third, although no obvious publication bias was observed, the selection bias may exist in the present meta-analysis. For example, Dang et al27 reported that HNF1A-AS1 was significantly downregulated in gastric cancer, and low HNF1A-AS1 expression was associated with tumor size and levels of serum CEA and CA199. Using high-throughput next-generation sequencing (NGS)-based technologies, Müller et al28 reported that HNF1A-AS1 was downregulated in pancreatic cancer. These two studies suggested a possible role of HNF1A-AS1 as a tumor suppressor. However, the lack of relevant data precluded them from the present meta-analysis. Finally, some HRs were extracted from the survival curves, which may lead to small statistical errors.
Conclusion
Our meta-analysis suggested that high expression of HNF1A-AS1 was significantly associated with poor outcomes in various cancers and could serve as a potential prognostic biomarker. However, considering the limitations of the present analysis, more studies with high quality and large sample size are needed to further confirm the prognostic role of HNF1A-AS1 in cancers.
Acknowledgment
This study was supported by the National Natural Science Foundation of China (No 81700514).
Disclosure
The authors report no conflicts of interest in this work.
References
Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. | ||
Siegel RL, Miller KD, Jemal A. Statistics C. CA Cancer J Clin. 2017;2017;67(1):7–30. | ||
Neal JW, Sledge GW. Decade in review-targeted therapy: successes, toxicities and challenges in solid tumours. Nat Rev Clin Oncol. 2014;11(11):627–628. | ||
Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. | ||
Jandura A, Krause HM. The New RNA World: Growing Evidence for Long Noncoding RNA Functionality. Trends in Genetics. 2017;33(10):665–676. | ||
Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA. and Cancer: A New Paradigm. Cancer Res. 2017;77(15):3965–3981. | ||
Serghiou S, Kyriakopoulou A, Ioannidis JPA. Long noncoding RNAs as novel predictors of survival in human cancer: a systematic review and meta-analysis. Mol Cancer. 2016;15(1):50. | ||
Yang X, Song JH, Cheng Y, et al. Long non-coding RNA HNF1A-AS1 regulates proliferation and migration in oesophageal adenocarcinoma cells. Gut. 2014;63(6):881–890. | ||
Cai L, Lv J, Zhang Y, Li J, Wang Y, Yang H. The lncRNA HNF1A-AS1 is a negative prognostic factor and promotes tumorigenesis in osteosarcoma. J Cell Mol Med. 2017;21(11):2654–2662. | ||
Fang C, Qiu S, Sun F, et al. Long non-coding RNA HNF1A-AS1 mediated repression of miR-34a/SIRT1/p53 feedback loop promotes the metastatic progression of colon cancer by functioning as a competing endogenous RNA. Cancer Lett. 2017;410:50–62. | ||
Liu Z, Wei X, Zhang A, Li C, Bai J, Dong J. Long non-coding RNA HNF1A-AS1 functioned as an oncogene and autophagy promoter in hepatocellular carcinoma through sponging hsa-miR-30b-5p. Biochem Biophys Res Commun. 2016;473(4):1268–1275. | ||
Ma YF, Liang T, Li CR, Li YJ, Jin S, Liu Y. Long non-coding RNA HNF1A-AS1 up-regulation in non-small cell lung cancer correlates to poor survival. Eur Rev Med Pharmacol Sci. 2016;20(23):4858–4863. | ||
Wang C, Mou L, Chai H-X, Wang F, Yin Y-Z, Zhang X-Y. Long non-coding RNA HNF1A-AS1 promotes hepatocellular carcinoma cell proliferation by repressing NKD1 and P21 expression. Biomed Pharmacother. 2017;89:926–932. | ||
Wang G, Zhao W, Gao X, et al. HNF1A-AS1 promotes growth and metastasis of esophageal squamous cell carcinoma by sponging miR-214 to upregulate the expression of SOX-4. Int J Oncol. 2017;51(2):657–667. | ||
Wu Y, Liu H, Shi X, Yao Y, Yang W, Song Y. The long non-coding RNA HNF1A-AS1 regulates proliferation and metastasis in lung adenocarcinoma. Oncotarget. 2015;6(11):9160–9172. | ||
Zhan Y, Li Y, Guan B, et al. Long non-coding RNA HNF1A-AS1 promotes proliferation and suppresses apoptosis of bladder cancer cells through upregulating Bcl-2. Oncotarget. 2017;8(44):76656–76665. | ||
Zhao H, Hou W, Tao J, et al. Upregulation of lncRNA HNF1A-AS1 promotes cell proliferation and metastasis in osteosarcoma through activation of the Wnt/beta-catenin signaling pathway. Am J Transl Res. 2016;8(8):3503–3512. | ||
Zhu W, Zhuang P, Song W, et al. Knockdown of lncRNA HNF1A-AS1 inhibits oncogenic phenotypes in colorectal carcinoma. Mol Med Rep. 2017;16(4):4694–4700. | ||
Zhuang K, Wu Q, Jin C-S, Yuan H-J, Cheng J-Z. Long non-coding RNA HNF1A-AS is upregulated and promotes cell proliferation and metastasis in nasopharyngeal carcinoma. Cancer Biomarkers. 2016;16(2):291–300. | ||
Zhang LF. Screening and Function Study of Aberrantly Expressed LncRNAs in Hepatocellular Carcinoma [Doctoral dissertation, in Chinese]. Hangzhou: Zhejiang University; 2017. | ||
Zhang Y, Cai P, Liang T, Wang L, Hu L. TIM-3 is a potential prognostic marker for patients with solid tumors: A systematic review and meta-analysis. Oncotarget. 2017;8(19):31705–31713. | ||
Wang Y, Huang Y, Xiang P, Tian W. LncRNA expression and implication in osteosarcoma: a systematic review and meta-analysis. Onco Targets Ther. 2017;10:5355–5361. | ||
Zhu J, Kong F, Xing L, Jin Z, Li Z. Prognostic and clinicopathological value of long noncoding RNA XIST in cancer. Clinica Chimica Acta. 2018;479:43–47. | ||
Chen T, Wang H, Yang P, He Z-Y, Zy H. Prognostic role of long noncoding RNA NEAT1 in various carcinomas: a meta-analysis. Onco Targets Ther. 2017;10:993–1000. | ||
Chen X, Lun L, Hou H, Tian R, Zhang H, Zhang Y. The Value of lncRNA HULC as a Prognostic Factor for Survival of Cancer Outcome: A Meta-Analysis. Cellular Physiology and Biochemistry. 2017;41(4):1424–1434. | ||
Zhang X, Xiong Y, Tang F, Bian Y, Chen Y, Zhang F. Long noncoding RNA HNF1A-AS1 indicates a poor prognosis of colorectal cancer and promotes carcinogenesis via activation of the Wnt/β-catenin signaling pathway. Biomed Pharmacother. 2017;96:877–883. | ||
Dang Y, Lan F, Ouyang X, et al. Expression and clinical significance of long non-coding RNA HNF1A-AS1 in human gastric cancer. World J Surg Oncol. 2015;13(1):302. | ||
Müller S, Raulefs S, Bruns P, et al. Next-generation sequencing reveals novel differentially regulated mRNAs, lncRNAs, miRNAs, sdRNAs and a piRNA in pancreatic cancer. Mol Cancer. 2015;14(1):94. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.